Abstract
Segregation distortion is the phenomenon whereby the observed genotypic frequencies of a locus fall outside the expected Mendelian segregation ratio, and it is increasingly recognised as a potentially powerful evolutionary force. The main reason for segregation distortion is a difference in the viability of gametes and zygotes caused by viability loci in the segregating progeny. However, the maternal cytoplasm may also be involved in the viability selection of gametes and zygotes. The objectives of this study were to map the segregation distortion loci (SDL) in maize and to test the hypothesis that the viability of gametes and zygotes may also be associated with the maternal cytoplasmic environment. In the present study, a reciprocal mating design was conducted to generate an F2-segregating population. A linkage map was constructed with 126 microsatellite markers. A whole-genome scan was performed to detect the SDL in segregating populations with different maternal cytoplasm environments. Altogether, 14 SDL with strong LOD (logarithm (base 10) of odds) supports were identified in the specifically designed F2 populations. Interestingly, we found dramatic changes in the genotypic frequencies of the SDL in the two maternal cytoplasmic backgrounds, which indicated a change in the viability of gametes and zygotes in different cytoplasmic environments. Furthermore, in the JB cytoplasmic background, most of the detected SDL and complete distortion markers exhibited similar bias patterns favouring the Y53 alleles. These results suggested that selfish cytoplasmic elements may have an important role in shaping the patterns of segregation distortion in F2 populations through selective viability of gametes and zygotes.
Similar content being viewed by others
Introduction
Segregation distortion, also called transmission ratio distortion, is the phenomenon whereby alleles at a locus deviate from the Mendelian expectation. This phenomenon has been described in many species, including maize (Lu et al., 2002), rice (Yamagishi et al., 2010), eggplant (Barchi et al., 2010), alfalfa (Li et al., 2011), mouse (Casellas et al., 2010) and drosophila (Orr and Irving, 2005; Phadnis, 2011). These alleles, also called segregation distorters, might alter the transmission ratio in specific cases and are considered selfish genetic elements that manipulate Mendelian transmission to their own advantage (Hurst and Werren, 2001; Phadnis and Orr, 2009; Werren, 2011). Evolutionary research suggests that segregation distorters may be involved in promoting population divergence and, ultimately, speciation (Phadnis and Orr, 2009; McDermott and Noor, 2010). A variety of physiological and genetic factors may explain the observed segregation distortion. For example, meiotic drive, the preferential selection that occurs during meiosis, may cause certain alleles to be overrepresented in the gametes (McDermott and Noor, 2010). As a result, distorted Mendelian ratios may be observed in offspring that have undergone genotypic segregation. Zygotic selection may also have an important role in the observed segregation distortion. Li et al. (2011) studied diploid alfalfa and concluded that zygotic and not gametic selection is the main cause of segregation distortion. In invertebrates, Sinkins (2011) found a new embryo-killing gene that causes post-segregation distortion. In addition, mapping populations, genetic transmission, non-homologous recombination, gene transfer, transposable elements, reproductive isolation, environmental agents and non-biological factors, such as sample size, marker type and genotyping errors, are all possible causes for the observed segregation distortion (Xu et al., 1997; Wang et al., 2009; Phadnis, 2011). However, the most promising explanation is viability selection occurring at the markers or loci linked to the markers observed to have distorted inheritance (Vogl and Xu, 2000). If the gametic or zygotic viability of an allele at a locus selectively diminishes, then that locus and other loci linked to it will deviate from the expected Mendelian segregation ratio. These loci are referred to as segregation distortion loci (SDL). It appears that segregation distortion is rather common. If segregation distortion is indeed caused by viability selection loci, these loci are themselves of interest because they may help elucidate the mechanism of natural selection and evolution. Mapping the SDL provides reliable statistical parameters that will help to define the viability loci for further characterisation at a molecular level.
In genetic mapping, the change of marker allelic frequencies is reflected by the departure from the Mendelian segregation ratio. The non-Mendelian segregation of markers has been used to map SDL along the genome. χ2-tests are commonly used to test segregation distortion and to detect the markers linked to SDL at each locus in a set of marker loci. In addition to this single-marker analysis, several methods have been developed for mapping the SDL using whole-genome scanning (Vogl and Xu, 2000; Luo and Xu, 2003; Zhu et al., 2007; Zhan and Xu, 2011). Based on the genotypic frequencies of observed markers, Luo et al. (2005) proposed a novel liability model for mapping the SDL by assuming that the viability of individuals is controlled by a continuous liability. Mapping the SDL can then be formulated as a problem of general quantitative trait loci (QTL) mapping using only the allelic frequency information of markers along the genome. Recently, the SDL mapping module was integrated into the PROC QTL software, making it user friendly for many disciplines (Hu and Xu, 2009).
Currently, many SDL have been identified in plant and animal studies. However, few studies have investigated the possible maternal cytoplasmic contribution in segregation distortion. The cytoplasm contains a variety of organelles, including mitochondria and chloroplasts, which contain their own DNA. The cytoplasm provides an environment for nuclear gene expression and cellular metabolic reactions. In addition, the close relationship between the nucleus and cytoplasm has attracted much attention from macro- and micro-evolutionary perspectives. Research on cytonuclear coevolution and coadaptation has suggested that cytonuclear epistasis substantially contributes to genetic expression (Rand et al., 2004; Wolf, 2009; Chou and Leu, 2010).
Therefore, we hypothesise that the cytoplasm may also be involved in viability selection of gametes and zygotes. To verify this hypothesis, we analysed reciprocal F2 populations in maize. We examined the segregation patterns along the genome. The objectives of the experiment were twofold as follows: (1) to map the SDL that trigger the viability of gametes and zygotes in the maize genome and (2) to find the SDL with varying distortion ratios that are dependent upon the maternal cytoplasmic environments, thereby confirming the inference that the viability of gametes and zygotes is associated with the maternal cytoplasmic environment.
Materials and methods
Maize population and marker assays
The two inbred lines used to develop the F2-segregating populations were JB and Y53, which are the parental lines of the maize hybrid, Suyu 16, which has strong heterosis. To investigate the role of the maternal cytoplasmic environment, we constructed a bulked F2-mapping population with 120 individuals from the JB (female parent) × Y53 (male parent) direct cross and 120 individuals from the Y53 (female parent) × JB (male parent) reciprocal cross. The benefit of this mating design is that the segregation ratios of each marker are observed in two different maternal cytoplasmic backgrounds under the same genomic background. The bulked F2 populations were planted in the field at an experimental station located at Yangzhou University during July 2007. Samples of total DNA from the parents and F2 individuals were extracted using the procedures described by Rogers and Bendich (1988). Primer sequences were obtained from the maize genome database (http://www.maizegdb.org). Each F2 individual was genotyped by non-denaturing polyacrylamide gel electrophoresis without considering the cytoplasmic source. In total, 126 markers were screened and used to construct the linkage map, which covered 10 chromosomes and 1418.1 cM of the maize genome with an average marker spacing of 12.2 cM. The missing proportion of the genotypes was 6.1%. The distribution of these SSR markers was consistent with publicly available published linkage maps (Lima et al., 2006; Frascaroli et al., 2007; Yang et al., 2008) and the integrated map in the maize genome database (www.maizegdb.org).
Detection of the SDL
To evaluate the association between the maternal cytoplasmic environment and the viability of gametes or zygotes, we performed the analysis on the direct and reciprocal crosses separately. A joint analysis of QTL and SDL employing the Expectation-Maximisation algorithm has been proposed to estimate QTL and SDL parameters simultaneously (Xu and Hu, 2009). This joint method incorporated in PROC QTL Version 2.0 was used to detect the underlying SDL with a step length of 1 cM along each linkage group (Hu and Xu, 2009). Details of this method have been described by Xu and Hu (2009) and can also be found in the PROC QTL (Version 2.0) manual (http://statgen.ucr.edu/download/software). In the output result table, the threshold to identify a significant SDL was set at an LOD of 3.0.
To test the different effect of the two cytoplasmic backgrounds on the level of segregation distortion, χ2-tests were implemented to compare the genotypic proportion in the two crosses. The selection coefficients and degree of dominance were estimated for each SDL according to the fitness model (Luo et al., 2005). The comparison of selection coefficients in the two cytoplasmic backgrounds was implemented by the Wilcoxon signed-rank test. In the present study, the effect size of the cytoplasm on segregation distortion was defined as the difference of the average selection coefficient for a given genotype in the two cytoplasmic backgrounds.
Results
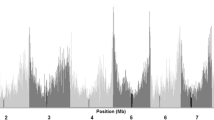
The top panels in Figures 1 and 2 show the frequencies of the three genotypes plotted against the maize genome. Theoretically, the genotype ratio should be 1:2:1. However, there were several severe distortions along the genome, such as at the end of chromosome 3, as shown in both figures. The interesting phenomenon was the difference of segregation patterns between the two reciprocal F2 populations (top panels of Figures 1 and 2), although the two populations were derived from the same two parental lines. For instance, the ratio of JB homozygotes, heterozygotes and Y53 homozygotes was 14.83:16.06:69.10% at the end of chromosome 6 with the JB cytoplasm. However, this ratio was 23.31:53.28:23.41% in the Y53 cytoplasm at the same position of chromosome 6.
The frequencies of the three genotypes and LOD score profiles of the maize genome under the cytoplasmic environment of JB, which was an F2 population derived from the cross of two inbred lines (JB × Y53). (a) The top panel shows the frequencies of the three genotypes with blue, red and green patterns representing the genotypes of the Y53 homozygotes, heterozygotes and JB homozygotes, respectively. (b) The bottom panel shows the LOD score profiles for the maize genome obtained from the interval mapping of segregation distortion. Vertical lines correspond to chromosome boundaries.
The frequencies of the three genotypes and LOD score profiles of the maize genome under the cytoplasmic environment of Y53, which was an F2 population derived from the cross of two inbred lines (Y53 × JB). (a) The top panel shows the frequencies of the three genotypes with blue, red and green patterns representing the genotypes of the Y53 homozygotes, heterozygotes and JB homozygotes, respectively. (b) The bottom panel shows the LOD score profiles for the maize genome obtained from the interval mapping of segregation distortion. Vertical lines correspond to chromosome boundaries.
The LOD profiles for the SDL detection are presented in the bottom panels of Figures 1 and 2 for the JB and Y53 cytoplasms, respectively. There were apparent differences between the two LOD profiles. The peaks with an LOD score >3.0 indicated the existence of an SDL. Table 1 summarises the detected SDL in the F2-segregating populations with either the JB or Y53 cytoplasmic environment. In the direct cross with the JB cytoplasm, the 10 SDL detected were distributed among chromosomes 1, 2, 3, 4, 5, 6, 7 and 8. In the reciprocal cross with the Y53 cytoplasm, the seven detected SDL were distributed among chromosomes 2, 3, 5 and 7. It should be noted that the SDL mapping results were dramatically changed when the F2-segregating population was put in a different maternal cytoplasmic environment. Of the 14 SDL, SDL2.2, SDL3.3 and SDL7 were located in the same chromosome region in both the JB and Y53 cytoplasms. However, these loci had different distortion ratios in the different cytoplasmic backgrounds. In addition, both the number of SDL and LOD value of these SDL suggested that the segregation distortion was more severe in the JB cytoplasm (Figure 1) than in the Y53 cytoplasm (Figure 2).
To test the different effect of the JB and Y53 cytoplasms on the level of segregation distortion, we performed a χ2-test to compare genotypic frequencies in the two cytoplasmic backgrounds. The results suggested that most SDL, excluding SDL3.2 and SDL3.3, had significant different distortion patterns in the two cytoplasmic backgrounds (Table 1). SDL2.2, SDL3.3 and SDL7 were detected in the same chromosome region in both the JB and Y53 cytoplasms. However, only SDL3.3 had the same distortion pattern in the two cytoplasmic backgrounds (P>0.05).
The direction of the distortion in different cytoplasmic backgrounds attracted our attention. We found that the 10 SDL identified in the F2 population with the JB cytoplasm trended in the same direction such that the distorted ratios favoured the Y53 homozygotes (Table 1). For the SDL detected in the JB cytoplasm, the average frequencies of Y53 homozygotes, heterozygotes and JB homozygotes were 53.59%, 29.93% and 16.48%, respectively. The average frequency of the Y53 allele was approximately 70%, and the average frequency of the JB allele was approximately 30%. However, among the SDL detected in the Y53 cytoplasm, the average frequencies of the Y53 homozygotes, heterozygotes and JB homozygotes were 32.52%, 42.56% and 24.92%, respectively. The average frequencies of the Y53 and JB alleles were approximately 54% and 46%, respectively. Importantly, the direction of the distortion biased to the Y53 allele in the F2 population with the JB cytoplasm, and this trend was mild in the F2 population with the Y53 cytoplasm.
Further Wilcoxon signed-rank tests were performed to compare the selection coefficients (Tables 1 and 2) within the cytoplasm and among cytoplasms for the level of segregation distortion based on the fitness model (Luo et al., 2005). In the JB cytoplasm, the average selection coefficients for the Y53 homozygote (sY53) and JB homozygote (sJB) were 0.01 and 0.60, respectively. The median for sY53 and sJB were 0.00 and 0.73, respectively. There was a significant difference between sY53 and sJB in the JB cytoplasm (P=0.0007). The average and median sY53 were 0.15 and 0.00 in the Y53 cytoplasm, respectively. The difference of sY53 in the JB and Y53 cytoplasmic backgrounds was also significant (P=0.0313). In addition, the average and median sJB were 0.22 and 0.05 in the Y53 cytoplasm, respectively. The median sJB did not show significant difference with the median sY53 in the Y53 cytoplasm (P=0.6698). However, it showed significant difference with the median sJB in the JB cytoplasm (P=0.0266). In present study, the effect size of the cytoplasm was listed in Table 2. For sJB, the median effect size of the cytoplasm was 0.53, and the median effect size of the cytoplasm for sY53was 0.00.
Furthermore, the 10 SDL detected in the F2 population with the JB cytoplasm exhibited three types of segregation patterns (Table 1). Selection type I showed a strong elimination of gametes and zygotes of the JB alleles, including SDL1, SDL3.2, SDL4, SDL5.3, SDL6 and SDL8. On average, these SDL had a segregation ratio with the JB homozygotes that was <15%. The frequency of the heterozygotes was approximately 25%, and the frequency of the Y53 homozygotes was >60%. The selection coefficients for JB homozygotes ranged from 0.62 to 0.92 with an average of 0.80. The degree of dominance of these SDL ranged from 0.81 to 1.33 with an average of 1.01. Selection type II showed complete elimination of gametes with the JB allele from one parent. This type only included one SDL, SDL3.1, which had a segregation ratio of 1.32:52.10:46.58% for JB homozygotes/heterozygotes/Y53 homozygotes. The ratio of the heterozygotes and Y53 homozygotes was close to the expected 1:1 ratio. Almost all JB homozygotes and half of the heterozygotes were not observed, which suggested a strong elimination of gametes with the JB alleles inherited from one parent. For SDL3.1, the selection coefficient for the JB homozygote reached 0.97 and the degree of dominance was 0.45. Selection type III showed the extreme elimination of heterozygotes, including SDL2.2, SDL3.3 and SDL7. These SDL had severe distortions due to the absolute deficiency of heterozygotes. It should be noted that these SDL also showed a mild selection of JB homozygotes and an excess of Y53 homozygotes. On average, the frequency of JB homozygotes was <25%, and the frequency of Y53 homozygotes was >75%. The selection coefficients for JB homozygotes ranged from 0.65 to 0.76 with an average of 0.70. All of the 10 SDL detected in the JB cytoplasm skewed towards the same direction with an excess of Y53 alleles. The degree of dominance of selection type III ranged from 1.11 to 1.52 with an average of 1.34.
However, the segregation patterns of SDL detected in the Y53 cytoplasm varied from each other. SDL2.1 belonged to selection type I with a strong elimination of the JB alleles in the gametes and zygotes. SDL3.3 was detected in both JB and Y53 cytoplasms, and it showed a similar distortion in the two cytoplasm background, which belonged to selection type III. SDL2.3 presented as a selection type IV with a strong elimination of gametes and zygotes with Y53 alleles. The frequency of Y53 homozygotes in SDL2.3 was <10%, and the ratio of heterozygotes/JB homozygotes was 52.16:38.07%, which was <2:1. The selection coefficient for Y53 homozygotes was 0.74, and the degree of dominance was 0.42. In addition, SDL5.2 and SDL7 detected in the Y53 cytoplasm showed mild elimination of heterozygotes, and the ratio of their genotypic frequencies closed to 1:1:1. The degrees of dominance for SDL5.2 and SDL7 were 2.76 and 2.83, respectively. They were categorised as selection type V. In contrast, SDL2.2 and SDL5.1 detected in the Y53 cytoplasm had a segregation distortion with mild elimination of both JB and Y53 homozygotes. The degrees of dominance for SDL2.2 and SDL5.1 were −0.65 and −1.16, respectively. They were categorised as selection type VI in the present study.
Discussion
Natural selection acts directly on the fitness of individuals in a population. Viability selection will change the gene frequencies at loci that control fitness. Consequently, the frequencies of marker loci linked to the viability loci will also change (Sukhodolets, 1986; Lessard, 1997; Mojica and Kelly, 2010). In genetic mapping, the change in marker allelic frequencies is reflected by a departure from the expected Mendelian segregation ratio, which is the observed segregation distortion of markers. SDL are the vital genetic factors affecting the viability of gametes and zygotes (Vogl and Xu, 2000; Luo and Xu, 2003). However, cytoplasm and its organelles, such as mitochondria, have been widely studied for their critical role in cellular metabolism, energy production and cell death (Newmeyer and Ferguson-Miller, 2003). Furthermore, cytonuclear interaction may have an important role as the evolutionary force driving speciation (Rand et al., 2004; Chou and Leu, 2010).
Based on the above-mentioned studies, we hypothesised that the cytoplasmic environment of maize may be involved in the viability selection of gametes and zygotes. To test this hypothesis, we constructed a segregating population experiment with two maternal cytoplasmic environments using a reciprocal mating design. Because there was little consideration for segregation distortion for the construction of the linkage map (Hackett and Broadfoot, 2003), a linkage map including 126 markers was constructed before the examination of distortion. We compared the patterns of segregation distortion under different maternal cytoplasmic environments to distinguish between distortion caused by cytoplasm-specific viability selection and general distortion caused by viability loci or SDL. The results demonstrated a dramatic change in the distortion patterns between the two different maternal cytoplasmic environments and suggested a significant association between the segregation distortion and maternal cytoplasmic environment, which directly verified our hypothesis.
To test our hypothesis in other species, we also analysed a similar data set of an F2 rat experiment performed by Solberg et al. (2004), which is freely available on the internet. These authors used Wistar–Kyoto (WKY) and Fisher 344 (F344) rat strains. Supplementary Figures S1 and S2 present the genotype frequencies and the LOD profile of the F2 populations along the rat genome under the WKY and F344 cytoplasms, respectively. Supplementary Table S1 summarises the detected SDL in the F2 population with WKY and F344 cytoplasmic environments. There were significant differences in the segregation distortion under the two cytoplasmic environments. In addition, Niehuis et al. (2008) also performed a reciprocal cross experiment to obtain male F2 hybrids in Nasonia giraulti and N. vitripennis cytoplasms. However, they found loci with different segregation patterns at three locations and at one location in N. vitripennis and N. giraulti cytoplasms, respectively. These results from maize, rats and Nasonia suggest that the maternal cytoplasmic environment has an important role in the viability selection of gametes and zygotes, resulting in the observed non-Mendelian segregation of genetic markers. Clearly, no single genetic mechanism can account for the patterns of distortion. The present data provided new insight into the role of the cytoplasm in viability selection and shaping the patterns of segregation, which raises further questions about the underlying molecular mechanisms.
In the present study, the interesting phenomenon was the direction of the distortion that selectively biased to the Y53 allele and homozygotes in the JB cytoplasm. Among the SDL detected, the average frequency of Y53 homozygotes was 54%, and the frequency of the Y53 allele reached 70%. However, for SDL detected in the Y53 cytoplasm, the average frequency of Y53 homozygotes was 33%, and the frequency of the Y53 allele was 54%. In addition, statistical tests suggested that the sY53 was significantly different from sJB in the JB cytoplasm (P=0.0007) and also from sY53 in the Y53 cytoplasm (P=0.0313). The relative fitness of Y53 homozygotes (1−sY53) was approximately 0.99 on average, indicating that the chance of survival for Y53 homozygotes was 99% on average in the JB cytoplasm, which was significantly greater than that of the JB homozygotes. These results suggested that the direction of the distortion selectively biased to the Y53 allele and homozygote in the JB cytoplasm.
Similarly, in the rat F2 population with the WKY cytoplasm, most distortions were caused by a mild deficiency of WKY homozygote and heterozygotes and an excess of F344 homozygotes (Solberg et al., 2004). The direction of the distortion was selectively biased to F344 homozygotes in the WKY cytoplasm. In rice, Wang et al. (2009) found that the japonica cytoplasm did not cause any distortion favouring a special parent, but that the indica cytoplasm favoured the maternal parent, which suggested that the indica cytoplasm was incompatible with a japonica nuclear background and that the japonica cytoplasm did not have such trouble with the indica nuclei. These results suggested that one of the cytoplasms in each case favoured the nucleus allele originating in the other inbred line and that the other cytoplasm showed no consistent bias.
We do not understand the reason for the selective bias of the patterns of distortion in the JB cytoplasm. Cytonuclear incompatibility may partially explain the skewed viability selection of gametes and zygotes in different cytoplasmic environments (Fishman and Willis, 2006; Niehuis et al., 2008). Evolutionary force might also be involved. In the history of evolution, cytoplasmic DNA (such as mitochondrial DNA and chloroplast DNA) and nuclear genes may evolve to form coadapted complexes even within reproductively isolated populations. As a consequence of coadaptation, the fitness of particular nuclear alleles may depend on special cytoplasmic DNA genotypes. Willett and Burton (2001) verified this evolutionary theory by tracking the genotypic ratios of the nuclear gene encoding cytochrome-c in different reciprocal crosses of Tigriopus californicus. These researchers suggested that the cytoplasm may have an important but often undetected role in the viability or fitness selection and in shaping the patterns of evolution and speciation.
It should be mentioned that JB and Y53 are the parental lines of the Suyu 16 maize hybrid, and they display strong heterosis only when the JB cytoplasm was selected by the cross direction (JB × Y53). Field experiments exhibited significant differences in growth, development, morphology and production between the direct and reciprocal hybrid F1 population. It is difficult to explain the association between the unidirectional bias in genotype frequencies and complex heterosis. Although the genetic basis of heterosis remains unclear, we infer that the cytoplasm may not only be involved in the viability selection of gametes and zygotes but may also interact with the nuclear genes from Y53 and strongly contribute to the heterosis of Suyu 16.
In the present study, we found 16 markers with an extreme segregation ratio (Table 3). These markers were not included in the linkage mapping because the effect of extreme segregation on the construction of linkage maps was not clear. In addition, the current version of PROC QTL cannot handle markers with extreme segregation ratios. Therefore, these markers were not involved in the above SDL mapping. χ2-tests were used to analysis the distortion degree of markers. These markers only were observed as single homozygotes for the marker genotype in one cytoplasm while presenting with the expected Mendelian segregation ratio or a mild segregation distortion in another cytoplasm. Similar to the patterns of bias direction exhibited by the SDL detected by the whole-genome scan, eight out of nine markers were extremely skewed towards Y53 homozygotes in the JB cytoplasm. However, in a reciprocal cross direction with the Y53 cytoplasm, these markers did not generate an unidirectional bias. There are many possible causes of such complete transmission ratio distortion in plant hybrids as follows: nonrandom segregation of chromosomes in female meiosis; differential male gamete survival or performance; differential female gamete survival or performance (including interactions with maternal nuclear and cytoplasmic genotypes); and differential seed survival. Fishman and Willis (2005) found a female-specific meiotic drive locus that exhibits extreme segregation distortion in Mimulus hybrids, and they suggested that severe distortion at the drive locus results from biased chromosomal segregation during female meiosis. At a molecular level, Borodin (2001) suggested that chromosomal competition during female meiosis drives a rapid turnover of centromeres associated with histone proteins and potentially accelerates the development of reproductive incompatibilities between species. In addition, cytoplasm-specific epigenetic influence may also be involved in the viability selection of gametes and zygotes. However, the population genetic theory shows that selfish genetic elements can trigger segregation and even act as extreme drivers, which may be an important force in promoting evolutionary change (Hurst and Werren, 2001; Werren, 2011). Although the present observations based on the reciprocal mating design still do not provide enough evidence for investigating the detailed mechanism of such extremely biased distortion, they do lend support to the hypothesis that the cytoplasm may be involved in viability selection of gametes and zygotes and the generation of different segregation ratios in the two cytoplasmic environments.
Data archiving
Genotype data have been submitted to Dryad: doi:10.5061/dryad.s1674.
References
Barchi L, Lanteri S, Portis E, Stagel A, Vale G, Toppino L et al (2010). Segregation distortion and linkage analysis in eggplant (Solanum melongena L.). Genome 53: 805–815.
Borodin PM (2001). Speciation and centromere evolution. Science 294: 2478–2480.
Casellas J, Farber CR, Verdugo RA, Medrano JF (2010). Segregation analysis of a sex ratio distortion locus in congenic mice. J Hered 101: 351–359.
Chou J-Y, Leu J-Y (2010). Speciation through cytonuclear incompatibility: insights from yeast and implications for higher eukaryotes. Bioessays 32: 401–411.
Fishman L, Willis JH (2005). A novel meiotic drive locus almost completely distorts segregation in mimulus (monkeyflower) hybrids. Genetics 169: 347–353.
Fishman L, Willis JH (2006). A cytonuclear incompatibility causes anther sterility in Mimulus hybrids. Evolution 60: 1372–1381.
Frascaroli E, Cane MA, Landi P, Pea G, Gianfranceschi L, Villa M et al (2007). Classical genetic and quantitative trait loci analyses of heterosis in a maize hybrid between two elite inbred lines. Genetics 176: 625–644.
Hackett CA, Broadfoot LB (2003). Effects of genotyping errors, missing values and segregation distortion in molecular marker data on the construction of linkage maps. Heredity (Edinb) 90: 33–38.
Hu Z, Xu S (2009). PROC QTL-A SAS procedure for mapping quantitative trait loci. Int J Plant Genomics 2009, (e-pub ahead of print 8 December 2012; doi:10.1155/2009/141234).
Hurst GD, Werren JH (2001). The role of selfish genetic elements in eukaryotic evolution. Nat Rev Genet 2: 597–606.
Lessard S (1997). Fisher's fundamental theorem of natural selection revisited. Theor Popul Biol 52: 119–136.
Li X, Wang X, Wei Y, Brummer EC (2011). Prevalence of segregation distortion in diploid alfalfa and its implications for genetics and breeding applications. Theor Appl Genet 123: 667–679.
Lima M, de Souza C, Bento D, de Souza A, Carlini-Garcia L (2006). Mapping QTL for grain yield and plant traits in a tropical maize population. Mol Breeding 17: 227–239.
Lu H, Romero-Severson J, Bernardo R (2002). Chromosomal regions associated with segregation distortion in maize. Theor Appl Genet 105: 622–628.
Luo L, Xu S (2003). Mapping viability loci using molecular markers. Heredity (Edinb) 90: 459–467.
Luo L, Zhang YM, Xu S (2005). A quantitative genetics model for viability selection. Heredity (Edinb) 94: 347–355.
McDermott SR, Noor MA (2010). The role of meiotic drive in hybrid male sterility. Philos Trans R Soc Lond B Biol Sci 365: 1265–1272.
Mojica JP, Kelly JK (2010). Viability selection prior to trait expression is an essential component of natural selection. Proc Biol Sci 277: 2945–2950.
Newmeyer DD, Ferguson-Miller S (2003). Mitochondria: releasing power for life and unleashing the machineries of death. Cell 112: 481–490.
Niehuis O, Judson AK, Gadau J (2008). Cytonuclear genic incompatibilities cause increased mortality in male F2 hybrids of Nasonia giraulti and N. vitripennis. Genetics 178: 413–426.
Orr HA, Irving S (2005). Segregation distortion in hybrids between the Bogota and USA subspecies of Drosophila pseudoobscura. Genetics 169: 671–682.
Phadnis N (2011). Genetic architecture of male sterility and segregation distortion in Drosophila pseudoobscura Bogota-USA hybrids. Genetics 189: 1001–1009.
Phadnis N, Orr HA (2009). A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science 323: 376–379.
Rand DM, Haney RA, Fry AJ (2004). Cytonuclear coevolution: the genomics of cooperation. Trends Ecol Evol 19: 645–653.
Rogers SO, Bendich AJ (1988). Extraction of DNA from plant tissues. Plant Molecular Biology Manual A6: 1–10.
Sinkins SP (2011). Invertebrate post-segregation distorters: a new embryo-killing gene. PLoS Biol 9: e1001114.
Solberg LC, Baum AE, Ahmadiyeh N, Shimomura K, Li R, Turek FW et al (2004). Sex- and lineage-specific inheritance of depression-like behavior in the rat. Mamm Genome 15: 648–662.
Sukhodolets VV (1986). The role of natural selection in evolution. Genetika 22: 181–193.
Vogl C, Xu S (2000). Multipoint mapping of viability and segregation distorting loci using molecular markers. Genetics 155: 1439–1447.
Wang S, Tan Y, Tan X, Zhang Z, Wen J, Kou S (2009). Segregation distortion detected in six rice F2 populations generated from reciprocal hybrids at three altitudes. Genet Res (Camb) 91: 345–353.
Werren JH (2011). Selfish genetic elements, genetic conflict, and evolutionary innovation. Proc Natl Acad Sci USA 108 ((Suppl 2)): 10863–10870.
Willett CS, Burton RS (2001). Viability of cytochrome c genotypes depends on cytoplasmic backgrounds in Tigriopus californicus. Evolution 55: 1592–1599.
Wolf JB (2009). Cytonuclear interactions can favor the evolution of genomic imprinting. Evolution 63: 1364–1371.
Xu S, Hu Z (2009). Mapping quantitative trait loci using distorted markers. Int J Plant Genomics 2009, (e-pub ahead of print 21 February 2010; doi:10.1155/2009/410825).
Xu Y, Zhu L, Xiao J, Huang N, McCouch SR (1997). Chromosomal regions associated with segregation distortion of molecular markers in F2, backcross, doubled haploid, and recombinant inbred populations in rice (Oryza sativa L.). Mol Gen Genet 253: 535–545.
Yamagishi M, Takeuchi Y, Tanaka I, Kono I, Murai K, Yano M (2010). Segregation distortion in F(2) and doubled haploid populations of temperate japonica rice. J Genet 89: 237–241.
Yang XJ, Lu M, Zhang SH, Zhou F, Qu YY, Xie CX (2008). QTL mapping of plant height and ear position in maize (Zea mays L.). Hereditas 30: 1477–1486.
Zhan H, Xu S (2011). Generalized linear mixed model for segregation distortion analysis. BMC Genet 12: 97.
Zhu C, Wang C, Zhang YM (2007). Modeling segregation distortion for viability selection. I. Reconstruction of linkage maps with distorted markers. Theor Appl Genet 114: 295–305.
Acknowledgements
We thank Dr Leah Solberg Woods and QTL Archive (http://qtlarchive.org/) for providing the rat data set. This work was supported by grants from the National Basic Research Program of China (2011CB100106), the National Natural Science Foundation of China (30971846, 31171187) and the Vital Project of Natural Science of Universities in Jiangsu Province (09KJA210002) to C Xu, the Agricultural Part of Technology Supported Program of Jiangsu Province (BE2011303) to D Deng, a Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions at Soochow Univerisity and Yangzhou Univerisity and the National Natural Science Foundation of China (31100882) to Z Tang.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Heredity website
Supplementary information
Rights and permissions
About this article
Cite this article
Tang, Z., Wang, X., Zhang, M. et al. The maternal cytoplasmic environment may be involved in the viability selection of gametes and zygotes. Heredity 110, 331–337 (2013). https://doi.org/10.1038/hdy.2012.89
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2012.89