Abstract
Metaproteomics is one of a suite of new approaches providing insights into the activities of microorganisms in natural environments. Proteins, the final products of gene expression, indicate cellular priorities, taking into account both transcriptional and posttranscriptional control mechanisms that control adaptive responses. Here, we report the proteomic composition of the < 1.2 μm fraction of a microbial community from Oregon coast summer surface waters, detected with two-dimensional liquid chromatography coupled with electrospray tandem mass spectrometry. Spectra corresponding to proteins involved in protein folding and biosynthesis, transport, and viral capsid structure were the most frequently detected. A total of 36% of all the detected proteins were best matches to the SAR11 clade, and other abundant coastal microbial clades were also well represented, including the Roseobacter clade (17%), oligotrophic marine gammaproteobacteria group (6%), OM43 clade (1%). Viral origins were attributed to 2.5% of proteins. In contrast to oligotrophic waters, phosphate transporters were not highly detected in this nutrient-rich system. However, transporters for amino acids, taurine, polyamines and glutamine synthetase were among the most highly detected proteins, supporting predictions that carbon and nitrogen are more limiting than phosphate in this environment. Intriguingly, one of the highly detected proteins was methanol dehydrogenase originating from the OM43 clade, providing further support for recent reports that the metabolism of one-carbon compounds by these streamlined methylotrophs might be an important feature of coastal ocean biogeochemistry.
Similar content being viewed by others
Introduction
Although many metagenomic studies of marine communities have been reported (Venter et al., 2004; Tringe et al., 2005; DeLong et al., 2006; Rusch et al., 2007; Wilhelm et al., 2007), so far there have been only a few surveys of gene expression in marine communities (Poretsky et al., 2005; Frias-Lopez et al., 2008; Sowell et al., 2008). The first proteomic analyses of marine systems detected only a few proteins, such as proteorhodopsin (Giovannoni et al., 2005a) and oxidoreductase (Kan et al., 2005); however, they paved the way for more comprehensive proteomic studies by establishing the feasibility of studying complex communities with these methods and by emphasizing the importance of understanding the protein composition of cells in nature (Sowell et al., 2008; Morris et al., 2010). An organism-centric analysis of the Sargasso Sea metaproteome revealed the priority of transport proteins, particularly those involved in phosphorus and amino acid uptake, for SAR11 survival in this oligotrophic environment. Similarly, nutrient acquisition was found to be central to the survival of oxygenic phototrophic organisms as proteins involved in transport, photosynthesis and CO2 fixation were frequently detected for Synechococcus and Prochlorococcus (Sowell et al., 2008). Using techniques that focused on membrane proteins, Morris and coworkers studied plankton in South Atlantic surface waters from a low-nutrient gyre to a highly productive coastal upwelling region, and reported variation in proteins for nutrient utilization and energy transduction along the environmental gradient (Morris et al., 2010).
The identification of proteins in complex samples by tandem mass spectrometry is dependent on the existence of good quality protein or gene databanks. The availability of large metagenomic data sets from marine environments, such as the data provided by the Global Ocean Sampling (GOS) expedition (Rusch et al., 2007), has not only improved our understanding of the diversity and genetic potential of marine microorganisms, but it has also enabled the identification of many of their expressed proteins using mass spectrometry-based proteomics methods.
The upwelling and mixing of nutrient-laden waters cause coastal ecosystems to be more productive than their open-ocean counterparts. Coastal ecosystems contribute as much as 40% to global carbon sequestration (Muller-Karger et al., 2005). Surface water concentrations of phosphate and nitrate off the coast of Oregon are about ∼1 μM and 5 μM, respectively (Park, 1967), and particulate organic carbon can reach levels of 20–63 μM C (Karp-Boss et al., 2004). The higher levels of available nutrients in coastal systems tend to favor the growth of larger eukaryotic phytoplankton over that of bacterial picophytoplankton, such as Prochlorococcus and Synechococcus. Phylogenetic analysis of 16S rDNA clones shows that many of the dominant bacterial groups, such as the SAR11, Roseobacter and SAR116 clades, are ubiquitous in their distribution in both open-ocean systems and coastal waters (Rappe et al., 2000; Giovannoni and Stingl, 2005). Conversely, members of OM43 clade of methylotrophic betaproteobacteria, and some members of the Oligotrophic Marine Gammaproteobacteria (OMG) clade, are predominantly found in coastal systems (Rappe et al., 2000; Cho and Giovannoni, 2004; Giovannoni and Stingl, 2005; Morris et al., 2006).
In this paper, we report a comprehensive metaproteomic analysis of the <1.2 μm fraction of the microbial community from surface waters on the Oregon shelf during an upwelling period in the late summer, 2006. Peptides from marine microorganisms were detected using two-dimensional high-performance liquid chromatography coupled with electrospray tandem mass spectrometry (MS/MS). Peptides were identified by spectral comparison with translated environmental protein-coding sequences (eCDSs) collected from coastal environments during the GOS metagenomics project (GOS02-11) (Rusch et al., 2007) as well as the genomes of two coastal Bacterial isolates, Candidatus Pelagibacter ubique strain HTCC1062 of the SAR11 clade (Giovannoni et al., 2005b) and the OM43 clade strain HTCC2181(Giovannoni et al., 2008). Our goal was to identify abundant proteins that could reveal the dominant metabolic processes occurring within marine bacterial communities.
Materials and methods
Sample collection
Microbial cells for proteomic analysis were collected on 26 September 2006 at station NH-5, located 5 miles off the coast of Newport, Oregon (44°39.1′N, 124°10.6′W). Although properties of the water column were not measured at the time of sampling, oceanographic measurements associated with a long-term study in this region were made on the preceding and following days at nearby sites. These measurements indicated that, at the time of sampling, the surface waters were of halocline origin, as is typical of upwelling water masses on the Oregon shelf. Nutrient concentrations are elevated in halocline water, but in situ chlorophyll fluorescent sensors in the region indicated that at the time of sampling, dense phytoplankton blooms had not developed (F Chan, personal communication).
Cells from ∼100 l of surface (10 m) seawater were prefiltered through an A/E glass fiber filter (Pall, West Chester, PA, USA) to enrich the sample for smaller organisms (<∼1.2 μm), mainly bacteria. The filtrate containing the small cells was then concentrated by tangential flow filtration with a Millipore Pellicon system (Millipore, Billerica, MA, USA), with a 30 kDa regenerated cellulose filter (filtration rate: ∼1 l min−1). The concentrated cells were sedimented by centrifugation at 48 400 × g for 1 h at 4 °C using a Beckman J2-21 centrifuge (Brea, CA, USA) with a JA-20 rotor. The resulting pellet was stored at −80 °C until proteomic analysis.
Sample preparation
The microbial cell pellet (47 mg wet weight) was resuspended in 6 M guanidine and 10 mM DTT to lyse cells and denature proteins (Thompson et al., 2008). The protein content of the pellet was estimated to be 10% (w/w), or 4.7 mg protein in the starting material. The guanidine concentration was diluted to 1 M with 50 mM Tris buffer and 10 mM CaCl2, and 40 μg sequencing grade trypsin (Promege, Madison, WI, USA) was added at two intervals 12 h apart to digest proteins to peptides (∼1:50 w/w, 80 μg trypsin total). The complex peptide solution was desalted by C18 solid phase extraction, concentrated and filtered through an Ultrafree-MC 0.45 μm spin filter (Millipore), generally 1/2 of the total protein is lost during processing. The estimated 2.3 mg of protein remaining was divided into four aliquots and frozen at −80 °C until analysis. For each 2D-LC-MS/MS analysis made, ∼1/4 of the total sample was used, resulting in an estimated 500 μg of digested protein loaded onto the two-dimensional high-performance liquid chromatography column for each analysis.
Two-dimensional high-performance liquid chromatography and mass spectrometry
The sample was analyzed by two-dimensional nano-LC MS/MS system with a split-phase column (RP-SCX-RP) (McDonald et al., 2002) on a LTQ-Orbitrap (Thermo Fisher Scientific, Waltham, MA, USA) with a 22 h run, which consisted of 11 salt pulses, followed by 2 h reverse phase gradients. The entire elute was electrosprayed from and integrated nanospray tip (Proxeon, Odense, Denmark). The Orbitrap settings were as follows: 30 K resolution on MS scans in Orbitrap, data-dependent MS/MS in the LTQ performed on the top five MS peaks, two microscans for both MS and MS/MS scans, centroid data for all scans and two microscans averaged for each spectrum, and dynamic exclusion set at 1. No charge state screening was employed. Liquid chromatography and mass spectrometry were performed as previously described (Ram et al., 2005; Lo et al., 2007; Verberkmoes et al., 2008).
Database and protein identification
To identify peptide sequences, MS/MS spectra were compared with a protein database consisting of translated eCDSs from the GOS expedition metagenomics project (Rusch et al., 2007). To decrease database size, only eCDSs from the sites deemed similar to the Oregon coast in bacterial distribution and environmental conditions were included (GOS2-11). Translated genomes of two known Oregon coastal isolates, Candidatus Pelagibacter ubique strain HTCC1062 and OM43 clade strain HTCC2181 were also included in the database, as were common contaminants such as trypsin and keratins. To estimate a false positive rate, exact reverse sequences of the GOS eCDSs were added to the database (Peng et al., 2003; Lo et al., 2007) and false positive levels were determined, as previously described (Lo et al., 2007; Verberkmoes et al., 2008). All MS/MS spectra were searched with the SEQUEST algorithm (Eng et al., 1994) (enzyme type, trypsin; parent mass tolerance, 3.0; fragment ion tolerance, 0.5; up to four missed cleavages allowed (internal lysine and arginine residues) and fully tryptic peptides only (both ends of the peptide must have arisen from a trypsin-specific cut, except N and C termini of proteins)) and filtered with DTASelect/Contrast (Tabb et al., 2002) at the peptide level (Xcorrs of at least 1.8 (+1), 2.5 (+2); 3.5 (+3)). Only proteins identified with two fully tryptic peptides from a 22 h run were considered for biological interpretation (Supplementary Table S2). Monoisotopic theoretical masses for all peptides identified by SEQUEST were generated and compared with observed masses. Observed high-resolution masses were extracted from .raw files from the full scan preceding the best identified spectra; parts per million (p.p.m.) calculations were made comparing each identified peptide observed and its theoretical mass. When quality MS/MS spectra did not have an observed mass (low intensity), a mass of 0 was reported and p.p.m. was calculated as infinity. We applied the accepted method of reverse database searching (Peng et al., 2003) to determine a false positive level for this data set. This test resulted in a false discovery rate of 0.2%. To prevent an underestimation of the false discovery rate due to the large number of non-unique peptides identified, for this analysis, each unique peptide sequence was only counted once regardless of the number of eCDSs to which it matched. Furthermore, over 80% of the identified peptides had high-mass accuracy measurements of the intact peptides from the Orbitrap mass spectrometer (less than 10 p.p.m.), this subgroup of identified peptides are virtually 100% correct when estimating false positive rates (Verberkmoes et al., 2008), the other ∼20% had passing Xcorrs and delCN values, but either inaccurate assignment of parent mass or no mass assignment at all. The protein database used in the search and the raw output results (DTASelect files), as well as every identified peptide MS/MS spectra with corresponding mass accuracies, can be accessed online at http://compbio.ornl.gov/Oregon_Coast_metaproteome/, and raw files are available upon request.
Data analysis
Each detected eCDS was queried against the NCBI-nr database (as of July 1, 2008), using BLASTP to identify a probable function and the closest matching relative. Best hits with a bit score <50 were annotated as ‘unknown’. Because many peptides matched to multiple eCDSs and because many eCDSs had similar functions, similar eCDSs were grouped together into the protein clusters outlined by Yooseph et al. (2007), by matching the detected peptides to the protein cluster database using BLASTP. The protein cluster database is available at https://portal.camera.calit2.net/gridsphere/gridsphere?cid=apply).
Results and discussion
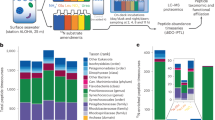
The Oregon shelf is a highly productive region because of seasonal wind patterns and currents that cause the upwelling of nutrient-rich halocline waters, supporting dense phytoplankton blooms. In previous work, we examined the metaproteome of highly oligotrophic microbial plankton communities collected from the Sargasso Sea. In this study, we analyzed in detail the metaproteome of a microbial plankton community from a surface water sample collected during a period of coastal upwelling (Figure 1).
The complex peptide mixture was characterized by matching 2D-LC-MS/MS mass spectra to peptide sequences from the translated GOS02-11 eCDS database and two genomes from coastal marine bacterioplankton isolates. With this database, mass spectra matching 7151 distinct peptide sequences (typically 5–25 amino acids in length) were detected. These peptide sequences mapped to 13 469 eCDSs (full- or nearly full-length translated environmental protein-coding sequences). At least two peptide matches per eCDS were required for a positive identification. For each identified eCDS, putative function and closest relative were determined by comparison with the National Center for Biotechnology Information non-redundant protein database (NCBI-nr; as of 1 July 2008) using BLASTP. Clustering similar eCDSs into the protein families outlined by Yooseph et al. (2007) resulted in the identification of 481 unique protein families (Table 1 and Supplementary Table S1). Owing to a high degree of subtle sequence variation, some of the eCDSs in the environmental database only differ by a few amino acids across their entire sequence. Although each reported peptide was unique, peptides often mapped to multiple eCDSs that were identical at the locus of the peptide. The subtle sequence variation also led to multiple eCDSs having the same putative function and closest relative using BLASTP. A study of SAR11 peptides found that of the 1146 peptides that mapped to SAR11 eCDSs, only 155 (14%) also mapped to eCDSs that had a different organism as closest relative. A taxonomic breakdown of these shared peptides shows that 11% overlapped at the Family level, suggesting that they are from conserved regions of common proteins; the remainder was shared between more distant relatives (4% Class, 2% Phylum, 0.2% Kingdom).
Frequently detected proteins
One of the objectives of measuring gene expression is to infer the metabolic status of cells in their natural environment. The importance of nutrient acquisition to the microbial community was shown by the prevalence of transport proteins: a total of 29% of all the detected spectra and 21% of all the eCDSs matched proteins with transport or putative transport functions (Figure 2). When protein families were ranked by their spectral count, 11 of the 25 most abundantly represented families were proteins involved in transport (Table 1). The eCDSs that were best matches to SAR11 proteins contributed a significant proportion to the total spectral count; however, even when SAR11 eCDSs were removed, nine families with transport proteins remained among the top ranking 25. These remaining transporter families were enriched with eCDS matches to other alphaproteobacteria, which correlates well with the overall distribution of eCDSs among bacterial classes (Figure 3b).
The distribution of detected spectra by their functional classification. Spectra for bacterial proteins with transport or putative transport functions were frequently detected, as were spectra for proteins involved in transcription, translation and protein folding. Viral proteins were also abundant in this data set.
The distribution of the detected eCDSs among the three domains of life and viruses (a) Each detected eCDS was used to query NCBI-nr, using BLASTP to determine its closest relative. Sequence matches with bit scores less than 50 were considered unknown. (b) The fraction of bacterial eCDSs best matching organisms from each class. The number of SAR11 eCDSs, as a fraction of alphabacterial sequences, is shown in red.
The abundance of transport proteins in the data suggests that successful scavenging of nutrients, even in nutrient replete coastal ecosystems, is a determining factor in microbial competition and survival. In a previous metaproteomic study of an ocean gyre ecosystem that focused on SAR11 and cyanobacteria, we reached similar general conclusions, but there were substantial differences between the transport proteins detected in that study and those found on the Oregon shelf. The abundantly detected transport proteins in the Oregon sample had predicted substrate specificities for nitrogen-, carbon- and sulfur-containing compounds, such as amino acids, glycine betaine, polyamines and taurine (Table 2). SAR11 phosphate and phosphonate transporters were among the most abundantly detected proteins in the ocean gyre ecosystem, and nearly absent from the Oregon shelf sample (Sowell et al., 2008). Proteins involved in phosphorus transport have been shown to be more abundant during periods of phosphate starvation and less abundant when phosphorus was abundant (Scanlan et al., 1997; Dyhrman and Haley, 2006; Martiny et al., 2006). Oregon shelf water is replete with phosphorus (1 μM) (Park, 1967), whereas summer surface waters in the Sargasso Sea are a well-known example of a phosphorus-limited system (< 5 nM) (Wu et al., 2000; Steinberg et al., 2001). Our data are also consistent with those of Morris et al. (2010) whose metaproteomic analyses of membrane proteins reveal a lack of proteins involved in phosphorus transport when measured phosphate was abundant. Thus, the relative scarcity of peptides matching proteins for phosphorous uptake in the coastal sample is reassuringly consistent with expectations and supports observations that metaproteomic analyses can help reveal the nutrient status of cells in marine environments.
Glutamine synthetase, which is involved in the assimilation of ammonium into amino acids, was also frequently detected, with 147 spectra matching 151 eCDSs identified. Hoch et al. (2006) noted that marine bacterioplankton express this enzyme when dissolved organic or inorganic nitrogen is limiting in the environment. The presence of glutamine synthetase in coastal seawater in conjunction with high levels of expression of transporters whose substrate specificities are for nitrogen-containing compounds indicates that, in the nutrient-repleted Oregon coastal summer surface water, nitrogen may be more limiting than phosphorus or perhaps even carbon.
Although the data that we report are consistent with knowledge of ocean conditions in the region, it is nonetheless a rare example of direct measurements, showing the in situ metabolic status of microbial plankton cells, and provides important, complementary support for hypotheses about the microbial ecology of this region that were originally based on chemical measurements. Comparisons of measured concentrations of carbon (20–63 μM), nitrogen (5 μM) and phosphorus (1 μM) in this environment to the Redfield ratio suggests that carbon and nitrogen may become limiting before phosphorus, and competition for carbon- and nitrogen-containing compounds may be of greater importance to the microbial community in the Oregon coastal environment where phosphorus is plentiful (Park, 1967; Karp-Boss et al., 2004).
By far, the most abundant class of proteins detected (64% of all the spectra) was periplasmic substrate-binding subunits of ABC or TRAP transporters. Similar findings were reported from the proteomic analysis that focused on the SAR11 metaproteome in the oligotrophic Sargasso Sea, where it was suggested that these cells might survive by allocating a large fraction of protein synthesis to nutrient acquisition (Sowell et al., 2008), and in both coastal and open-ocean samples analyzed by Morris et al. (2010). Adaptations to ocean oligotrophy include an increased surface area-to-volume ratio and high levels of expression of periplasmic substrate-binding proteins that contribute to growth efficiency by balancing the periplasm's capacity to import nutrients with the cytoplasm's capacity to utilize them (Button and Robertson, 2000). The majority of the bacterioplankton community consists of ultramicrobial cells (Lee and Fuhrman, 1987); therefore, this observation of high expression of multiple periplasmic substrate-binding proteins, in conjunction with the large surface area-to-volume ratios of these cells, indicates that adaptations for effective nutrient scavenging are generally important in pelagic marine environments, whether they are oligotrophic gyres or productive coastal ecosystems. Morris et al. (2010) also detected transport proteins with high frequency in their coastal samples. However, their study focused on the isolated membrane fraction of cells and the majority of the transporters they detected were of the TonB type (TBDT), of Shewanella origin. These accounted for only 1.2% of all the transport protein spectra in this study. This contrast could be due to differences in sample preparation techniques (membrane specific versus total protein) or to differences in the microbial makeup of the environments studied (predominantly gammaproteobacteria and Bacteriodetes/Chlorobi in the South Atlantic study versus predominantly alphaproteobacteria in Oregon shelf samples).
Overall, 37% of all the spectra attributed to the SAR11 clade were matches to transport proteins. This is lower than the 67% seen in SAR11 cells from the Sargasso Sea (Sowell et al., 2008) and, thus, suggests that, although competition for nutrients is vital in nutrient-repleted environments, a greater proportion of protein expression is directed towards proteins involved in other functions when nutrients are readily available. Similar to what was seen with transporters from the general marine microbial community outlined above, substrate specificity for carbon- and nitrogen-containing compounds predominated among the detected transport proteins.
As seen in this study and others (Morris et al., 2010), viral proteins are a significant fraction of the marine microbial plankton proteome. Viruses are widely recognized as significant agents of microbial plankton mortality and consequently in the cycling of dissolved organic matter, and, not surprisingly, viral infection of bacterial cells in Oregon coastal surface water is common (Bouvier and del Giorgio, 2007). The prevalence of spectra for viral capsid proteins (8% of all the spectra) in this study strongly supports broad scientific conclusions about the importance of viruses in microbial plankton ecology. Although the most abundant viral proteins were best matches to the Prochlorococcus phage P-SSM2, Prochlorococcus is known to be in very low abundance or absent from marine coastal environments (Zubkov et al., 2000; Sherr et al., 2005). This, in combination with a relative scarcity of marine bacterial phage genomes in the NCBI-nr database, suggests the likely existence of a phage related to Prochlorococcus phage P-SSM2.
The detection of peptides matching hypothetical proteins suggests that many of the dominant biological processes occurring in marine environments are carried out by proteins with unknown functions. Cluster 7053 was among the most frequently detected protein families, but has no corresponding Gene Ontology classification and all of the eCDSs in the family only matched conserved hypothetical proteins from the NCBI-nr database (Table 1). This is true for 19% of the detected protein families (5% of all the eCDSs, 6% of all the spectra, Supplementary Table S1). This observation is in agreement with data from previous gene expression and proteomic analyses (Frias-Lopez et al., 2008; Morris et al., 2010) where abundant hypothetical genes/proteins were also detected.
As was the case in our metaproteomic analysis of the Sargasso Sea (Sowell et al., 2008), no proteorhodopsin peptides were detected in this study. Proteorhodopsin peptides were previously detected in Oregon coastal seawater with specialized sample preparation methods that targeted membrane proteins from small cells (Giovannoni et al., 2005a), and later by Morris et al. who used similar procedures (Morris et al., 2010).
Distribution of detected eCDSs between dominant marine bacterial groups
Each detected eCDS was queried against NCBI-nr using BLASTP to determine the phylogenetic classification of the most closely related protein (Figure 2). The partitioning of the eCDSs between the three domains of life, viruses and sequences without any matches is depicted in Figure 3a. This experiment was designed to focus on free-living bacterial proteins, so the sample was prefiltered to remove larger, eukaryotic organisms that might mask the detection of bacterial proteins. For this reason, the contribution from eukaryotic organisms is low (0.3%). As expected, bacterial proteins most closely matched the majority of the eCDSs, accounting for ∼95% of the total. Because of their smaller size, Archaea and viruses were not excluded by prefiltration, as is evident by their contribution of 1% and 2.5%, respectively, of the total of the eCDSs detected. Large numbers of eCDS from bacterial groups that are known to be dominant in Oregon coastal surface water were detected. The number of eCDSs that were most closely related to sequences from each bacterial class can be seen in Figure 2b, with the fraction of alphaproteobacterial sequences represented by SAR11 proteins (57%) highlighted in red. It is not surprising that 36% of all the bacterial eCDSs were most closely matched to SAR11 proteins, as SAR11 cell counts in Oregon coastal surface water range from 17% to 38% of the total bacteria enumerated (Morris et al., 2002). Other groups that are abundant in coastal communities (Rappe et al., 2000) and were also detected in this study include the Roseobacter clade (17% of the eCDSs), OMG group (6%) and OM43 clade (1%).
In addition to an overall abundance of SAR11 eCDS detections, SAR11 peptides were also well represented among the most frequently detected protein families (Table 1), accounting for 58% of the eCDSs in the 25 most abundant protein clusters. This reflects not only the numerical dominance of SAR11 cells but also solidifies their position as major participants in the dominant metabolic processes in oceanic ecosystems.
The majority of the eCDSs closely matching proteins from the Roseobacter clade were best hits to strain HTCC2255 (29%), which is an Oregon coastal isolate. Members of the Roseobacter clade have been isolated from a wide variety of habitats and are physiologically diverse (Brinkoff et al., 2008), with marine strains having unique traits in carbon and sulfur sequestration, such as aerobic anoxygenic photosynthesis, carbon monoxide oxidation and dimethylsulfoniopropionate degradation (Allgaier et al., 2003; Moran et al., 2003, 2004; Miller and Belas, 2004). Although the most abundantly detected Roseobacter proteins, such as GroEL, TufB and ribosomal proteins, tended to have housekeeping functions, proteins involved in amino acid, sugar and proline/glycine betaine transport were also frequently observed. Proteomic evidence for the use of alternative carbon and energy sources was also observed, including single eCDS matches to a putative carbon monoxide dehydrogenase protein and an aminomethyl transferase family protein from Rhodobacterales bacterium HTCC2150, and a membrane protein involved in aromatic hydrocarbon degradation from Jannaschia sp. CCS1.
The OMG group is a diverse group of gammaproteobacteria made up of multiple clades (Cho and Giovannoni, 2004), which are common in coastal surface water. Genomic and proteomic analysis of some OMG group isolates indicates these organisms contribute to carbon cycling using aerobic anoxygenic photosynthesis (Cho et al., 2007; Fuchs et al., 2007). The majority of detected proteins from this group were best hits to OM60 clade isolates HTCC2080 and Congregibacter litoralis KT 71, which have been quantified, respectively, at 3.4% and up to 11% of the bacterial cells in coastal surface water (Cho et al., 2007; Fuchs et al., 2007). Proteins from the SAR92 clade isolate HTCC 2207 (Stingl et al., 2007) and the BD1-7 clade isolate HTCC2143 (Cho and Giovannoni, 2004) were also detected. Although not particularly abundant, eCDSs best matching to aerobic anoxygenic phototrophy protein, PuhA, from HTCC2080 were identified, supporting previous observations of alternate photosynthetic mechanisms among marine gammaproteobacteria (Beja et al., 2002). As seen for the Roseobacter clade, the most frequently detected OMG peptides matched housekeeping proteins, such as GroEL, TufB and ribosomal proteins.
The large subunit of the OM43 clade methanol dehydrogenase (MDH) (XoxF aka MxaF) was the best match to 2.3% of all the spectra identified, lending further support to recent findings that implicate methanol and other one carbon compounds as important substrates for bacterioplankton in coastal ecosystems (Giovannoni et al., 2008). A total of 168 spectra matching 33 eCDSs were closest matches to the OM43 clade MDH (Figure 4). The betaproteobacterial OM43 clade is commonly found at low levels in coastal ecosystems and has been observed to increase in abundance to ∼2% of bacterial cells during phytoplankton blooms (Morris et al., 2006). Phylogenetic and genome analysis of the OM43 isolate HTCC2181 placed this organism among type I methylotrophs that can use methylated compounds as a sole carbon and energy source (Giovannoni et al., 2008). It was also shown that cell growth yield was proportional to the amount of methanol or formaldehyde added to sterile seawater media. Although the source of methanol in the environment is currently uncertain, this observation of the MDH proteins in relatively high abundance suggests that oxidation of methanol is providing a significant source of carbon and energy for the OM43 clade in coastal ecosystems.
Peptide coverage and phylogenetic position of large subunit MDH eCDSs. The data illustrate the specificity of the peptides detected for xoxF, a large subunit MDH found in genomes from the OM32 clade. RAxML was used to infer the phylogenetic tree from an alignment of full-length homologous amino-acid sequences from reference genomes. For each sequence in the tree, the colored bars show peptides detected by mass spectrometry. Amino acids are heat mapped according to the prevalence of the given residue at that position in the consensus sequence, with red indicating the most common and green indicating the least common amino acid. Gray indicates undetected residues.
Concluding remarks
In an earlier metaproteomic study of an oligotrophic ocean region, we focused on the proteomes of specific organisms, and addressed issues of peptide specificity and diversity (Sowell et al., 2008). Those issues were a concern because of the high sequence divergence commonly observed in marine systems, but we found relatively low ambiguity in the identification of proteins and their assignments to species. Therefore, in this study, we applied more general methods for peptide identification, and expanded the analysis to include a larger metagenomic database that included many species.
Although there were differences between the procedures used for this analysis and the preceding analysis of an open-ocean site, some general conclusions are clear. Like their open-ocean counterparts, the metaproteome of the Oregon shelf SAR11 population was dominated by transport proteins, but the main targets for transport in this productive system were predominantly carbon- and nitrogen-containing compounds, rather than phosphate, supporting the developed ecological theory for this region, which predicts that nitrogen, rather than phosphorous, is ultimately the nutrient that limits productivity (Wheeler et al., 2003).
Perhaps one of the most important characteristics of proteomics is that it provides taxon-specific confirmation of the expression of genomic potential. For example, serving this role, metaproteomics was used previously to confirm the expression of SAR11 proteorhodopsins in seawater. Thus, it was particularly significant that strong evidence emerged in this study of the expression of OM43 clade MDH, supporting the unexpected finding, reported in earlier studies, that these common coastal betaproteobacteria are obligate oxidizers of C1 compounds. Also, interesting was the metaproteomic evidence of energy acquisition by one-step bacteriochlorophyl photosystems, which appear to play a larger role in biogeochemical processes over productive shelves. Although the expression of these systems has been reported previously from coastal ecosystems, based on bacteriochlorophyll measurements (Goericke, 2002), metaproteomic data not only showed the expression of this system, but also demonstrated that the major source of these proteins was gammaproteobacteria of the OM60 clade.
LC-MS/MS studies of proteins, such as this one, are a powerful complement to other tools being developed for microbial ecology. In practice, multiple factors affect protein detection, leading to the common caveat that failure to detect a given protein is not proof of its absence from the sample. Notwithstanding this issue, LC-MS/MS methods are rapidly progressing, and the advantages of seeing the final products of gene expression assure that metaproteomics will assume a valuable role in the advancement of microbial ecology.
References
Allgaier M, Uphoff H, Felske A, Wagner-Dobler I . (2003). Aerobic anoxygenic photosynthesis in Roseobacter clade bacteria from diverse marine habitats. Appl Environ Microbiol 69: 5051–5059.
Beja O, Suzuki MT, Heidelberg JF, Nelson KE, Preston CM, Hamada T et al. (2002). Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature 415: 630–633.
Bouvier T, del Giorgio PA . (2007). Key role of selective viral-induced mortality in determining marine bacterial community composition. Environ Microbiol 9: 287–297.
Brinkoff T, Giebel H-A, Simon M . (2008). Diversity, ecology, and genomics of the Roseobacter clade: a short overview. Arch Microbiol 189: 531–539.
Button DK, Robertson B . (2000). Effect of nutrient kinetics and cytoarchitecture on bacterioplankter size. Limnol Oceanogr 45: 499–505.
Cho JC, Giovannoni SJ . (2004). Cultivation and growth characteristics of a diverse group of oligotrophic marine Gammaproteobacteria. Appl Environ Microbiol 70: 432–440.
Cho JC, Stapels MD, Morris RM, Vergin KL, Schwalbach MS, Givan SA et al. (2007). Polyphyletic photosynthetic reaction centre genes in oligotrophic marine Gammaproteobacteria. Environ Microbiol 9: 1456–1463.
DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard N-U et al. (2006). Community genomics among stratified microbial assemblages in the ocean's interior. Science 311: 496–503.
Dyhrman ST, Haley ST . (2006). Phosphorus scavenging in the unicellular marine diazotroph Crocosphaera watsonii. Appl Environ Microbiol 72: 1452–1458.
Eng J, McCormack AL, Yates III JR . (1994). An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5: 976–989.
Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW et al. (2008). Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci USA 105: 3805–3810.
Fuchs BM, Spring S, Teeling H, Quast C, Wulf J, Schattenhofer M et al. (2007). Characterization of a marine gammaproteobacterium capable of aerobic anoxygenic photosynthesis. Proc Natl Acad Sci USA 104: 2891–2896.
Giovannoni SJ, Bibbs L, Cho JC, Stapels MD, Desiderio R, Vergin KL et al. (2005a). Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature 438: 82–85.
Giovannoni SJ, Stingl U . (2005). Molecular diversity and ecology of microbial plankton. Nature 437: 343–348.
Giovannoni SJ, Hayakawa DH, Tripp HJ, Stingl U, Givan SA, Cho JC et al. (2008). The small genome of an abundant coastal ocean methylotroph. Environ Microbiol 10: 1771–1782.
Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D et al. (2005b). Genome streamlining in a cosmopolitan oceanic bacterium. Science 309: 1242–1245.
Goericke R . (2002). Bacteriochlorophyll a in the ocean: Is anoxygenic bacterial photosynthesis important? Limnol Oceanogr 47: 290–295.
Hoch MP, Jeffrey WH, Snyder RA, Dillon KS, Coffin RB . (2006). Expression of glutamine synthetase and glutamate dehydrogenase by marine bacterioplankton: assay optimization and efficacy for assessing nitrogen to carbon metabolic balance in situ. Limnol Oceanogr 4: 308–328.
Kan J, Hanson TE, Ginter JM, Wang K, Chen F . (2005). Metaproteomic analysis of Chesapeake Bay microbial communities. Saline Systems 1: 7.
Karp-Boss L, Wheeler PA, Hales B, Covert P . (2004). Distributions and variability of particulate organic matter in a coastal upwelling system. J Geophys Res 109: C09010.
Lee S, Fuhrman JA . (1987). Relationships between biovolume and biomass of naturally derived marine bacterioplankton. Appl Environ Microbiol 53: 1298–1303.
Lo I, Denef VJ, Verberkmoes NC, Shah MB, Goltsman D, DiBartolo G. et al. (2007). Strain-resolved community proteomics reveals recombining genomes of acidophilic bacteria. Nature 446: 537–541.
Martiny AC, Coleman ML, Chisholm SW . (2006). Phosphate acquisition genes in Prochlorococcus ecotypes: evidence for genome-wide adaptation. Proc Natl Acad Sci USA 103: 12552–12557.
McDonald WH, Ohi R, Miyamoto DT, Mitchison TJ, Yates JR . (2002). Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. 219: 245–251.
Miller TR, Belas R . (2004). Dimethylsulfoniopropionate metabolism by Pfiesteria-associated Roseobacter spp. Appl Environ Microbiol 70: 3383–3391.
Moran MA, Buchan A, Gonzalez JM, Heidelberg JF, Whitman WB, Kiene RP et al. (2004). Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432: 910–913.
Moran MA, Gonzalez JM, Kiene RP . (2003). Linking a Bacterial taxon to sulfur cycling in the sea: studies of the marine Roseobacter group. Geomicrobiol J 20: 375–388.
Morris RM, Longnecker K, Giovannoni SJ . (2006). Pirellula and OM43 are among the dominant lineages identified in an Oregon coast diatom bloom. Environ Microbiol 8: 1361–1370.
Morris RM, Nunn BL, Frazar C, Goodlett DR, Ting YS, Rocap G . (2010). Comparative metaproteomics reveals ocean-scale shifts in microbial nutrient utilization and energy transduction. ISME J 4: 673–685.
Morris RM, Rappe MS, Connon SA, Vergin KL, Siebold WA, Carlson CA et al. (2002). SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420: 806–810.
Muller-Karger FE, Varela R, Thunell R, Luerssen R, Hu C, Walsh JJ . (2005). The importance of continental margins in the global carbon cycle. Geophys Res Lett 32: L01602.
Park K . (1967). Nutrient regeneration and preformed nutrients off oregon. Limnol Oceanogr 12: 353–357.
Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP . (2003). Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res 2: 43–50.
Poretsky RS, Bano N, Buchan A, LeCleir G, Kleikemper J, Pickering M et al. (2005). Analysis of microbial gene transcripts in environmental samples. Appl Environ Microbiol 71: 4121–4126.
Ram RJ, Verberkmoes NC, Thelen MP, Tyson GW, Baker BJ, Blake RC et al. (2005). Community proteomics of a natural microbial biofilm. Science 308: 1915–1920.
Rappe MS, Vergin K, Giovannoni SJ . (2000). Phylogenetic comparisons of a coastal bacterioplankton community with its counterparts in open ocean and freshwater systems. FEMS Microbiol Ecol 33: 219–232.
Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S et al. (2007). The sorcerer II global ocean sampling expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol 5: e77.
Scanlan DJ, Silman NJ, Donald KM, Wilson WH, Carr NG, Joint I et al. (1997). An immunological approach to detect phosphate stress in populations and single cells of photosynthetic picoplankton. Appl Environ Microbiol 63: 2411–2420.
Sherr EB, Sherr BF, Wheeler PA . (2005). Distribution of coccoid cyanobacteria and small eukaryotic phytoplankton in the upwelling ecosystem off the Oregon coast during 2001 and 2002. Deep Sea Res Part 2 Top Stud Oceanogr 52: 317–330.
Sowell SM, Wilhelm LJ, Norbeck AD, Lipton MS, Nicora CD, Barofsky D et al. (2008). Transport functions dominate the SAR11 metaproteome at low nutrient extremes in the Sargasso Sea. ISME J 3: 93–105.
Steinberg DK, Carlson CA, Bates NR, Johnson RJ, Michaels AF, Knap AH . (2001). Overview of the US JGOFS Bermuda Atlantic Time-series Study (BATS): a decade-scale look at ocean biology and biogeochemistry. Deep-Sea Research II 48: 1405–1447.
Stingl U, Desiderio RA, Cho JC, Vergin KL, Giovannoni SJ . (2007). The SAR92 clade: an abundant coastal clade of culturable marine bacteria possessing proteorhodopsin. Appl Environ Microbiol 73: 2290–2296.
Tabb DL, McDonald WH, Yates III JR . (2002). DTASelect and contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res 1: 21–26.
Thompson MR, Chourey K, Froelich JM, Erickson BK, VerBerkmoes NC, Hettich RL . (2008). Experimental approach for deep proteome measurements from small-scale microbial biomass samples. 80: 9517–9525.
Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW et al. (2005). Comparative metagenomics of microbial communities. Science 308: 554–557.
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA et al. (2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science 304: 66–74.
Verberkmoes NC, Russell AL, Shah M, Godzik A, Rosenquist M, Halfvarson J et al. (2008). Shotgun metaproteomics of the human distal gut microbiota. ISME J 3: 179–189.
Wheeler PA, Huyer A, Fleischbein J . (2003). Cold halocline, increased nutrients and higher chlorophyll off Oregon in 2002. Geophys Res Lett (online) 30: 8021; doi:10.1029/2003GL017395.
Wilhelm LJ, Tripp HJ, Givan SA, Smith DP, Giovannoni SJ . (2007). Natural variation in SAR11 marine bacterioplankton genomes inferred from metagenomic data. Biol Direct 2: 27.
Wu J, Sunda W, Boyle EA, Karl DM . (2000). Phosphate depletion in the western North Atlantic Ocean. Science 289: 759–762.
Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K et al. (2007). The sorcerer II global ocean sampling expedition: expanding the universe of protein families. PLoS Biol 5: e16.
Zubkov MV, Sleigh MA, Burkill PH . (2000). Assaying picoplankton distribution by flow cytometry of underway samples collected along a meridional transect across the Atlantic Ocean. Aquat Microb Ecol 21: 13–20.
Acknowledgements
We thank the crew of the Elaka and Joshua Kitner for their help in sample collection, and Francis Chan for data and discussions about oceanographic conditions. This work was supported in part by a Marine Microbiology Initiative Investigator Award from the Gordon and Betty Moore Foundation. Additional sponsorship was received from the US Department of Energy under contract DE-AC05-00OR22725 with Oak Ridge National Laboratory, managed and operated by UT-Battelle, LLC.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Sowell, S., Abraham, P., Shah, M. et al. Environmental proteomics of microbial plankton in a highly productive coastal upwelling system. ISME J 5, 856–865 (2011). https://doi.org/10.1038/ismej.2010.168
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2010.168
Keywords
This article is cited by
-
Compartment-related aspects of XoxF protein functionality in Methylorubrum extorquens DM4 analysed using its cytoplasmic targeting
Antonie van Leeuwenhoek (2023)
-
Metabolic tuning of a stable microbial community in the surface oligotrophic Indian Ocean revealed by integrated meta-omics
Marine Life Science & Technology (2022)
-
Protist diversity and community complexity in the rhizosphere of switchgrass are dynamic as plants develop
Microbiome (2021)
-
Bacterial community variations in the South China Sea driven by different chemical conditions
Ecotoxicology (2021)
-
Metaproteomics reveal that rapid perturbations in organic matter prioritize functional restructuring over taxonomy in western Arctic Ocean microbiomes
The ISME Journal (2020)