Abstract
Encephalitozoon cuniculi is a model microsporidian species with a mononucleate nucleus and a genome that has been extensively studied. To date, analyses of genome diversity have revealed the existence of four genotypes in E. cuniculi (EcI, II, III and IV). Genome sequences are available for EcI, II and III, and are all very divergent, possibly diploid and genetically homogeneous. The mechanisms that cause low genetic diversity in E. cuniculi (for example, selfing, inbreeding or a combination of both), as well as the degree of genetic variation in their natural populations, have been hard to assess because genome data have been so far gathered from laboratory-propagated strains. In this study, we aim to tackle this issue by analyzing the complete genome sequence of a natural strain of E. cuniculi isolated in 2013 from a steppe lemming. The strain belongs to the EcIII genotype and has been designated EcIII-L. The EcIII-L genome sequence harbors genomic features intermediate to known genomes of II and III lab strains, and we provide primers that differentiate the three E. cuniculi genotypes using a single PCR. Surprisingly, the EcIII-L genome is also highly homogeneous, harbors signatures of heterozygosity and also one strain-specific single-nucleotide polymorphism (SNP) that introduces a stop codon in a key meiosis gene, Spo11. Functional analyses using a heterologous system demonstrate that this SNP leads to a deficient meiosis in a model fungus. This indicates that EcIII-L meiotic machinery may be presently broken. Overall, our findings reveal previously unsuspected genome diversity in E. cuniculi, some of which appears to affect genes of primary importance for the biology of this pathogen.
Similar content being viewed by others
Introduction
Encephalitozoon cuniculi is a microsporidian species that infects a wide range of vertebrate hosts from birds to humans (Katinka et al., 2001; Corradi, 2015). Microsporidia are obligate intracellular parasites that are characterized by a unique invasion apparatus (the polar tube) and a loss (or severe degeneration) of mitochondrial genomes (Keeling and Fast, 2002). Other seemingly simplified cellular features in this group include the presence of ultrashort and diverged ribosomal RNA (rRNA) genes, and a lack of flagella and peroxisomes (Vavra and Lukes, 2013). The entire lineage has been recently associated with the Cryptomycota, a phylum that sits at the base of the fungal branch of the tree of Life (James et al., 2013). To date, many microsporidian genomes have been sequenced and all have been found to be very gene poor—that is, harboring a maximum of 3500 open reading frames (Corradi, 2015). Homologs of genes involved in conserved biochemical pathways are few in microsporidia compared with other eukaryotes; a feature that highlights their dependence on host cells for metabolic supplies (Corradi and Selman, 2013). Finally, most genome studies of species with mononucleated spores have revealed evidence of diploidy in microsporidia (Katinka et al., 2001; Cuomo et al., 2012; Selman et al., 2013; Desjardins et al., 2015; Watson et al., 2015), although recent analyses of diplokaryotic species have suggested that polyploidy can also occur in this group (Pelin et al., 2015).
The genome of E. cuniculi was the first to be sequenced in this phylum, and has been widely acknowledged as a model of reduction and adaptation (Katinka et al., 2001). Indeed, this genome is not only gene poor but is also extremely small (2.9 Mb in size) and compressed, harboring ~2000 genes that locate within minute intergenic regions (mean intergenic region is 80 bp; Katinka et al., 2001). To date, four E. cuniculi genotypes have been recognized to exist (referred to as EcI, II, III and IV) and are differentiated on the basis of repeats located in their rRNA internal transcribed spacers (Talabani et al., 2010; Pombert et al., 2013). Genomes representatives of EcI, II and III acquired from laboratory strains have been sequenced and found to be not only very divergent in sequence but also identical in gene content (Pombert et al., 2013). Evidence of recombination among genotypes is absent (Pombert et al., 2013), but the presence of low levels of genetic diversity, with putative heterozygous single-nucleotide polymorphism (SNP) ranging from 21 to 23 in all strains, suggested that a sexual diploid–haploid cycle leading to novel genetic diversity via outcrossing exist in E. cuniculi (Selman et al., 2013).
Low genetic diversity in E.cuniculi has been proposed to result from self-reproduction (selfing), inbreeding, mitotic recombination or a combination of all (Selman et al., 2013). The first two processes involve sexual reproduction, but these have been hard to differentiate because passage in culture for decades could theoretically lead to inbreeding and ultimately loss of diversity (Saul et al., 1999; Wang et al., 2012). Mitotic recombination is an asexual alternative to reduce genetic diversity in E. cuniculi (LaFave and Sekelsky, 2009), but its frequency must be unusually high to produce highly homogeneous genomes (Esquissato et al., 2014). To understand how low diversity is generated and maintained in E. cuniculi, genome analyses of new strains isolated from the field (that is, natural populations) may be required. In particular, their inspection could reveal if low genetic diversity is the norm in this species. Assuming that a conventional microsporidian sexual cycle exist in E. cuniculi (Lee et al., 2014), the presence of a highly heterozygous (assuming diploidy) or genetically diverse population of spores in natural strains could indicate that E. cuniculi strains occasionally outcross in the field (Selman et al., 2013). Genome sequence data from new natural strains are also a requisite to identify the extent and nature of genetic diversity that exist in field samples of these vertebrate parasites.
In the present study, we provide the complete genome of a natural strain we refer to as EcIII-L (genotype III), isolated in 2013 in the Czech Republic (Hofmannova et al., 2014). This genome sequence was annotated and compared with similar data from lab strains, revealing important insights into the natural genetic diversity present in this group and their mode of reproduction.
Materials and methods
Culturing, spore purification and DNA extraction of ECIII-L
The spores of E. cuniculi strain ECIII-L were originally isolated from a naturaly infected steppe lemming (Lagurus lagurus; Hofmannova et al., 2014), passaged through severe combined immunodeficient mice infected perorally with a suspension made from steppe lemming-homogenized brain, and spores acquired from peritoneal lavage of these mice made 21 days post infection were then grown in vitro in Green monkey kidney cells (VERO, line E6) maintained in RPMI-1640 medium (Sigma) supplemented with 2.5% heat-inactivated fetal bovine serum. The spores were collected weekly from infected cell line by collecting supernatants of the cultures and stored in phosphate-buffered saline supplemented with antibiotics (Sigma, 100 U/ml penicillin, 100 μg/ml streptomycin and 2.5 μg/ml amphotericin B) at 4 °C. Prior DNA isolation, the spores were purified from cell debris by centrifugation over 50% Percoll (Sigma) at 1100 g for 30 min and washed three times in sterilized deionised water. DNA was extracted from Percoll-purified spores using MasterPure Complete DNA and RNA purification kit from Epicentre Biotechnologies (Madison, WI, USA).
Genome sequencing, assembly and annotation
Extracted DNA was subjected to deep sequencing. Libraries were constructed and sequenced using Illumina HiSeq 2500 technology by Fasteris SA (Geneva, Switzerland). Sequencing resulted in 28 870 687 paired-end reads with a Q30 of 92.62% and a length of 125 bp. Raw reads have been deposited in the SRA database under accession SRR2105612. Adapters were trimmed and overlapping paired reads were merged using SeqPrep (github.com/jstjohn/SeqPrep). Merged reads were treated as single-end reads in downstream analysis.
Two independent denovo assemblies were run using both merged and paired-end unmerged reads. Initially, we used Ray denovo assembler v2.3.1 (Boisvert et al., 2010) with a k value of 123 to generate contiguous sequences. Contigs were validated by comparing with existing E. cuniculi assemblies, as well as by mapping paired-end reads back to them using mapping algorithms implemented in Geneious Pro. Because some chromosomes were represented by multiple contigs, we have tried to assemble the data set with SPAdes v3.5.0 (Bankevich et al., 2012). By manually merging both assemblies using overlap-layout-consensus algorithms implemented in Geneious Pro. The resulting contigs were then screened for misassemblies by mapping paired-end reads and visually inspecting coverage.
The final assembly was annotated by identifying open reading frames (ORFs) using ‘Find ORFs’ function in Geneious Pro. These were then blasted using blastp homology searches against the nr database. Whole-genome alignment using MAUVE (Darling et al., 2004) revealed the genome to be in perfect synteny with other E. cuniculi isolates. RNA features such as rRNA and transfer RNA were transferred from E. cuniculi GB-M1 using reciprocal blast.
Read processing, mapping and coverage analysis
Quality trimming minimizes downstream artefacts (Minoche et al., 2011) and was performed using the PERL script trim-fastq.pl from the PoPoolation toolkit (Kofler et al., 2011). Quality trimmed reads were used for any downstream analysis. By mapping reads back to the assembled genome using mapping algorithms implemented in Geneious Pro, we have determined the average sequencing depth of the genome to be 2332 ×. In order to exclude paralagous regions from SNP discovery, we have excluded areas of the genome that had a depth 25% higher than the average sequencing depth.
Polymorphism discovery and SNP calling
Trimmed reads were mapped against our final assembly to quantify variation. Mapping was done using mapping algorithms implemented in Geneious Pro (Kearse et al., 2012), which in turn use ‘low sensitivity’ parameters that allow up to a 10% disagreement between reads and reference. To identify heterozygous loci, we have used the ‘Find Variations/SNPs’ function implemented in Geneious Pro. We have set a minimum allele frequency of 35% as previously described (Selman et al., 2013). All potential heterozygous loci were verified using PCR, followed by direct Sanger sequencing of the resulting products and manual inspection of the sequencing chromatograms (Supplementary Figure S1). By repeating this analysis with a smaller threshold for allele frequency, we identified lower frequency SNPs that we could not validate using Sanger Sequencing.
In order to compare our novel strain with other available sequences of E. cuniculi, we have aligned sequenced of individual chromosomes together using the MAUVE algorithm (Darling et al., 2004) implemented in Geneious Pro. Alignment blocks containing sequences from all isolates (ECI, ECII, ECII-CZ, ECIII and ECIII-L) were analyzed for variation using the ‘Find Variations/SNPs’ function implemented in Geneious Pro.
PCR for SNP and genotyping validation
PCRs performed to validate SNP calls were carried out in 20 μl final volume containing a mixture of 10.0 μl of EconoTaq DNA polymerase (Lucigen, Middleton, WI, USA), 0.5 mm each primer and 1.0 μl of DNA. The thermal cycling conditions included an initial step of 94 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 2 min and final step of 72 °C for 12 min.
In order to evaluate the divergent region on chromosome 6 as a potential genotyping locus, we have designed primers annealing to flanking conserved regions. Primers CH06_Geno_F 5p-AATACTTGGCCAGGTGTATGTC-3p and CH06_Geno_R 5p-AGCAGTTCAGTTTCCTCTCCATG-3p were used on DNA extracted from cultured spores successfully (Figure 4a) with the following thermal cycling conditions: an initial step of 95 °C for 3 min, followed by 35 cycles of 95 °C for 30 s, 54.8 °C for 30 s, and 72 °C for 1 min and final step of 72 °C for 10 min. Obtaining PCR products from other sources, however, required a nested PCR approach (Figure 4b). In this case, the first PCR was carried out using the same primers and conditions except for the annealing step, which was carried out at 50 °C instead of 54.8 °C. The second PCR used the product of the first PCR as template, and was carried out with the same forward primer CH06_Geno_F 5p-AATACTTGGCCAGGTGTATGTC-3p and a different reverse primers CH06_Geno_R2 5p-CACTGGACCGGCGATCT-3p and again an annealing step at 50 °C. Ladders used in the Figures 4a and b gels are ExcelBand 1 kb (0.25–10 kb) DNA Ladder (Diamed, Mississauga, ON, Canada) and 100 bp DNA Ladder (Solis BioDyne, Tartu, Estonia), respectively, with expected PCR product sizes observed: ECI 1467 bp, ECII 1270 bp and ECIII/ECIII-L 1030 bp.
Heterologous expression of SPO11 in yeast
S. cerevisiae strains used in this study included MATα and MATa strains from BY4741 background (ura3Δ0 leu2Δ0 his3Δ1 met15Δ0). S. cerevisiae SPO11 gene was knocked out in MATα strain using directed PCR transformation approach (Omidi et al., 2014). Diploid spo11Δ cells were generated by crossing two haploid strains: MATa containing kanamycin-resistance cassette and MATα containing clonNAT-resistance cassette. SPO11 overexpression plasmid was obtained from the yeast ORF collection (Gelperin et al., 2005). pBI-880 plasmid was used to facilitate the directional cloning of ECIII and ECIII-L genes (Kohalmi et al., 1998).
PCR product containing ECIII and ECIII-L flanked with NotI and SalI was generated and cloned into pBI-880 plasmid. Colony PCR followed by sequencing was used to confirm the identity of the new constructs.
Diploid spo11Δ S. cerevisiae cells carrying the appropriate plasmids were grown at 30°C from single colonies to near-stationary phase in synthetic complete media with -leucine or -uracil dropout (Samanfar et al., 2014). Strains were washed and resuspended in sporulation medium (Tong et al., 2001) to an optical density at 600 nm of 1.0 (Gerke et al., 2006). After 3 days at room temperature the number of sporulated cells was counted as colony-forming unit and presented as percentage of spores (McCusker and Haber, 1977). Selection based on both resistant markers (kanamycin and clonNAT) was used to account for the presence of diploid cells. Galactose was used to induce gene expression of the plasmids.
Results and Discussion
Genome acquisition annotation of ECIII-L
The microsporidian spores were isolated from a naturally infected steppe lemming (Lagurus lagurus) originating from private breeding suffering from lethal microsporidiosis (Hofmannova et al., 2014). Genotyping was performed using PCR amplification of a partial sequence of the 18S rRNA operon using modified microsporidia-specific primers described by De Bosschere et al. (2007) (Katzwinkel‐Wladarsch et al., 1996). This revealed the Encephalitozoon cuniculi belonged to genotype III. DNA extracted from ~108 spores of EcIII-L was used to construct a 2 × 125 bp Illumina library, which was sequenced using the HiSeq 2500 platform. Sequencing resulted in 28 870 687 pairs of paired-end reads with a Q30 of 92.62%. Reads were assembled in parallel with the SPAdes v3.5.0 assembler (Bankevich et al., 2012) as well as with the Ray v2.3.1 assembler (Boisvert et al., 2010), and the resulting assemblies manually inspected and merged using the overlap-layout-consensus algorithm implemented in Geneious Pro version R8.1.2 (http://www.geneious.com, Kearse et al., 2012). The final assembly size reaches 2.3 Mb, and is separated into 15 contigs with an N50 of 201956 bp. The assembly size and the number of contigs compare favorably with those previously obtained by others on other isolates, but is lower than that of GB-M1 (2.50 Mb). Ten contigs represent nearly complete E. cuniculi chromosomes. As previously observed in Encephalitozoon spp chromosome 9 is fragmented into five pieces, reflecting the repetitive nature of this particular region of the genome (Pombert et al., 2013).
Genome annotation was performed manually using ORF identification and BLAST (Altschul et al., 1990) procedures included in Geneious Pro, resulting in the identification of 1896 genes (Table 1), 1834 coding regions (coding sequences), 11 pseudogenes, 46 transfer RNA, 3 rRNA and 2 non-coding RNA features. Blast homology searches failed to reveal novel genes with known function and the EcIII-L genome is perfectly syntenic with the other E. cuniculi assemblies available (GCA_000091225.1, GCA_000221245.2, GCA_000221265.2 and GCA_000221285.2). We found that EC1-GB-M1 (Katinka et al., 2001) has a slightly higher ORF count compared with all other isolates, probably a result of a more complete genome assembly. Indeed, EC1-GB-M1 has been sequenced by Sanger and better covers duplicated regions. All sequence data analyzed in the study, including sequencing reads, assembly and annotation, have been deposited on NCBI (BioProject PRJNA210874; Annotation LFTZ01000000; Reads SRR2105612). Genome statistics and comparisons with other available strains are reported on Table 1.
The ECIII-L spores are genetically very homogeneous
The presence of genome diversity among spores of ECIII-L was investigated by mapping high-quality Illumina reads against the assembled reference genome. SNP located in regions that significantly deviate from average coverage were discarded to avoid false positives. This approach allowed us to score a total of 247 SNPs where at least five reads with an alternate allele mapped against the genome (5% of reads at a given location; Supplementary Table S1; Li et al., 2008; Venturini et al., 2013). Sanger sequencing of regions encompassing SNP with variable frequencies (0.1–0.5, n=29) could never validate SNP with frequencies <35% (Supplementary Figure S1). Instead, these procedures showed that all SNP present with frequencies >35% are at a 50/50 ratio in the genome (Supplementary Figure S2; also see Material and methods for additional details). These PCR based validations assume that there are no biases towards one of the alternative alleles, but are supported by independent analyses of Illumina sequence quality (see below).
In total, these procedures retrieved a total of 23 putative heterozygous SNPs in ECIII-L, a number almost identical to those previously reported from laboratory isolates (see Figure 1, Table 2; Selman et al., 2013). As for other isolates, the location of all SNP is unique to ECIII-L and does not appear to affect particular pathways, and the level and nature of intragenomic diversity of EcIII-L is virtually identical to that of lab strains. The total amount of SNP found in all strains is always extremely low compared with distant species (that is, SNP affect between 0.003–0.007% of each E. cuniculi genome, as opposed to 0.99%–1.24% for Nematocida and Nosema, respectively). Importantly, we found that the variation in amount of SNP is well correlated with the average quality of Illumina sequencing used to generate the data. Specifically, only 80% of the reads used to map strains with much higher SNP counts (EC1, EC2 and EC3, sequenced by the Broad Institute) have a Q30 value or higher, but this number goes up to 94% for those strains with lower SNP counts ((EC2-CZ and EC3-L, this study; Supplementary Figure S3). This supports the notion that many low-frequencies SNP identified through mapping are probably the result of sequencing errors, although some of these could also result from intra-population diversity.
Interestingly, we found that very low-genome diversity also extents to the related species Encephalitozoon hellem and E. romaleae, which harbor 33 and 27 SNPs at a 50/50 ratio, respectively (Figure 1). Overall, these analyses demonstrate that genetic homogeneity is high and common in isolates of Encephalitoon cuniculi and related species, and confirms that nuclei in this lineage, referred to as monokaryons (Didier et al., 1991), are genetically very homogeneous, and possibly diploid like all mononucleate microsporidian species with sequenced genomes (Cuomo et al., 2012; Selman et al., 2013; Desjardins et al., 2015; Watson et al., 2015). An alternative explanation for the existence of a conserved 50/50 SNP would assume a balanced mixture of equally frequent homozygous genotypes in several independent mixed infections (and strains), a situation that sounds improbable.
Comparative genomics and inter-strain divergence
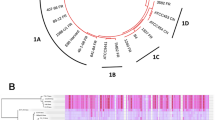
Genome comparisons uncovered SNPs specific to each strain (Figure 2) and confirmed that ECI represents the most divergent isolate (Pombert et al., 2013). We confirm that the internal transcribed spacers region of ECIII-L harbors the typical repeat signature of genotype III (Figure 3a) and that this isolate shares more indels with ECIII than it does with other strains (Figure 3b). Surprisingly, however, ECIII-L shares substantially more substitutions with ECII and ECII-CZ (443 SNPs) than it does with the other ECIII isolate (39 SNPs; Figure 2a).
Characteristics of polymorphisms found in our novel ECIII-L strain. (a) Venn diagram of shared and unique SNPs of all currently sequenced E. cuniculi strains. (b) Phylogeny based on SNPs only extracted from genome alignment of all strains. (c) Distribution along all 11 chromosomes of SNPs unique to ECIII-L (top), shared between ECIII-L and ECII (middle), and those shared between ECIII and ECIII-L (bottom).
Genotype identification of a novel Encephalitozoon cuniculi strain. (a) Alignment of ribosomal RNA intergenic spacer region of different genotype of Encephalitozoon cuniculi. Repeated GTTT motifs indicating the genotype of a E. cuniculi strain are noted by gray box. (b) Large indels, 50 bp and more are shown. These indels are shared features between our novel strain and previously sequenced ECIII.
Our investigations also uncovered a region that can be readily used to genotype E. cuniculi strains (that is, EcI, II, III) without the need for sequencing. This region is found on chromosome 6, and in isolates ECI and ECIII this region harbors ORFs with conserved homeodomain motifs and top blast hits (Supplementary Figure S4,Supplementary Table S2) to yeast Yarrowia lipolytica homeodomain genes (ECI) and A2 mating type protein in the basiodiomycete Phanerochaete chrysosporium (ECII; James et al., 2011).
Interestingly, this ORF is absent in ECIII and ECIII-L. PCR using corresponding flanking primers produces specific bands for each genotype (Figure 4a). The use of present primers represent a good alternative to a marker based on the repeat-rich SWP gene previously proposed for genotyping, and can be readily used to genotype DNA extracted from spleen, liver, brain, kidney and feces (Figure 4b). This proposed method is the first to reliably identify E. cuniculi genotypes without the need for Sanger sequencing.
(a) Agarose gel showing PCR products for two sets of primers ran on all available Encephalitozoon cuniculi isolates. First PCR primer set shown on the left allows discrimination of E. cuniculi genotypes based on size only. Second PCR primer set shown on the right using Spore Wall Protein (SWP) as a target for genotyping. (b) Agarose gel showing PCR products of our genotyping locus on samples originating from spleen, liver, brain, kidney and feces from different E. cuniculi genotypes.
A stop codon in the 5′region of a key meiosis gene in ECIII-L: identification and functional analysis
Unsurprisingly, sequence divergence affects mostly the coding regions of this gene-dense genome (87.1% of all substitutions), but it can sometimes result in pseudogenization (see Supplementary Table S3 for a list of ECIII-L pseudogenes). As previously reported, rapid divergence does not seem to affect a particular pathway in E. cuniculi strains ((Pombert et al., 2013); see Supplementary Table S4 for a list of the 30 E. cuniculi most rapidly diverging genes) or in other microsporidian species (Pelin et al., 2015). Intriguingly, we also identified one specific SNP in ECIII-L located 153 bp downstream the start codon of the gene Spo11, a key regulator of meiotic recombination (Inagaki et al., 2010). This SNP creates a stop codon that should theoretically result the loss of an essential domain (TopoIIB, Figure 5) conserved in other E. cuniculi isolates and distantly related microsporidia. The presence of the mutation was validated using PCR and Sanger sequencing (Figure 5c). The potential effects of this SNP on Spo11 function was tested using a heterologous Saccharomyces cerevisiae system (Figure 6).
Verification of spo11 mutation by PCR. (a) Schematic representation of the role of spo11 in meiosis. (b) Alignment of spo11 gene (ECU04_1110) from E. cuniculi I/II/III/III-L, showing position of stop codon in E. cuniculi III-L. (c) PCR validation of spo11 mutation in Encephalitozoon cuniculi III-lemming and II. Mutation site is identified by a black arrow. (d) Alignment consisting of different phyla of Spo11 protein sequence.
Relative spore formation for different S. cerevisiae strains. Total number of spores formed after three days of incubation is normalized to that produced by WT. Each experiment was repeated at least four times. Error bars represent s.d. Deletion of S. cerevisiae spo11 resulted in reduced spore formations. This reduction was compensated by introduction of a plasmid harboring spo11 for either S. cerevisiae or ECIII, but not for ECIII-L or an empty vector. The presence of the mutation was validated using PCR and Sanger sequencing.
It was observed that ECIII Spo11 containing the domain was able to restore the function of S. cerevisiae Spo11 in SPO11 gene deletion strain (spo11Δ), but not the ECIII-L version with a premature stop codon. As expected (Klapholz et al., 1985; Keeney, 2001) in S. cerevisiae, deletion of SPO11 resulted in a significant reduction (over 80%) in spore formation. Reintroduction of S. cerevisiae and ECIII restored the ability of spo11Δ to generate spores, but not the introduction of ECIII-L Spo11.
Discovery of new Encephalitozoon genome diversity in the field
Studies of intra-species genome diversity in E. cuniculi have so far been limited to what is available—that is, strains propagated under laboratory conditions for decades. As a result, our knowledge of their genome evolution in the field is non-existent. The present study fills this gap by reporting the first genome analysis of an E. cuniculi strain recently isolated from the field (Hofmannova et al., 2014). Our findings revealed that all strains analyzed to date evolve in very similar ways, regardless of their origin. Specifically, they are all genetically homogenous, most likely diploid, very divergent (59% of SNP in the ECIII-L are specific to this strain) and are also prone to pseudogenizations. The EcIII-L genome also revealed that this strain is transitional to known EcIII and EcII strains, sharing important genomic characteristics with both—that is, indels/repeats link shared with EcIII, but genome sequence related to EcII. Clearly, techniques commonly used to genotype E. cuniculi strains reveal only a small portion of their evolutionary relationships. Unfortunately, we could not find a genetic marker that easily distinguishes all five strains analyzed in this study, as our PCR approach can only distinguish the three fully sequenced genotypes. Nonetheless, our study clearly demonstrates that inter-strain genetic diversity in E. cuniculi is quite high, a finding that warrants additional investigations of natural samples of these parasites.
What drives genome homogenization in Encephalitozoon?
Previous studies of genome diversity in E.cuniculi revealed evidence that the spores isolated from lab cultures are genetically highly homogeneous (Selman et al., 2013). However, the mechanisms that homogenize the E. cuniculi genomes remained unclear. Here we report that ECIII-L exhibits little genetic diversity (247 SNP scored, of which 23 are validated as potentially heterozygous) on par with values reported from lab strains. This demonstrates that reduction in genetic diversity is common in this species and not linked to long-term culturing. Selfing is one mechanism that produces homozygous genomes, but this process requires a meiotic machinery (that is, sex) that may be broken in EcIII-L. Indeed, all E. cuniculi strains harbors complete sequences of most meiosis-specific genes (Lee et al., 2008, 2014), but EcIII-L harbors a frameshift mutation that cannot fully restore meiosis in a model fungus. A priori, this suggests that ECIII-L (and to certain extent, also other species that potentially lack portions of Spo11, such as the Mitosporidium daphnia, Rozella allomycis and Nosema bombycis, Figure 5) may not be capable of sexual reproduction and that genome homogenization in this strain (and possibly all strains) must be driven by asexual mechanisms. One of these could be mitotic recombination, as this known asexual driver of genome homogenization in many microbial eukaryotes, including many pathogens (Butler et al., 2009; Cuomo et al., 2012; Rosenblum et al., 2013), has been shown to increase in frequency in the absence of a functional Spo11 (Lario et al., 2015; Sun and Heitman, 2015).
As a sexual alternative, ECIII-L could undergo meiosis without the need for Spo11, a situation that would be analogous to what is seen in the distant microbial lineage Dyctiostelium sp (Goodenough and Heitman, 2014), or perhaps, the SNP we identified is too recent to have impacted the genome of ECIII-L in significant ways (that is, EcIII-L has always been a ‘selfer’, and the recent Spo11 mutation we found will only affect the mutational patterns of ECIII-L down the road). Indeed, besides the one SNP we report, and the rest of Spo11 in ECIII-L is completely identical to other isolates, suggesting that this frameshift is recent. The effect of this mutation on the genome will soon be tested by propagating EcIII-L under laboratory conditions, and by screening SNP at different time intervals.
One last, provocative hypothesis for the lack of genetic diversity is that the putative diploid monokaryons of E. cuniculi spores (and possibly in the spores of allied species) are not homologous to diploid nuclei. Perhaps, these must first fuse and form tetraploid diplokaryons (Bernander et al., 2001; Lee et al., 2014) to trigger meiosis and create genetic diversity? This hypothesis is supported by evidence of diploidy in mononucleate species (Katinka et al., 2001; Cuomo et al., 2012; Selman et al., 2013; Desjardins et al., 2015; Watson et al., 2015) and tetraploidy in Nosema spp. diplokaryons—that is,the stage that triggers meiosis and formation of unikaryons in this genus (Pelin et al., 2015). To be fully supported, however, this hypothesis will require the identification of diplokaryotic (or uni-diplokaryotic) populations of E. cuniculi. Such analyses would also greatly benefit from cytogenetic observations and/or estimates of nuclear DNA content, which are currently lacking in the field of microsporidian research. In any case, it is expected that further explorations of genome ploidy and diversity in the field and crossing experiments performed in the lab will shed light into this unknown aspect of the E. cuniculi biology.
Data Archiving
All data analyzed and discussed within the manuscript is publicly available on GenBank though the genome project PRJNA210874 (http://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA210874). The project contains: (1) the complete genome sequence; (2) the genome annotation; and (3) the sequencing reads used to assemble the genome and identify SNP.
Accession codes
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ . (1990). Basic local alignment search tool. J Mol Biol 215: 403–410.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19: 455–477.
Bernander R, Palm JE, Svard SG . (2001). Genome ploidy in different stages of the Giardia lamblia life cycle. Cell Microbiol 3: 55–62.
Boisvert S, Laviolette F, Corbeil J . (2010). Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol 17: 1519–1533.
Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA et al. (2009). Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459: 657–662.
Corradi N . (2015). Microsporidia: eukaryotic intracellular parasites shaped by gene loss and horizontal gene transfers. Ann Rev Microbiol 69: 167–183.
Corradi N, Selman M . (2013). Latest progress in microsporidian genome research. J Eukaryot Microbiol 60: 309–312.
Cuomo CA, Desjardins CA, Bakowski MA, Goldberg J, Ma AT, Becnel JJ et al. (2012). Microsporidian genome analysis reveals evolutionary strategies for obligate intracellular growth. Genome Res 22: 2478–2488.
Darling AC, Mau B, Blattner FR, Perna NT . (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14: 1394–1403.
De Bosschere H, Wang Z, Orlandi P . (2007). First diagnosis of Encephalitozoon intestinalis and E. hellem in a European brown hare (Lepus europaeus) with kidney lesions. Zoonoses Public Health 54: 131–134.
Desjardins CA, Sanscrainte ND, Goldberg JM, Heiman D, Young S, Zeng Q et al. (2015). Contrasting host-pathogen interactions and genome evolution in two generalist and specialist microsporidian pathogens of mosquitoes. Nature Commun 6: 7121.
Didier PJ, Didier ES, Orenstein JM, Shadduck JA . (1991). Fine structure of a new human microsporidian, Encephalitozoon hellem, in culture. J Protozool 38: 502–507.
Esquissato GN, Sant'anna JR, Franco CC, Rosada LJ, Santos PA, Castro-Prado MA . (2014). Gene homozygosis and mitotic recombination induced by camptothecin and irinotecan in Aspergillus nidulans diploid cells. An Acad Bras Ciênc 86: 1703–1710.
Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ et al. (2005). Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev 19: 2816–2826.
Gerke JP, Chen CT, Cohen BA . (2006). Natural isolates of Saccharomyces cerevisiae display complex genetic variation in sporulation efficiency. Genetics 174: 985–997.
Goodenough U, Heitman J . (2014). Origins of eukaryotic sexual reproduction. Cold Spring Harb Perspect Biol 6: a016154.
Hofmannova L, Sak B, Jekl V, Mináriková A, Skorič M, Kváč M . (2014). Lethal Encephalitozoon cuniculi genotype III infection in Steppe lemmings (Lagurus lagurus). Vet Parasitol 205: 357–360.
Inagaki A, Schoenmakers S, Baarends WM . (2010). DNA double strand break repair, chromosome synapsis and transcriptional silencing in meiosis. Epigenetics 5: 255–266.
James TY, Lee M, van Diepen LT . (2011). A single mating-type locus composed of homeodomain genes promotes nuclear migration and heterokaryosis in the white-rot fungus Phanerochaete chrysosporium. Eukaryot Cell 10: 249–261.
James TY, Pelin A, Bonen L, Ahrendt S, Sain D, Corradi N et al. (2013). Shared signatures of parasitism and phylogenomics unite Cryptomycota and microsporidia. Curr Biol 23: 1548–1553.
Katinka MD, Duprat S, Cornillot E, Méténier G, Thomarat F, Prensier G et al. (2001). Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414: 450–453.
Katzwinkel‐Wladarsch S, Lieb M, Helse W, Löscher T, Rinder H . (1996). Direct amplification and species determination of microsporidian DNA from stool specimens. Trop Med Int Health 1: 373–378.
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649.
Keeling PJ, Fast NM . (2002). Microsporidia: biology and evolution of highly reduced intracellular parasites. Ann Rev Microbiol 56: 93–116.
Keeney S . (2001). Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol 52: 1–53.
Klapholz S, Waddell CS, Esposito RE . (1985). The role of the SPO11 gene in meiotic recombination in yeast. Genetics 110: 187–216.
Kofler R, Orozco-terWengel P, De Maio N, Pandey RV, Nolte V, Futschik A et al. (2011). PoPoolation: a toolbox for population genetic analysis of next generation sequencing data from pooled individuals. PLoS One 6: e15925.
Kohalmi SE, Reader LJV, Samach A, Nowak J, Haughn GW, Crosby WL . (1998) Identification and characterization of protein interactions using the yeast 2-hybrid system In Gelvin SB, Schilperoort RA (eds) Plant Molecular Biology Manual. Springer: Amsterdam, Netherlands. pp 95–124.
LaFave MC, Sekelsky J . (2009). Mitotic recombination: why? when? how? where? PLoS Genet. 5: e1000411.
Lario LD, Botta P, Casati P, Spampinato CP . (2015). Role of AtMSH7 in UV-B-induced DNA damage recognition and recombination. J Exp Bot 66: 3019–3026.
Lee SC, Heitman J, Ironside JE . (2014). Sex and the Microsporidia. In: Weiss LM, Becnel J (eds) Microsporidia: Pathogens of Opportunity First Edition. Wiley Blackwell: West Sussex, UK, pp 231–243.
Lee SC, Corradi N, Byrnes EJ 3rd, Torres-Martinez S, Dietrich FS, Keeling PJ et al. (2008). Microsporidia evolved from ancestral sexual fungi. Curr Biol 18: 1675–1679.
Li H, Ruan J, Durbin R . (2008). Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858.
McCusker JH, Haber JE . (1977). Efficient sporulation of yeast in media buffered near pH6. J Bacteriol 132: 180–185.
Minoche AE, Dohm JC, Himmelbauer H . (2011). Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and genome analyzer systems. Genome Biol 12: R112.
Omidi K, Hooshyar M, Jessulat M, Samanfar B, Sanders M, Burnside D et al. (2014). Phosphatase complex Pph3/Psy2 is involved in regulation of efficient non-homologous end-joining pathway in the yeast Saccharomyces cerevisiae. PloS One 9: e87248.
Pelin A, Selman M, Aris-Brosou S, Farinelli L, Corradi N . (2015). Genome analyses suggest the presence of polyploidy and recent human‐driven expansions in eight global populations of the honeybee pathogen Nosema ceranae. Environ Microbiol 17: 4443–4458.
Pombert J-F, Xu J, Smith DR, Heiman D, Young S, Cuomo CA et al. (2013). Complete genome sequences from three genetically distinct strains reveal high intraspecies genetic diversity in the microsporidian Encephalitozoon cuniculi. Eukaryot Cell 12: 503–511.
Rosenblum EB, James TY, Zamudio KR, Poorten TJ, Ilut D, Rodriguez D et al. (2013). Complex history of the amphibian-killing chytrid fungus revealed with genome resequencing data. Proc Natl Acad Sci USA 110: 9385–9390.
Samanfar B, Tan le H, Shostak K, Chalabian F, Wu Z, Alamgir M et al. (2014). A global investigation of gene deletion strains that affect premature stop codon bypass in yeast, Saccharomyces cerevisiae. Mol Biosyst 10: 916–924.
Saul DJ, Reeves RA, Morgan HW, Bergquist PL . (1999). Thermus diversity and strain loss during enrichment. FEMS Microbiol Ecol 30: 157–162.
Selman M, Sak B, Kváč M, Farinelli L, Weiss LM, Corradi N . (2013). Extremely reduced levels of heterozygosity in the vertebrate pathogen Encephalitozoon cuniculi. Eukaryot Cell 12: 496–502.
Sun S, Heitman J . (2015). From two to one: unipolar sexual reproduction. Fungal Biol Rev 29: 118–125.
Talabani H, Sarfati C, Pillebout E, van Gool T, Derouin F, Menotti J . (2010). Disseminated infection with a new genovar of Encephalitozoon cuniculi in a renal transplant recipient. J Cin Microbiol 48: 2651–2653.
Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Pagé N et al. (2001). Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368.
Vavra J, Lukes J . (2013). Microsporidia and 'the art of living together'. Adv Parasitol 82: 253–319.
Venturini L, Ferrarini A, Zenoni S, Tornielli GB, Fasoli M, Dal Santo S et al. (2013). De novo transcriptome characterization of Vitis vinifera cv. Corvina unveils varietal diversity. BMC Genomics 14: 41.
Wang L, Shi X, Su Y, Meng Z, Lin H . (2012). Loss of genetic diversity in the cultured stocks of the large yellow croaker, Larimichthys crocea, revealed by microsatellites. Int J Mol Sci 13: 5584–5597.
Watson AK, Williams TA, Williams BA, Moore KA, Hirt RP, Embley TM . (2015). Transcriptomic profiling of host-parasite interactions in the microsporidian Trachipleistophora hominis. BMC Genomics 16: 983.
Acknowledgements
We are grateful to Karen Haag and Jean-François Pombert for their critical comments on an earlier version of this manuscript. NC is a Fellow of the Canadian Institute for Advanced Research. NC and AG’s work is supported by the Discovery program from the Natural Sciences and Engineering Research Council of Canada (NSERC-Discovery) and an Early Researcher Award from the Ontario Ministry of Research and Innovation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Heredity website
Supplementary information
Rights and permissions
About this article
Cite this article
Pelin, A., Moteshareie, H., Sak, B. et al. The genome of an Encephalitozoon cuniculi type III strain reveals insights into the genetic diversity and mode of reproduction of a ubiquitous vertebrate pathogen. Heredity 116, 458–465 (2016). https://doi.org/10.1038/hdy.2016.4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2016.4
This article is cited by
-
Encephalitozoon cuniculi infection in farmed rabbits in Egypt
Acta Veterinaria Scandinavica (2020)
-
Genetic and Genome Analyses Reveal Genetically Distinct Populations of the Bee Pathogen Nosema ceranae from Thailand
Microbial Ecology (2019)