Abstract
Recent research has convincingly documented cases of mitochondrial heteroplasmy in a small set of wild and cultivated plant species. Heteroplasmy is suspected to be common in flowering plants and investigations of additional taxa may help understand the mechanisms generating heteroplasmy as well as its effects on plant phenotypes. The role of mitochondrial heteroplasmy is of particular interest in plants as cytoplasmic male sterility is controlled by mitochondrial genotypes, sometimes leading to co-occurring female and hermaphroditic individuals (gynodioecy). Paternal leakage may be important in the evolution of mating systems in such populations. We conducted a genetic survey of the gynodioecious plant Plantago lanceolata, in which heteroplasmy has not previously been reported, and estimated the frequencies of mitochondrial genotypes and heteroplasmy. Sanger sequence genotyping of 179 individuals from 15 European populations for two polymorphic mitochondrial loci, atp6 and rps12, identified 15 heteroplasmic individuals. These were distributed among 6 of the 10 populations that had polymorphisms in the target loci and represented 8% of all sampled individuals and 15% of the individuals in those 6 populations. The incidence was highest in Northern England and Scotland. Our results are consistent with geographic differences in the incidence of paternal leakage and/or the rates of nuclear restoration of male fertility.
Similar content being viewed by others
Introduction
In genetic studies of flowering plants, it is often assumed that most individuals carry a single complement of mitochondrial genes inherited from the maternal parent. However, increasing numbers of reports from crop (see, for example, Bragin et al., 2012; Szklarczyk et al., 2014) and wild species (see, for example, McCauley et al., 2005; Mandel and McCauley, 2015) have documented the co-occurrence of multiple mitochondrial genotypes within plant cells (termed mitochondrial heteroplasmy), suggesting that it might be common in many angiosperms (Kmiec et al., 2006). Mitochondrial heteroplasmy has important implications for evolution under nuclear and organellar genomic conflict and for the evolution of plant male sterility (Wade and McCauley, 2005; McCauley, 2013; Christie et al., 2015), but very little is known about its effects on plant phenotypes in natural populations. Despite its likely prevalence, heteroplasmy has been documented in populations of only a few plant species (see, for example, Pearl et al., 2009; Mandel and McCauley, 2015). To empirically test the theoretical models of such evolutionary processes we require investigations of heteroplasmy in natural populations.
Two main processes can produce mitochondrial heteroplasmy, mutation and paternal leakage, and once generated heteroplasmy can potentially allow recombination between mitochondrial genotypes (Kmiec et al., 2006). Flowering plants generally exhibit low mitochondrial mutation rates (Wolfe et al., 1987); therefore, paternal leakage is considered the predominant mechanism generating heteroplasmy in angiosperms (McCauley, 2013; Christie et al., 2015). However, species in some genera, Plantago included, appear to have very high mitochondrial synonymous substitution rates (Mower et al., 2007) that may reflect high mutation rates. The co-occurrence of nonfunctional mutant cytoplasmic genotypes and functional (nonmutant) genotypes can also arise because nonmutant mitochondrial genomes are necessary for viability. For example, highly pathogenic point mutations in human mitochondrial genes often remain heteroplasmic (Ye et al., 2014). Such heteroplasmy could also be deleterious because of mitochondrial competition, either between functional mitotypes or between functional and nonfunctional mitotypes that could produce metabolic dysfunction (Sharpley et al., 2012; Greiner et al., 2014).
Under strict maternal inheritance, mutations causing loss of male functions are likely to be neutral or beneficial to female fitness and thus can increase mitochondrial transmission to future generations (Lewis, 1942; Greiner et al., 2014). Cytoplasmic male sterility (CMS) polymorphisms can therefore be established (Chase, 2007), and are known in many plant species (reviewed by Schnable and Wise, 1998). The CMS mutant phenotype can be negated by nuclear ‘restorer’ alleles; within a species, appearance of a restorer can result in a return to hermaphroditism or lead to long-term gynodioecy with male sterile (female) and hermaphroditic individuals coexisting within populations (Lewis, 1942; Gouyon et al., 1991; Chase, 2007; Touzet and Delph, 2009). Paternal leakage, causing mitochondrial heteroplasmy, can influence the evolution of CMS in two ways: it allows selection on mitochondrial effects on male fitness (Frank, 1989) and can potentially restore male function in the offspring of a CMS mother plant, thereby increasing nuclear gene fitness (Andersson-Ceplitis and Bengtsson, 2002; McCauley, 2013). In the latter case, genotypes that lead to heteroplasmy would be predicted to be favored in populations where rates of nuclear restoration are low. However, little is currently known about the natural incidence of mitochondrial heteroplasmy and its role in the evolution and maintenance of CMS.
We therefore conducted a survey of mitochondrial sequence variants throughout Western Europe in order to estimate frequencies of polymorphism in natural populations of a gynodioecious flowering plant, ribwort plantain (Plantago lanceolata), and test for heteroplasmy that has not previously been examined in the species. By combining analyses of sequences with information on sex phenotypes, we also tested whether heteroplasmy is more common in gynodioecious P. lanceolata lacking restorers of male fertility than in populations with restorers.
Materials and methods
Study system
P. lanceolata is a herbaceous flowering plant with a worldwide distribution. The species is obligately outcrossing because of self-incompatibility, and populations are usually gynodioecious (Ross, 1973). Controlled crossing experiments have demonstrated cytonuclear control of sex phenotypes in P. lanceolata; three CMS types were identified, with evidence that mitochondrial genotypes differ between them (Van Damme and Van Delden, 1982). Alleles that restore male fertility in the presence of specific CMS types are found at multiple nuclear loci, and interact epistatically (de Haan et al., 1997a). The estimated frequency of male restorer alleles varies among populations and CMS types, being lowest for CMS type-1 (de Haan et al., 1997a).
Plant collecting and phenotyping
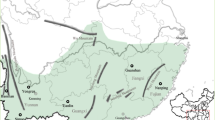
Living plants, leaf material or seeds were collected from 15 wild populations of P. lanceolata in Western Europe (Table 1 and Figure 1). Sex phenotypes were recorded in the wild at the time of collection or under greenhouse conditions following germination. All individuals represented by dried leaf or living material were genotyped, whereas wild collected seeds were germinated and one plant per seed family was chosen for downstream analyses. Seedlings and living plants were maintained in the Newcastle University growth facilities. Male sterility was identified as the absence of pollen producing anthers on all sexually mature inflorescences. When the phenotype was unambiguous, the trait values ‘male sterile’ or ‘hermaphrodite’ were assigned; individuals with ambiguous or unknown phenotypes were categorized as ‘undetermined’.
Population locations and mitochondrial genotype frequencies of Western European P. lanceolata. The relative proportion of single-nucleotide polymorphism (SNP) genotypes at atp6 and rps12 (listed in Table 2) are represented in pie charts by color: ‘T’ (black), ‘C’ (orange) and heteroplasmic ‘C/T’ (pink).
Sequencing, trace accuracy and genotype calling
DNA was extracted from leaf tissue samples using a modified CTAB (cetyl trimethylammonium bromide) protocol (Wolff, 1996). Partial mitochondrial genome assemblies (constructed from Illumina, San Diego, CA, USA; paired-end reads; N Levsen and K Wolff, unpublished) of eight geographically disparate European P. lanceolata individuals were used to design PCR and sequencing primers (Supplementary Table S1) for fragments of two mitochondrial genes (atp6 and rps12). PCR amplification was conducted in a 20 μl volume consisting of 4.2 μl of 5 × MyTaq Reaction Buffer (Bioline, London, UK), 0.5 μl (10 pM) of each primer, 13.4 μl H2O, 0.4 μl MyTaq DNA polymerase and 1 μl of template DNA. The temperature cycling protocol was as follows: 95 °C initial denaturation for 3 min; 35 cycles at 95 °C for 15 s, 52 °C for 15 s and 72 °C for 30 s; a final extension step at 72 °C for 5 min. Sanger sequencing of both DNA strands was performed at the Edinburgh Genomics facility (Edinburgh, UK). The sequence reads were quality trimmed and aligned to the Mimulus guttatus mitochondrial reference sequence (GenBank: JN098455.1; Mower et al., 2012) in CLC Genomics Workbench v8.0.2 (CLCbio, Arhus, Denmark). The annotated M. guttatus sequence provided a published guide to determining site position and category (for example, synonymous).
Identifying heteroplasmy
Sanger DNA sequencing at heterozygous or heteroplasmic sites produces two electropherogram trace peaks: one for each segregating nucleotide. The height ratio of those peaks indicated the proportion of each nucleotide in the underlying sample (Roy and Schreiber, 2014). To assess our ability to detect heteroplasmy, we generated mixtures of two alleles of the mitochondrial atp6 gene and sequenced them. PCR cloning of atp6 (CloneJET PCR cloning kit, Thermo Scientific, Waltham, MA, USA) was performed on an individual heteroplasmic for these variants, revealing a single polymorphism (C/T) at position 443 relative to the M. guttatus atp6 start position. Cleaned, quantified (10–20 ng μl−1) PCR products representing two clones of each type were mixed to generate the series ranging from 100% T to 100% C, with 10 intermediate mixtures (5:95, 10:90, 20:80, 30:70, 40:60, 60:40, 70:30, 80:20, 90:10 and 95:5). Each mix was bidirectionally sequenced with two replicates. Linear regression of the original allele mix proportions and mean trace peak heights for forward and reverse reads was performed using SigmaPlot v12.5 (Systat Software Inc., San Jose, CA, USA). Based on the results of the allelic mixtures, we set the Sanger trace peak threshold indicating heteroplasmy at a site to >10% of the site’s total peak height. Peak height was determined using the ‘Secondary Peak Calling’ tool in CLC.
Genotypes at atp6 position 443 and rps12 position 138 (also polymorphic for C and T) were used to identify heteroplasmic individuals. Individuals with significant peaks for both C and T in both the forward and reverse reads were classified as heteroplasmic.
Experimental crosses
We performed controlled crosses within P. lanceolata to test: (1) whether allele transmission was predominantly maternal; (2) whether mitochondrial heteroplasmy can be inherited; and (3) whether paternal leakage generates new cases of heteroplasmy. The first of these tests was done to check that our sequences were not from nuclear mitochondrial (numt) pseudogenes that are common in plant genomes (Hazkani-Covo et al., 2010). Homoplasmic maternal and paternal plants, as well as one heteroplasmic mother, were chosen from previously genotyped individuals. The plants chosen as parents were grown in adjacent pots and flowering shoots from each were entwined and covered with a paper bag to prevent pollen contamination. The resulting seeds were germinated on moist filter paper and mature progeny were genotyped as above for the atp6 and rps12 variants.
Recombination and tests for genotype–phenotype associations
To estimate linkage disequilibrium between atp6 and rps12 and test for historical recombination between them, we calculated r2 and the minimum number of recombination events (RM) in DnaSP (Librado and Rozas, 2009). Estimates of r2 and RM were generated for each population that was polymorphic at both loci, for the geographic region in which both polymorphisms were found (Northern England and Scotland, see Results) and for the entire data set (excluding individuals with missing data). Associations between atp6 and/or rps12 genotypes and male sterility (excluding plants with ‘undetermined’ sex phenotypes) were tested using two-tailed Fisher’s exact tests of independence in SigmaPlot. Based on our results, tests were also done separately for individuals from the populations of Northern England and Scotland and for individuals from all populations outside of that region.
Results
Sanger sequencing was used to assess the incidence of heteroplasmy at two mitochondrial loci: a nonsynonymous Ser/Leu variant in atp6 and a synonymous variant in rps12. Sequences were generated for 179 wild plants for atp6 and 178 individuals for rps12. The sequences are in GenBank under accession numbers KU596577–KU596971. Most populations from mainland Europe were monomorphic at both loci, but all Northern English and Scottish populations (hereafter referred to as NES) were polymorphic for both atp6 and rps12 sequence variants (Table 2 and Figure 1). The atp6 with the thymine nucleotide (hereafter denoted by the T-allele) was restricted to the NES region and was generally rarer than the C-allele (except in the CGR population). The rps12 C-allele was rare in non-NES populations, although it was the major allele (66.7%) in population SMY/SVL and the commoner allele in one (CGR) of the seven NES populations.
Sanger trace accuracy
Trace peak height ratios for the atp6 thymine nucleotide at position 443 were significantly correlated with the proportions of the T-allelic haplotype in the mixtures sequenced (Supplementary Figures S1a and b for forward and reverse sequencing, respectively) and generally agreed between the two sequencing directions. A single false positive for heteroplasmy was detected (in one reverse read). However, the r2 values (0.55 and 0.62 for forward and reverse reads, respectively) do not suggest precise allelic quantification. To avoid falsely inferring heteroplasmy when the individual was, in fact, homoplasmic, the threshold for declaring a false peak was set at a minor peak height of 10% of the total for the site being examined. False negatives for heteroplasmy were detected in 26 reads (out of 80 sequences of heteroplasmic mixtures).
Frequency of heteroplasmy
With the conservative (high) detection threshold just explained, mitochondrial heteroplasmy was detected in 6 of our 15 sampled populations (Table 2 and Figure 1). Heteroplasmy for one or other of the loci was generally found whenever the locus was polymorphic; the only exceptions in Table 2 are populations CGR and VIK. High percentages of plants heteroplasmic for both loci were found in three NES populations. Given that the proportions of mitochondrial genotypes in heteroplasmic plants can vary among cells of a single individual (Woloszynska, 2010), as well as our conservative detection threshold and probable high false negative rate (Supplementary Figure S1), our results probably must underestimate the prevalence of heteroplasmy in these natural populations. Considering only those populations in which heteroplasmy was detected, 15% of plants were heteroplasmic on average.
Inheritance of mitochondrial alleles in experimental crosses
The atp6 and rps12 genotypes in the progeny of five crosses between P. lanceolata individuals with different mitochondrial genotypes support maternal transmission of the loci (Table 3), confirming that the variants are in mitochondrial genes. In addition, heteroplasmy at atp6 and rps12 was transmitted from the maternal parent of the CGR18 × IJM11.2 cross to three of the four progeny (the reciprocal cross was not performed). Our Illumina paired-end read data (see section on sequencing methods) for atp6 in this maternal plant gives an allele ratio estimate of ∼76% T-allele (53T-allele reads versus 17 C).
Recombination
In the NES populations as a whole, both loci are polymorphic and all four haplotypes were found, whereas the non-NES populations included only the C-T and C-C (atp6-rps12) haplotypes at very different frequencies from those in the NES populations (Table 4). Shared haplotypes exhibited very different frequencies between the two data sets. Linkage disequilibrium between the atp6 and rps12 genes was significant in the pool of NES individuals (in these populations, both loci are polymorphic) and in five of the six individual NES populations that could be tested. Estimates from the NES population data set yield at least one historical recombination event between the genes (RM=1), but all individual populations included only three of the four possible haplotypes and, therefore, yielded zero RM estimates.
Genotype–phenotype association
Male sterility data were obtained for 89 individuals, including 7 heteroplasmic individuals (out of 15 in the total data set); all 7 were from 2 NES populations, TYN and CGR (4 were hermaphroditic and 3 were male sterile, see Supplementary Table S2). The male sterile individuals could be identified phenotypically as either CMS type-1 or type-3 (see Supplementary Table S2; Van Damme and Van Delden, 1982). Associations between the mitochondrial genotypes and sex phenotypes were tested separately for atp6 and rps12, as there is evidence that the loci recombine (Table 4). In the NES data, both loci showed associations, with the atp6 C and rps12 T-alleles significantly overrepresented among male sterile individuals (Table 5). In contrast, in the non-NES sample, with only rps12 polymorphic, no association was found with the T-allele.
Discussion
Our results show that gynodioecious populations of P. lanceolata contain single-nucleotide polymorphism sequence variants in mitochondrial genes, including both synonymous and nonsynonymous variants. Previous studies of mitochondrial genotypes in this species used anonymous variants detected by restriction enzymes (Rouwendal et al., 1987; de Haan et al., 1997b). We also confirmed, in further populations of this plant, these authors' tentative previous observations of associations between mitochondrial genotypes and male sterility in Dutch populations. Finally, we describe the first evidence of mitochondrial heteroplasmy in this plant.
Even with our moderate sample sizes, and just two loci, we identify at least ~8% (15 of 179) of the sampled individuals as heteroplasmic at one of the mitochondrial loci sequenced or, in 9 individuals, both (Table 2 and Supplementary Table S2). This percentage is lower than detected in natural populations of Silene vulgaris (~15%; Pearl et al., 2009) and Daucus carota (30%; Mandel and McCauley, 2015), possibly because the quantitative real-time PCR detection method used in those studies is more sensitive.
Heteroplasmic P. lanceolata individuals were found in 6 of the 15 populations sampled, including 4 populations from NES (Table 2 and Figure 1). Recombinant haplotypes were found in all NES populations but CGR, even in the two (DUN and CAR) in which heteroplasmy was not detected, indicating that recombination (Table 4) occurs, or has occurred, in their history. One cannot therefore assume that the P. lanceolata mitochondrial genome is nonrecombining when studying its sequence diversity (McCauley, 2013).
Plant mitochondrial heteroplasmy probably results primarily from paternal leakage and this has indeed been detected in family studies of S. vulgaris (Bentley et al., 2010) and in wild populations of Daucus (Mandel and McCauley, 2015); however, in Daucus a consistent deficit of the T variant in the atp9 gene is unexpected unless other processes also operate. Our experimental crosses (Table 3) provided no direct evidence for paternal leakage for either atp6 or rps12 alleles in P. lanceolata, but larger families should be tested. We did, however, find indirect evidence of paternal leakage: all four two-locus haplotype combinations were present in natural populations (Table 4). The generation of all four haplotype combinations between two two-allele loci requires either recombination between two divergent co-occurring mitochondrial genomes (Hudson and Kaplan, 1985) or else convergent evolution (McCauley and Olson, 2008). The independent appearance of the same variant by mutation occurring multiple times within a population is much less likely than a recombination event, particularly if the mutation rate in plant mitochondria is low. Although an elevated ancestral mitochondrial synonymous substitution rate has been inferred in the Plantago lineage (Mower et al., 2007), rates vary greatly among Plantago species and the rate estimated for P. lanceolata is among the lowest in the genus (Cho et al., 2004). However, we cannot exclude convergent mutation as an explanation for our results, given that none of our local population samples included all four haplotype combinations and that the T-T haplotype was represented by only one individual in the data set (Tables 2 and 4).
Once heteroplasmy is established, it may persist for multiple generations by maternal transmission, which we detect in P. lanceolata (Table 3). Nevertheless, it is expected to diminish over the generations because of the bottleneck in the egg cells each generation, and this could explain the absence of heteroplasmy in one offspring of CGR18 (Table 3). Stochastic losses may account for the highly variable frequencies of heteroplasmy in the different populations, with much higher frequencies in the CGR and TYN populations than in the other populations that exhibit polymorphisms. Alternatively, the variable frequencies could be because of selection.
A strong association between a mitochondrial genotype and the sex phenotype is evidence that nuclear genes restoring male fertility are not common in a population. We observe associations with the variants in both genes studied here, atp6 and rps12, in the NES P. lanceolata populations (Table 5). In contrast, despite a similar abundance of male sterile plants in our samples from the non-NES populations (both ∼40%, see Supplementary Table S2), the atp6 gene is not polymorphic, and the presence of the C versus T variant of the rps12 gene does not predict plants’ sex phenotypes in these populations (Table 5). Moreover, the rps12 C variant is rare in most populations in both sex types among the plants we phenotyped (Table 2). Nuclear restoration of male fertility therefore probably occurs oftener in those populations than in the NES region.
If one or more mitochondrial haplotypes carry a sterility factor, paternal leakage of a nonsterility haplotype could be an alternative mechanism that restores male fertility (Andersson-Ceplitis and Bengtsson, 2002; Wade and McCauley, 2005). Genotypes that are transmitted via paternal leakage and favored for their ability to restore male fertility should then occur most frequently in populations in which the rate of nuclear male restoration is low for at least one sterilizing element. However, given the absence of any mitochondrial variant associated with CMS in the non-NES populations, the lower frequency of mitochondrial heteroplasmy may simply be because of low diversity rather than a low rate of nuclear male restoration.
The differences between the two sets of populations studied here suggest the loss of a male fertile mitochondrial type (with the T-C haplotype) from the non-NES populations, rather than the spread of the male fertile haplotype in NES populations. Table 5 shows that the allele frequencies at both loci are similar in females in both sets of populations, whereas in hermaphrodites, the atp6 T variant has a much higher frequency in the NES populations (and is much more common than in females); the same is true for the rps12 C variant. This suggests a loss of the T-C haplotype, specifically in non-NES population hermaphrodites (see Table 4), and fixation of a male sterility haplotype with the C variant in atp6 and the synonymous rps12 T variant (the most common haplotype in females in NES populations, see Table 5). Supplementary Table S2 shows that, of the 24 NES females genotyped, 19 had the C-T haplotype, 3 were heteroplasmic (genotypes C/T-C/T) and only 2 had T-C. In contrast, of the 29 NES hermaphrodites genotyped, 14 had T-C, only 10 had C-T, 3 were heteroplasmic at both loci and the C-C and T-T haplotypes were each carried by one individual. The T-C haplotype is thus associated with male fertility and C-T with male sterility, although the association is not complete (consistent with our evidence above that recombination occurs in this plant’s mitochondrial genome). The possibility that the C-T haplotype (associated with male sterility in the NES region where restoration is infrequent) has become fixed in the non-NES populations conforms to the theoretical prediction (Frank, 1989) that CMS polymorphisms may often be ephemeral, that polymorphism may be lost once a restorer appears, and that this causes fixation of the CMS mitochondrial genotype, in an ‘epidemic’ or ‘arms race’ scenario. Uneven rates of restorer activity across populations have been previously documented in P. lanceolata (de Haan et al., 1997a); however, the differences reported here may have been caused or exacerbated by long-term gene flow restriction between mainland Europe and the northern British Isles (Searle et al., 2009). Whether the ‘epidemic’ scenario is likely to have happened in the P. lanceolata populations should be further tested in the future by comparing diversity of mitochondrial sequences with that of nuclear genes, including sequences from related outgroup species, in order to take account of possible mutation rate differences (Ingvarsson and Taylor, 2002).
At present, the high detection threshold and low precision of the Sanger genotyping method precludes additional investigations of allele ratios within individual P. lanceolata plants or allelic dosage effects (Stewart and Chinnery, 2015) on sex phenotypes. Neither the atp6 nor the rps12 locus was associated with a restriction site and the flanking sequence for both loci made allele-specific priming unreliable. However, now that it is known that at least some P. lanceolata populations are heteroplasmic, it should be possible to identify heteroplasmic variants in other loci that are amenable to more sensitive approaches such as restriction site genotyping, quantitative real-time PCR or quantitative genotyping via deep sequencing. Further investigations of mitochondrial heteroplasmy in P. lanceolata will depend on sensitive and consistent allele quantification and include: direct tests of paternal leakage using expanded experimental crossing designs, of gene-by-gene and whole mitochondrial genome associations with male sterility and of the threshold heteroplasmy ratio for phenotypic effects.
Data archiving
DNA sequences are available at GenBank (KU596577—KU596971).
Accession codes
References
Andersson-Ceplitis H, Bengtsson BO . (2002). Transmission rates and phenotypic effects of mitochondrial plasmids and cytotypes in Silene vulgaris. Evolution 56: 1586–1591.
Bentley KE, Mandel JR, McCauley DE . (2010). Paternal leakage and heteroplasmy of mitochondrial genomes in Silene vulgaris: evidence from experimental crosses. Genetics 185: 961–968.
Bragin AG, Ivanov MK, Fedoseeva LA, Dymshits GM . (2012). Analysis of mitochondrial DNA heteroplasmy of fertile and male-sterile sugar beet plants (Beta vulgaris. Russ J Genet Appl Res 2: 53–57.
Chase CD . (2007). Cytoplasmic male sterility: a window to the world of plant mitochondrial-nuclear interactions. Trends Genet 23: 81–90.
Cho Y, Mower JP, Qiu Y-L, Palmer JD . (2004). Mitochondrial substitution rates are extraordinarily elevated and variable in a genus of flowering plants. PNAS 101: 17741–17746.
Christie JR, Schaerf TM, Beekman M . (2015). Selection against heteroplasmy explains the evolution of uniparental inheritance of mitochondria. PLoS Genet 11: e1005112.
de Haan AA, Koelewijn HP, Hundscheid MPJ, Van Damme JMM . (1997a). The dynamics of gynodioecy in Plantago lanceolata L. II. Mode of action and frequencies of restorer alleles. Genetics 147: 1317–1328.
de Haan AA, Mateman AC, van Dijk PJ, van Damme JMM . (1997b). New CMS types in Plantago lanceolata and their relatedness. Theor Appl Genet 94: 539–548.
Frank SA . (1989). The evolutionary dynamics of cytoplasmic male sterility. Am Nat 133: 345–376.
Gouyon P-H, Vichot F, van Damme JMM . (1991). Nuclear-cytoplasmic male sterility: single-point equilibria versus limit cycle. Am Nat 137: 498–514.
Greiner S, Sobanski J, Bock R . (2014). Why are most organelle genomes transmitted maternally? Bioessays 37: 80–94.
Hazkani-Covo E, Zeller RM, Martin W . (2010). Molecular poltergeists: mitochondrial DNA copies (numts in sequenced nuclear genomes. PLoS Genet 6: e1000834.
Hudson RR, Kaplan NL . (1985). Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111: 147–164.
Ingvarsson PK, Taylor DR . (2002). Genealogical evidence for epidemics of selfish genes. PNAS 99: 11265–11269.
Kmiec B, Woloszynska M, Janska H . (2006). Heteroplasmy as a common state of mitochondrial genetic information in plants and animals. Curr Genet 50: 149–159.
Lewis D . (1942). The evolution of sex in flowering plants. Biol Rev 17: 46–67.
Librado P, Rozas J . (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452.
Mandel JR, McCauley DE . (2015). Pervasive mitochondrial sequence heteroplasmy in natural populations of wild carrot, Daucus carota spp. carota L. PLoS One 10: e0136303.
McCauley DE . (2013). Paternal leakage, heteroplasmy, and the evolution of plant mitochondrial genomes. New Phytol 200: 966–977.
McCauley DE, Bailey MF, Sherman NA, Darnell MZ . (2005). Evidence for paternal transmission and heteroplasmy in the mitochondrial genome of Silene vulgaris, a gynodioecious plant. Heredity 95: 50–58.
McCauley DE, Olson MS . (2008). Do recent findings in plant mitochondrial molecular and population genetics have implications for the study of gynodioecy and cytonuclear conflict? Evolution 62: 1013–1025.
Mower JP, Touzet P, Gummow JS, Delph LF, Palmer JD . (2007). Extensive variation in synonymous substitution rates in mitochondrial genes of seed plants. BMC Evol Biol 7: 135.
Mower JP, Case AL, Floro ER, Willis JH . (2012). Evidence against equimolarity of large repeat arrangements and a predominant master circle structure of the mitochondrial genome from a monkeyflower (Mimulus guttatus lineage with cryptic CMS. Genome Biol Evol 4: 670–686.
Pearl SA, Welch ME, McCauley DE . (2009). Mitochondrial heteroplasmy and paternal leakage in natural populations of Silene vulgaris, a gynodioecious plant. Mol Biol Evol 26: 537–545.
Ross MD . (1973). Inheritance of self-incompatibility in Plantago lanceolata. Heredity 30: 169–176.
Rouwendal GJA, van Damme JMM, Wessels JGH . (1987). Cytoplasmic male sterility in Plantago lanceolata L.: differences between male-sterile cytoplasms at the DNA- and RNA-level. Theor Appl Genet 75: 59–65.
Roy S, Schreiber E . (2014). Detecting and quantifying low level gene variants in Sanger sequencing traces using the ab1 Peak Reporter tool. J Biomol Tech 25 (Suppl): S13–S14.
Schnable PS, Wise RP . (1998). The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci 3: 175–180.
Searle JB, Kotlík P, Rambau RV, Marková S, Herman JS, McDevitt AD . (2009). The Celtic fringe of Britain: insights from small mammal phylogeography. Proc R Soc B 276: 4287–4294.
Sharpley MS, Marciniak C, Eckel-Mahan K, McManus M, Cimi M, Waymire K et al. (2012). Heteroplasmy of mouse mtDNA is genetically unstable and results in altered behavior and cognition. Cell 151: 333–343.
Stewart JB, Chinnery PF . (2015). The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat Rev Genet 16: 530–542.
Szklarczyk M, Szymański M, Wójcik-Jagła M, Simon PW, Weihe A, Börner T . (2014). Mitochondrial atp9 genes from petaloid male-sterile and male-fertile carrots differ in their status of heteroplasmy, recombination involvement, post-transcriptional processing as well as accumulation of RNA and protein product. Theor Appl Genet 127: 1689–1701.
Touzet P, Delph LF . (2009). The effect of breeding system on polymorphism in mitochondrial genes of Silene. Genetics 181: 631–644.
Van Damme JMM, Van Delden W . (1982). Gynodioecy in Plantago lanceolata L I. polymorphism for plasmon type. Heredity 49: 303–318.
Wade MJ, McCauley DE . (2005). Paternal leakage sustains the cytoplasmic polymorphism underlying gynodioecy but remains invisible by nuclear restorers. Am Nat 166: 592–602.
Wolfe KH, Li W-H, Sharp PM . (1987). Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA 84: 9054–9058.
Wolff K . (1996). RAPD analysis of sporting and chimerism in chrysanthemum. Euphytica 89: 159–164.
Woloszynska M . (2010). Heteroplasmy and stoichiometric complexity of plant mitochondrial genomes – though this be madness, yet there’s method in’t. J Exp Bot 61: 657–671.
Ye K, Lu J, Ma F, Keinan A, Gu Z . (2014). Extensive pathogenicity of mitochondrial heteroplasmy in healthy human individuals. Proc Natl Acad Sci USA 111: 10654–10659.
Acknowledgements
This work was supported by the Natural Environment Research Council (Grant Number NE/J011444/1). The majority of the DNA sequencing was carried out at Edinburgh Genomics facility at the University of Edinburgh. Edinburgh Genomics is partly supported through core grants from NERC (R8/H10/56), MRC (MR/K001744/1) and BBSRC (BB/J004243/1). We thank Edinburgh Genomics Staff, and in particular K Gharbi and H Gunter, for their support and assistance with sequencing. We thank E Nilsson for plant collection assistance. We also thank H Martin, M Peake and M Bendall for help with plant growth, DNA extraction and PCR cloning.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Heredity website
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Levsen, N., Bergero, R., Charlesworth, D. et al. Frequent, geographically structured heteroplasmy in the mitochondria of a flowering plant, ribwort plantain (Plantago lanceolata). Heredity 117, 1–7 (2016). https://doi.org/10.1038/hdy.2016.15
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2016.15