Abstract
Recombinant adeno-associated viral vectors (rAAVs) are used for therapeutic gene transfer in skeletal muscle, but it is unclear if immune reactivity to gene transfer and persistence of transgene are affected by pathologic conditions such as muscular dystrophy. Thus, we compared dystrophic mice devoid of α-sarcoglycan with healthy mice to characterize immune cell activation and cellular populations contributing to the loss of gene-modified myofibers. Following rAAV2/1 delivery of an immunogenic α-sarcoglycan reporter transgene in the muscle, both strains developed strong CD4 and CD8 T-cell-mediated immune responses in lymphoid organs associated with muscle CD3+ T and CD11b+ mononuclear cell infiltrates. Selective cell subset depletion models revealed that CD4+ T cells were essential for transgene rejection in both healthy and pathologic mice, but macrophages and CD8+ T cells additionally contributed as effector cells of transgene rejection only in dystrophic mice. Vectors restricting transgene expression in antigen-presenting cells showed that endogenous presentation of transgene products was the sole mechanism responsible for T-cell priming in normal mice, whereas additional and protracted antigenic presentation occurred in dystrophic animals, leading to secondary CD4+ T-cell activation and failure to maintain transgene expression. Therefore, the dystrophic environment diversifies cellular immune response mechanisms induced by gene transfer, with a negative outcome.
Similar content being viewed by others
Introduction
Recombinant adeno-associated viral vectors (rAAVs) are being used to treat genetic defects because of their low inflammatory effects and propensity to transduce specific tissues based on the serotype used. Among all currently available vectors, rAAV2/1 is particularly efficient for gene delivery into muscles and has been exploited through intramuscular (i.m.) injections to treat muscular dystrophies and non-muscular diseases.1,2 However, the i.m. route of administration, which is also used for vaccines, is strongly immunogenic and the induction of immune responses to the transgene product can limit long-term efficacy of the therapeutic gene.3, 4, 5, 6, 7 Among the parameters affecting the immunogenicity of the transgene product, accessibility to professional antigen-presenting cells (APC) is paramount. We and others have shown that the amount of myeloid cells and of potential APCs is increased in dystrophic versus healthy muscles8 (and Boisgerault, unpublished data) and this may impact on the antigenic presentation of the transgene to T cells after rAAV gene transfer. While cross-presentation was initially favored as the mechanism of antigenic presentation used to immunize by rAAV gene transfer,9, 10, 11 it is now clear that rAAV can also directly transduce APCs that become capable of direct antigenic presentation. Indeed, adoptive transfer of AAV-infected dendritic cells (DCs) constitutes an effective vaccine against the encoded proteins12 and is used for cancer immunotherapy.13 Furthermore, our own and recently published results suggest that direct antigenic presentation is the main mechanism of transgene-specific immunization following rAAV2/1 gene delivery in normal muscle.14,15 Understanding the relative contribution of these presentation pathways is needed to design strategies aiming to reduce T-cell immunization following gene transfer. It is therefore important to determine if principles established in normal mice also apply to pathologic conditions.
Dystrophic muscles are characterized by leaky fibers and by the presence of an inflammatory environment that may modulate antigenic presentation and provide immunostimulatory signals. While inflammatory cells are absent in normal muscles, they can reach 100 000 cells per mm3 in regenerative muscle tissue.16 Degenerative–regenerative myogenic processes and inflammation dominated by a myeloid cell infiltrate are prominent features in several progressive muscle diseases including Duchenne muscular dystrophy,17 and limb girdle muscular dystrophy type 2B, 2D18 and 2L.19 Although mast cells, neutrophils, eosinophils and cytotoxic CD8+ T cells can contribute to the pathogenesis in these chronic myopathies, different sub-populations of macrophages are involved either in the pathogenesis or in muscle regeneration.20 In the context of gene therapy performed on normal muscles, it was postulated that the loss of transgene-expressing muscle fibers is caused by effector cytotoxic CD8+ T cells4 even though it has been reported elsewhere that CD8+ T cells were not functional locally.21 Therefore, based on these observations, it was necessary to re-evaluate the mechanisms of antigenic presentation and the functional role of T cells and macrophages in the destruction of transgene-modified fibers in the context of muscular dystrophy when a therapeutic transgene is expressed.
We have previously developed a model to study cellular immune responses induced by rAAV gene transfer in the muscle of naive mice. In this model, the persistence of transgene and anti-transgene T-cell immune responses were differently controlled in dystrophic mice devoid of α-sarcoglycan (Sgca-/-) compared with normal congenic C57BL/6 mice.14 Here we use this model to show that CD4+ T cells are required to reject gene-modified cells in either normal or dystrophic mice, whereas CD8+ T cells and macrophages contribute to the rejection of gene-modified cells only in dystrophic mice. We also demonstrate that the dystrophic environment broadens the antigenic presentation pathways as compared with normal muscles, making the control of the immune response more difficult.
Results
T cells and CD11b+ cells are found in muscles undergoing transgene rejection
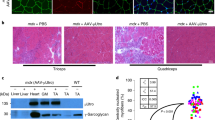
The chimeric SGCA-HY transgene was developed to stably express a modified human α-sarcoglycan protein (SGCA) at the membrane of skeletal myofibers. This transgenic protein carries in its intracytoplasmic domain an HY polypeptide tag allowing the measure of transgene-specific antigenic processing and T-cell immunity in female mice. This model is used here to understand the impact of pathologic conditions on immune rejection of gene-modified muscle fibers following rAAV gene delivery. In this model, anti-AAV capsid immune responses are not analyzed. As reported previously, the i.m. administration of rAAV2/1-expressing SGCA-HY from a cytomegalovirus (CMV) promoter (rAAV2/1_CMV_SGCA-HY) is immunogenic and leads to the loss of transgene-expressing myofibers after 14 days in both normal and in Sgca-/- dystrophic mice.14 Muscle immunohistology was performed to characterize the type of cellular infiltrates at the time of transgene rejection in these two strains of mice. The presence of a strong infiltrate composed of CD3+ T cells (red) and of CD11b+ myeloid cells (green) was found both in normal and dystrophic muscles following gene transfer (Figure 1). Both CD4+ and CD8+ T cells were present as depicted in Supplementary Figure S1a. The population of CD11b+ cells was composed almost entirely of F4/80+ macrophages (Supplementary Figure S1b). The levels of F4/80 marking defined two sub-populations of CD11b+ cells, which appeared to be differentially represented in normal or dystrophic muscle. At the time point analyzed, cells of C57BL/6 mice contained an approximately equal proportion of F4/80low M1 macrophages (CD206− and iNOShigh) and of F4/80high M2 macrophages (CD206+, Arg-1high, iNOSlow), whereas cells of Sgca-/- dystrophic mice contained essentially F4/80high cells with an M2 phenotype (Supplementary Figure S1b). In both C57BL/6 and Sgca-/- mice, the loss of transgene in muscles was documented at the mRNA level (Figures 2a and c) and confirmed by the absence of SGCA protein in treated Sgca-/- mice (Figure 2d). Extensive muscle tissue damage and regeneration was reflected by a strong cellular invasion and by evidence of regenerating centronucleated fibers as shown in C57BL/6 mice (Figure 2b). Thus, different types of leukocytes infiltrate the muscle at the time of rejection and may differentially accompany the loss of transgene-expressing muscle cells following rAAV2/1 gene transfer.
Analysis of immune cell infiltrates in the injected muscles. Cross-sections of the left TA muscles from C57BL/6 and Sgca-/- mice injected i.m. 14 days earlier with 5 × 109 vg of the rAAV1_CMV_SGCA-HY vector or PBS were immunostained for the lymphocyte T-cell-specific marker CD3 (red), and for inflammatory cells marker CD11b (green). Nuclei are counterstained with 4',6-diamidino-2-phenylindole (DAPI) (blue). Images are representative of four mice analyzed per condition (scale bar=100 μm). The upper right insert shows a magnification of selected areas.
Role of CD4+ T cells in transgene rejection. C57BL/6, CD4-/- mice (a and b) and Sgca-/- and CD4-/- Sgca-/- mice (c and d) were injected i.m. with PBS or with 5 × 109 vg of rAAV1_CMV_SGCA-HY vector in the left TA and transgene expression, and the muscle integrity were analyzed over time. Levels of SGCA mRNA expression in the TA of normal (a) and dystrophic (c) mice were measured at different time points by quantitative RT-PCR and normalized to PO levels. The reference of 1.0 corresponds to SGCA mRNA levels known to be therapeutic in Sgca-/- mice. Data show the average value obtained in eight mice per condition. (b) Cross-sections of the injected C57BL/6 and CD4-/- TAs were analyzed by microscopy after HE staining. (d) Immunostaining of the α-sarcoglycan protein was performed on cryosections from Sgca-/- and CD4-Sgca-/- injected muscles. Arrows indicate SGCA-positive fibers. In (b and d), images are representative of one experiment out of three, with three mice per group (scale bar=150 μm). ***P<0.001.
CD4+ T cells are mandatory in both normal and dystrophic strains for transgene rejection through the recruitment of effector cells
To determine if CD4+ T cells contribute to the loss of SGCA-HY, we tested the outcome of gene transfer into several strains of mice deficient in CD4+ T cells. Transgenic mice with a knock-out of the CD4 gene (CD4-/-) or of the major histocompatibility complex (MHC) class II locus (MHC II-/-) were injected with the rAAV2/1_CMV_SGCA-HY vector. In both of these CD4+ T-cell-deficient strains, the levels of transgene mRNA were maintained at high levels in the injected muscles over time compared with C57BL/6 control mice, which rapidly lost transgene expression (Figure 2a and Supplementary Figure S2a). At the histologic level, transgene expression was well tolerated in muscles of CD4-/- mice. Muscle integrity was preserved for at least 6 weeks after vector administration as shown by cohesive myofibers, little centronucleation and lack of cellular invasion, sharply contrasting with C57/BL6 controls (Figure 2b). In CD4-/- mice or in MHC II-/- mice, there was no evidence of either CD4 or CD8 T-cell-mediated anti-transgene immune responses (Supplementary Figures S2b and S3a). Such data argue for a strong implication of CD4+ T-cell activation in the loss of transgene-modified muscles. We ruled out a critical contribution of CD4+ T cells via the induction of anti-transgene antibodies generated upon CD4+ T/B-cell cooperation, as transgene rejection and T-cell responses were as efficient in μMT-/- mice devoid of Ig-secreting B cells as in controls (Supplementary Figure S2).
To investigate the contribution of CD4+ T cells in dystrophic mice, we crossed Sgca-/- mice with CD4-/- mice to obtain CD4-/-Sgca-/- mice. Muscles of those mice have regenerating centronucleated myofibers characteristics of a dystrophic phenotype (Supplementary Figure S3b). Similar to what was observed in normal mice, CD4+ T cells contributed in a major way to the rejection of modified dystrophic myofibers. Upon gene transfer, CD4-/- Sgca-/- mice maintained the expression of transgene mRNA (Figure 2c) and protein on myofibers (Figure 2d) for at least 6 weeks, in contrast to Sgca-/- mice (Figure 2c).
In the dystrophic background, the lack of CD4+ T cells also markedly reduced anti-transgene CD4 and CD8 T-cell responses (Supplementary Figure S3a) and limited the destruction and regeneration of myofibers that was so prominently induced by gene transfer in Sgca-/- mice (Supplementary Figure S3b). The lack of CD4+ T cells in the dystrophic background also had a strong impact to reduce the cell infiltrate as vector-injected muscles contained fewer CD3+ and CD11b+ cells (Supplementary Figure S3c).
These data clearly showed that CD4+ T cells are required for the loss of transgene-modified myofibers both in healthy and dystrophic mice, and furthermore that CD4+ T cells have a central role to mobilize effector cells locally.
Differential contribution of effector CD8+ T cells and macrophages for transgene rejection in normal and dystrophic models
Cytolytic CD8+ T cells are usually the focus of attention in cell-mediated immune rejection and our previous studies have confirmed that i.m. administration of rAAV2/1_CMV_SGCA-HY leads to CD8+ T cell-mediated cytotoxicity against the transgene product in vivo.14 However, to determine if CD8+ T cells are critically needed for the rejection of transgene-expressing cells, SGCA-HY gene transfer was tested in CD8+ T-cell-deficient murine models: CD8-/- mice and β2-microglobulin (β2M)-/- mice.
Unexpectedly, the lack of CD8+ T cells did not prevent the loss of transgene (Figure 3a and Supplementary Figure S2a). Injection of the vector in muscles of CD8-/- mice caused destruction and regeneration with a marked cellular infiltration only marginally reduced compared with C57/BL6 controls (Figure 3c). Similar results were obtained in β2M-/- mice (data not shown). As expected, anti-transgene CD8 T-cell responses were completely abolished in β2M-/- mice or in CD8-/- mice while anti-transgene CD4+ T-cell responses were maintained (Supplementary Figures S2b and S3a). These data suggest that CD8+ T cells are not the central effector cells associated with transgene rejection in our model. To evaluate the impact of a dystrophic context on CD8+ T-cell responses, we crossed CD8-/- mice with Sgca-/- mice to obtain CD8-/- Sgca-/- mice. Those double-knockout mice also displayed early signs of dystrophy characterized by a regenerating phenotype (Supplementary Figure S3b). Such mice failed to develop transgene-specific CD8+ T-cell response but retained the same level of CD4+ T-cell response as Sgca-/- mice (Supplementary Figure S3a). In the dystrophic background, the lack of CD8+ T cells did not prevent transgene rejection but mitigated this loss. The disappearance of transgene was documented at the mRNA level, but yet significantly higher levels of transgene mRNA remained after gene transfer in CD8-/- Sgca-/- mice compared with Sgca-/- mice (Figure 3b). In agreement with this finding, about 15% of SGCA+ myofibers were detected in muscles of CD8-/- Sgca-/- mice, while transgene marking was completely lost in Sgca-/- mice (Figure 3d). Thus, CD8+ T cells contribute to muscle destruction without being major effector cells.
Contribution of effector CD8+ T cells and macrophages to transgene rejection. C57BL/6, CD8-/-, Sgca-/- and CD8-/- Sgca-/- mice were injected i.m. with PBS or with 5 × 109 vg of rAAV1_CMV_SGCA-HY vector in the left TA and transgene expression, and the muscle integrity were analyzed at day 14. Levels of SGCA mRNA expression in the TA of normal (a) and dystrophic (b) mice were measured by quantitative RT-PCR (qRT-PCR) and normalized to PO levels. The reference of 1.0 corresponds to therapeutics levels of SGCA. Results show average values obtained from six mice per group. (c) HE stain and histologic analysis of C57BL/6- and CD8-/- injected TA muscles. (d) Immunostaining of the α-sarcoglycan protein on cross-sections of Sgca-/- and CD8-/- Sgca-/- injected TA muscles. Images are representative from one experiment out of three, with three mice per group (scale bar=150 μm). In (e and f), C57BL/6 or Sgca-/- mice received a single i.m. injection of PBS or of 5 × 109 vg of rAAV1_CMV_SGCA-HY vector in the left TA and four injections intravenously of Clodrosome (lipos. clodronate) or of Encapsome as control (lipos.ctl). Levels of SGCA mRNA expression in the TA of normal (e) and dystrophic (f) mice were measured at different time points by qRT-PCR and normalized to PO levels. The reference of 1.0 corresponds to levels of SGCA known to be therapeutic in Sgca-/- mice. Data are representative of the average value obtained in six mice per group. (g) HE histologic analysis of the TA of C57BL/6 mice injected 14 days earlier with the vector. (h) Immunohistochemical analysis of α-sarcoglycan expression in the injected TA of Sgca-/- mice. Images are representative from one experiment out of three, with three mice per group (scale bar=150 μm). ***P<0.001, **P<0.01, *P<0.05, NS, not statistically significant.
In the absence of an apparent robust involvement of CD8+T cells in the rejection of gene-modified myofibers and given the strong infiltrate of mononuclear CD11b+ F4/80+ cells in the muscle, we hypothesized that macrophages could be effector cells of this immune-mediated destruction of muscle. To analyze whether or not macrophages were functionally involved in the loss of transgene-modified muscle, we injected mice intravenously with clodronate-containing liposomes to deplete systemically macrophages 5, 7, 9 and 12 days after rAAV administration. Macrophage depletion by clodronate was verified in lymphoid organs such as the spleen with a 70% loss of the F4/80 marker among the CD11b+ cell population as compared with controls while the irrelevant Gr1high granulocyte population was unaffected (Table 1). Removal of macrophages did not restore transgene expression in C57BL/6 mice (Figure 3e) but significantly improved transgene expression in muscles of Sgca-/- mice (Figure 3f). These findings were coherent with the histologic analysis. The depletion of macrophages by clodronate did not seem to affect muscle destruction/regeneration in C57BL/6 mice (Figure 3g) but augmented SGCA protein levels in Sgca-/- muscles (Figure 3h). Thus, macrophages do not appear to have a role in transgene rejection in normal mice, but clearly contribute to the loss of transgene-modified myofibers in dystrophic mice.
Taken together, these data revealed that the cellular composition and the cellular effector mechanisms involved in the destruction of transgene-modified myofibers are different in normal and dystrophic muscles.
Inefficient control of immune cells infiltrate with Mir142T-regulated vector results in the loss of transgene expression in dystrophic mice
Loss of transgene expression owing to cell-mediated immune rejection is an early event driven by the priming of transgene-specific CD4+ T cells, suggesting that endogenous presentation of the transgene in professional APCs such as DCs is the triggering event both in normal and dystrophic mice. To prevent endogenous antigenic presentation by professional APCs, we inhibited transgene expression in hematopoietic cells by the insertion of four repeats of the hematopoietic-specific Mir142.3p target (Mir142T) sequence in the 3′-untranslated region of the SGCA-HY transgene cassette as described previously.14 The Mir142T-regulated vector induced stable SGCA mRNA in C57BL/6 mice for at least 30 days (Figure 4a), elicited only a small CD3+ T-cell infiltrate at day 14 without affecting CD11b+ cells (Figure 4b) and preserved muscle integrity (Figure 4c). On the contrary in Sgca-/- mice, transgene expression decreased over time following administration of the Mir142.3p-regulated vector (Figure 4a). In these mice, the number of CD11b+ as well as CD3+ cells in the muscle was increased compared with phosphate-buffered saline (PBS) (Figure 4b), preceding the loss of SGCA-expressing myofibers seen at day 21 by immunostaining (Figure 4d).
Transgene expression and inflammatory infiltrates using Mir142T-regulated rAAV2/1 vectors. C57BL/6 and Sgca-/- mice were injected i.m. with PBS or with 5 × 109 vg of rAAV1_CMV_SGCA_HY-Mir142T vector in the left TA, and then transgene expression and muscle integrity were analyzed over time. (a) Levels of SGCA mRNA expression in the TA were measured at the indicated time points by quantitative RT-PCR (qRT-PCR) and normalized to PO levels. Data are representative of at least six mice per condition. (b) Cell infiltrates were estimated on cross-sections of C57BL/6- and Sgca-/- TA injected 14 days earlier after immunostaining for CD3 (red), and of CD11b+ cells (green). Nuclei were stained with 4',6-diamidino-2-phenylindole (DAPI) (blue). (c) HE histologic analysis of cryosections of the treated C57BL/6 TAs 14, 21 and 30 days after injection. (d) Immunohistologic analysis of α-sarcoglycan expression on cryosections of Sgca-/- muscles 14, 21 or 30 days postinjection after staining of the nuclei with Mayer’s hemalun. Images are representative of one experiment with three mice analyzed per group, repeated ones (scale bar=100 μm). ***P<0.001, **P<0.01, NS, not statistically significant.
While Mir142T regulation of transgene efficiently prevents the local immune infiltrate in normal muscle, this strategy is not fully effective in dystrophic muscles to maintain SGCA expression; therefore, T-cell response mechanisms were investigated.
Incomplete control of anti-transgene CD4+ T-cell responses by Mir142T-regulated vectors in dystrophic mice
Administration of the Mir142T-regulated vector to C57BL/6 mice failed to induce CD4-mediated immune responses to the transgene in a durable manner compared with the non-regulated vector (Figure 5a). In contrast, in Sgca-/- mice, the Mir142T-regulated vector induced a potent CD4+ T-cell-mediated immune response reaching after 14 days the same levels of response as the non-regulated vector (Figure 5b). The levels of transgene-specific T cells in the animal can be tracked over time following the injection of congenic (CD45.1+) transgene-specific CD4+ T cells before gene transfer vector administration. With the non-regulated vector, the kinetics of transgene-specific blood CD4+ T-cell levels were similar in C57BL/6 (Figure 6a) and Sgca-/- mice (Figure 6b), starting day 6, peaking around 8 days even though the retraction phase was slightly different. Administration of the Mir142T-regulated vector in C57BL/6 mice did not result in a significant amplification of T cells as compared with PBS. On the contrary, in Sgca-/- mice T-cell responses were not controlled by the Mir142.3p-regulated vector, which induced a striking expansion of CD4+ T cells that peaked at day 11, beyond the peak time of the non-regulated immune response.
Transgene-specific CD4+ T-cell responses using Mir142T-regulated rAAV2/1 vectors. (a) C57BL/6 and (b) Sgca-/- mice were injected i.m. into the TA with PBS, 5 × 109 vg of rAAV1_CMV_SGCA-HY or rAAV1_CMV_SGCA-HY-Mir142T vectors. After 14, 21 and 30 days, spleen cells were harvested to measure Dby-specific CD4+ T-cell response by interferon-γ enzyme-linked immunospot assays following Dby peptide in vitro stimulation. Data represent four independent experiments in C57BL/6 mice and three independent experiments in Sgca-/- mice, each including three mice per group. Dots represent individual mouse data and horizontal bars indicate the average value. ***P<0.001, **P<0.01, *P<0.05, NS, not statistically significant.
Adoptive transfer of HY-specific transgenic CD4+ T cells. (a) C57BL/6 mice or (b) Sgca-/- mice were injected with 2 × 106 splenocytes from Marylin mice (CD45.1) (day 0 on graph). One day later, mice were injected i.m. with 5 × 109 vg of rAAV1_CMV_SGCA-HY or rAAV1_CMV_SGCA-HY-Mir142T vectors and the levels of HY-specific CD4+ CD45.1+ T cells among total CD4+ blood T cells were measured by FACS over time. Each curve represents the average value obtained with six mice per group.
T-cell immune responses to the transgene are therefore influenced by the muscle environment. The use of Mir142.3p-regulated vectors shows that endogenous expression of the transgene is of major importance to trigger anti-transgene CD4+ T-cell responses in normal mice, but additional mechanisms exist in dystrophic mice leading to protracted CD4+ T-cell activation.
Delayed antigenic presentation of the transgene by DCs additionally occurs in dystrophic mice
We hypothesized that the divergent anti-transgene CD4+ T-cell responses observed in C57BL/6 and Sgca-/- mice following injection of Mir142T-regulated rAAV2/1 vectors were due to differences in the type of antigenic presentation of the transgene by DC. Indeed, dystrophic muscles undergoing degeneration might provide exogenous sources of antigens to DC that could escape the Mir142T control strategy. To investigate this aspect, CD11c+ DCs were purified at different time points from mice injected with vectors regulated or not, and the purified DCs were tested ex vivo for their ability to present the transgene to T cells. Antigenic presentation was measured through the induction of proliferation of carboxyfluorescein succinimidyl ester (CFSE)-stained anti-transgene CD4+ T cells in DC:T-cell cocultures. The FACS (fluorescence-activated cell sorter) cell gating strategy is shown in Figure 7 dot blots. At day 5, DCs harvested following rAAV2/1 injection induced the proliferation of CFSE-stained T cells (Figure 7, gray histogram lines) as expected. In contrast, DCs harvested from healthy or from Sgca-/- mice injected with the Mir142T-regulated vector induced no T-cell proliferation (Figure 7, black histogram lines), indicating a complete control of antigenic presentation by the Mir142T sequences at this time point regardless of the dystrophic context. However, 3 days later, at day 8 following vector administration, DC harvested from Sgca-/- mice administered with the Mir142T-regulated vector induced a strong T-cell proliferation, whereas those collected from C57BL/6 mice still did not, demonstrating that different mechanisms of antigenic presentation occur in those strains.
MHC II antigenic presentation by DCs to specific CD4+ T cells. C57BL/6 mice and Sgca-/- mice were injected i.m. with 5 × 109 vg of rAAV1_CMV_SGCA-HY (gray histogram) or rAAV1_CMV_SGCA-HY-Mir142T (black histogram) vectors into both TA. After 5 or 8 days, popliteal and inguineal lymph node were harvested from five mice per group to select CD11c+ cells by magnetic cell sorting and culture them with CFSE-stained Dby-specific CD4+ T cells for 3 days at 37 °C. Cell division was analyzed by FACS in live CD4+ T cells after sequentially gating on lymphoid cells in the culture (upper left scatter plot), and then on live CD4+ 7-actinomycin D (7AAD)-negative cells (upper right scatter plot). Lower panel histograms show the levels of CFSE in CD4+ T cells in one representative experiment out of three.
Thus, the inhibition of endogenous expression of the transgene is sufficient to fully block antigenic presentation and therefore to prevent T-cell responses in normal mice but not in Sgca-/- mice in which a protracted exogenous antigenic presentation also occurs and leads to late-phase T-cell responses.
Overall, rAAV-mediated SCGA-HY gene transfer provided a model to test the impact of a muscle dystrophic environment on immune responses against gene-modified cells and to determine the role of various cellular contributors and mechanisms controlling transgene persistence and tissue integrity. Experimental results obtained in the different strains of mice are summarized in Table 2.
Discussion
Our data provide the first set of evidences that the muscular dystrophic environment diversifies the mechanisms of immune reactivity following rAAV2/1-mediated gene transfer by generating more complex antigenic presentation pathways involved in CD4+ T cells activation and by enlarging the panel of effectors cells recruited to orchestrate the loss of transgene-modified myofibers.
Previously, we and others had focused on the importance of anti-transgene CD8+ T cells that were detected in blood and lymphoid organs following rAAV2/1 i.m. administration, associated with the production of large amount of interferon-γ and cytotoxicity.14 However, the role and importance of these CD8+ T cells had never been established even though we found them able to migrate to the transduced muscle. We observed a lack of requirement of CD8+ T cells for muscle destruction in normal mice, which could result from a local silencing of their activity, as it has been reported that CD8+ T cells are eliminated through programmed death at the site of antigen production in healthy muscles.21 In contrast, in dystrophic mice CD8+ T cells contribute to the elimination of transgene-modified muscles at early time points, meaning that at least a fraction of those CD8+ T cells are functional. Possibly, apoptotic CD8+ T lymphocytes could be rescued by the presence of activated CD4+ T cells, although we did not see any major differences between normal or dystrophic mice in terms of initial CD4 activation or CD4-dependency. Inhibitory molecules that normally contribute locally to the inhibition of CD8+ T-cell effector function such as PD-L122 may be differentially expressed upon gene transfer in a dystrophic environment. In this context, fibers transduced with rAAV might also express more MHC I molecules than normal and become more prone to CD8-mediated cytotoxicity. In that sense, muscle injury activates CD8+ T cells to endogenous expressed antigens.23 All these reasons could explain the particular importance of CD8+ T cells for transgene rejection in dystrophic muscle but not in normal conditions.
Large infiltrates of myeloid cells were detected in muscles at the time of rejection, particularly in Sgca-/- mice. In Sgca-/- muscles, we detected a prominent representation of F4/80high CD206+ M2 macrophages with regenerative potential20 while C57BL/6 muscles contained both proinflammatory F4/80low iNOShigh M1 macrophages and non-inflammatory and regenerative M2 F4/80high macrophages, pointing out to an important difference in the models. However, the process of immune-mediated tissue destruction and repair, as well as macrophage transition from an M1 to an M2 phenotype in an injured dystrophic muscle, is a dynamic process controlled by many factors.20 Thus, M1 macrophages could possibly be found in dystrophic muscle at an earlier time point. A small number of M1 and M2 macrophages were detected in Sgca-/- muscles not undergoing immune rejection (Supplementary Figure S1b), suggesting that both populations of cells can coexist in this dystrophic environment. The recruitment of CD4+ T cells seems paramount for the activation of macrophages in tissues. As most myofibers do not naturally express MHC II molecules, cooperation between CD4+ T cells and macrophages could be an alternative way to eliminate MHC II-negative cells as reported in cancer.24 The recruitment and activation of M1 macrophages is favored by T-helper type 1 immune responses induced against the transgene as described in a Duchenne muscular dystrophy model;25 therefore, the local release of cytokines from CD4+ T cells is a key factor. All these features may explain the important contribution of macrophages to the loss of transgene-modified myofibers following gene therapy in a dystrophic environment.
The critical role of CD4+ T cells in the loss of transgene expression in the muscle is obvious in both normal and dystrophic models but several mechanisms can be debated. As discussed above with macrophages, the functional contribution of CD4+ T cells could be indirect but a direct participation to tissue rejection may be envisioned. In CD8-knockout mice, we did not detect any in vivo cytolytic activity against target cells (data not shown), meaning that CD4+ T cells did not replace cells secreting the classical granzyme and perforin mediators. However, it has been previously shown that CD4+ T cells cause muscle injury through Fas and Fas-L interactions in inflammatory myopathies leading to apoptosis.26 In the context of local interferon-γ production and inflammatory cell recruitment leading to upregulation of MHC II expression, the direct killing of transgene-modified muscle cells by activated CD4+ T effector cells could be a favored hypothesis as Fas-L is overexpressed after activation.
The dystrophic environment not only affected effector cells but also modified the antigenic presentation pathways involved in the activation of anti-transgene CD4+ T cells following rAAV gene transfer. Endogenous presentation of the transgene by professional APCs occured in both strains, as the Mir142T-regulated vector was able to control the early expansion of anti-transgene CD4+ T cells and the early MHC II presentation of the transgene by DC. However, we show for the first time that exogenous presentation of the transgene by DC also occurs at later time points only in dystrophic mice, inducing a protracted expansion and activation of CD4+ T cells. This event requires the expression of the transgene in non-hematopoietic cells, and transfer of antigenic component to APC. The possibility that exogenous presentation of a transgene by MHC II+ cells occurs following rAAV gene transfer in non-hematopoietic cells has already been shown in vitro, but clearly depends on high antigenic amounts and death of transduced cells.10 It is therefore likely that once transduced, Sgca-/- myofibers release sufficient amounts of antigen to allow exogenous presentation by DC, which is obviously favored in a dystrophic environment in which fiber strength is reduced.18 As a consequence, controlling only the endogenous presentation of the transgene in dystrophic mice is not sufficient to prevent rejection and requires other strategies.
Restricting T-cell priming and reducing effector functions are two complementary approaches known to improve the persistence of transduced cells in normal muscles.27 It might be interesting to test this option in a dystrophic environment in which different pathways are involved to prime CD4+ T cells that are central to recruit multiple effector cells involved in the rejection of the transgene. Specific studies in dystrophic models are therefore not only essential to design gene-corrective approaches but also to adapt those strategies to limit the immune-mediated loss of the transgene. Finally, our findings also suggest that i.m. gene transfer approaches, which are developed for various indications ranging from hereditary diseases to vaccination, should consider the quality of the muscle tissue to be injected as a main factor in treatment outcome.
Materials and methods
Animals
Six- to eight-week-old C57BL/6 (CD45.2) mice were purchased from Charles River (L'abresle, France). Marilyn (CD45.1 Rag2-/-) mice carrying a transgenic T-cell receptor that recognizes products of the male HY gene, the peptide NAGFNSNRANSSRSS from the DBY protein presented by I-Ab (referred as the Dby peptide), were kindly provided by Lantz et al.28 The Sgca-null mice obtained from K Campbell (Howard Hughes Medical Institute, Iowa City, IA, USA) were backcrossed onto C57BL/6 background (I Richard, Genethon, Evry, France) to obtain Sgca-/- further bred in our animal facility.29,30 Strains B6.129S2-CD4tm1Mak/J (CD4-/-), B6.129S2-CD8atm1Mak/J (CD8-/-), B6.129S2-H2dlAbl-Ea/J (MHC II-/-), B6.129P2-B2mtm1Unc/J (β2m-/-) and B6.129S2-Igh-6tm1Cgn/J (μMT-/-) were purchased from The Jackson Laboratory (Bar Harbor, ME, USA) and bred in our animal facility. CD4-/- Sgca-/- and CD8-/- Sgca -/- mice were obtained at Genethon after selection of the double-knockout mice resulting from the intercross of Sgca-/- mice with CD4-/- and CD8-/- mice, respectively. All animal experiments were performed according to institutional and international guidelines for animal care and use.
Peptides and rAAV vectors
The Dby and Uty peptides were synthesized by Genepep (Montpellier, France). Adenovirus-free preparations of the rAAV2/1_CMV_SGCA_HY and rAAV2/1_CMV_SGCA_HY-Mir142T vectors were generated by transfection of HEK293 cells with three plasmids: the plasmid encoding the serotype 1 capsid (pLTR CO2), the plasmid helper (pXX6), the plasmid encoding the transgene with or without the Mir142.3p target sequence (pGG1 SGCA HY, pGG1 SGCA HY-miR142T, respectively) constructed as described previously.14 rAAV2/1 vectors were purified by centrifugation over a CsCl gradient and tittered in viral genome (vg) by real-time PCR as described previously.31 All the vectors were tested for endotoxin levels.
I.m. injection, immunofluorescence and histology
For i.m. injection, animals were anesthetized by intraperitoneal injection of xylazine (10 mg kg−1) and ketamine (100 mg kg−1) and the left tibialis anterior (TA) was injected with 25 μl of the viral preparation adjusted at the specified concentration. Mice were killed at indicated time points and the injected muscles were harvested and snap frozen in liquid nitrogen.
Transverse cryosections (8 μm thick) of the muscles were either stained with hematoxylin and eosin (HE) or used for immunostaining. In such case, expression of the human α-sarcoglycan was investigated by immunohistochemistry using a rabbit polyclonal antibody directed against amino acids 366–379 of the human α-sarcoglycan sequence (AC-ahSarco57) as described previously.18 In parallel, detection of cell infiltrates was performed using double indirect immunofluorescent staining with rabbit anti-CD3 (Epitomics, Burlingame, CA, USA) to identify T cells and rat monoclonal antibodies (mAbs) anti-CD11b (Pharmingen, BD Biosciences, San Jose, CA, USA) to detect inflammatory cells in particular macrophages and neutrophils. A second sequential incubation with anti-species secondary antibodies was performed using Alexa-594 goat anti-rabbit Ab (Invitrogen, Carlsbad, CA, USA) and Alexa-488 goat anti-rat Ab (Invitrogen). 4',6-Diamidino-2-phenylindole staining of the nuclei was added to this staining.
The slides were examined with a Nikon microscope (Champigny sur Marne, France). Digital images were captured using CCD camera (Sony) and processed using Cartograph 5.5 (Microvision Software; Microvision, Evry France).
Depletion of macrophages in vivo
Liposomal clodronate called Clodrosome was used to deplete macrophages as well as its related liposome control devoid of clodronate, called Encapsome (both from Encapsula, Nanosciences, Brentwood, TN, USA). Mice were injected intravenously with 0.1 ml per 10 g body weight according to the advices of the provider at days 5, 7, 9 and 12 postvector administration.
Measure of transgene expression by real-time RT-PCR
Total RNA was extracted from muscles using Trizol (Invitrogen). Residual DNA was removed from the samples using the Free DNA Kit (Ambion, Courtaboeuf, France). cDNA was synthesized from 1 μg of RNA using random hexamers according to the protocol Superscript II first-strand synthesis system for reverse transcription-PCR (RT-PCR) (Invitrogen). Real-time PCR was performed using ABI PRISM 7700 system (Life Technology, Saint Aubin, France) with 0.2 mM of each primer and 0.1 mM of the probe according to the protocol Absolute QPCR Rox Mix (ABgene, Cambridge, UK). The primer pairs and Taqman probes used for amplification of either the ha-sarcoglycan transgene (SGCA) or the ubiquitous acid ribosomal phosphoprotein (PO) normalizer were described previously.14
Determination of the immune response to the transgene
Interferon-γ enzyme-linked immunospot assays were performed as described previously,14 by culturing 106 spleen cells per well with or without 1 μM of peptide. As a positive control, cells were stimulated with concanavalin A (5 μg ml−1). After 24 h, spots were revealed and counted using a Bioreader 2000 (BIO-SYS, Karben, Germany). Spot-forming units are represented after subtraction of background values obtained with non-pulsed splenocytes.
In vivo expansion of transgene specific CD4+ T cells
C57BL/6 mice and Sgca-/- mice were injected with 106 splenocytes from congenic Marilyn mice for an enrichment of the anti-transgene-specific CD4+ T cells, and the following day, the injected mice received the indicated dose of given rAAV2/1 vector. The level of expansion of adoptively transferred cells in blood was investigated after erythrocyte lysis by hypotonic shock with PharMLysis buffer (BD Biosciences) and monitoring of the CD45.1+ CD4+ cells among the total CD4+ population by FACS over time. All reagents used for flow cytometry were purchased from BD Biosciences. Cell suspensions were first incubated with anti-FcγRIII/II (2.4G2) mAbs for 15 min at 4 °C and then stained for 30 min at 4 °C in PBS with 0.1% bovine serum albumin using saturating amounts of the following mAbs: pacific blue-conjugated anti-CD4, biotinylated-anti-CD45.1 and APC-conjugated streptavidin. Dead cells were excluded using 7-actinomycin D (Sigma Chemical Co., St Louis, MO, USA) staining. Analyses were performed on a FACSCalibur or LSRII using CELLQuest or Diva software (BD Biosciences).
Ex vivo antigenic presentation of the transgene to specific CD4+ T cells
The draining lymph nodes were harvested from four to five mice injected in the left and right TA 5 or 10 days before with the different rAAV2/1 vectors (and controls). CD11c+ cells were obtained from these lymph nodes digested first with collagenase IV (Invitrogen), DNAse I (Roche Diagnostics, Meylan, France), incubated with anti-CD11c magnetic beads (Miltenyi Biotec, Paris, France) and purified by automatic magnetic cell sorting (Miltenyi Biotec). The purity was about 95%. To analyze the antigenic presentation to specific CD4+ T cells, T cells were purified from Marilyn mice with anti-CD90.2 magnetic beads and then stained with CFSE (Molecular Probes, Cambridge, UK) (2 μM) in RPMI-1640 (10 × 106 cells per ml) at room temperature for 8 and 2 min at 37 °C. The reaction was stopped by adding an equal volume of RPMI-1640 10% fetal calf serum followed by 5 min incubation at 4 °C. Cells were extensively washed, resuspended in PBS and put together with the CD11c+ cells at different ratios in a 96-well plate. Three days later, the CD4+ T-cell proliferation was assessed by FACS by estimating the CFSE dilution in CD4+-stained cells. Analyses were performed on a FACSCalibur or LSRII using CELLQuest or Diva software (BD Biosciences). Negative (CD11c+ with PBS) and positive (CD11c+ with DBY) controls were always included to assess the proliferation level.
Statistical analysis
Two-group comparisons were performed using a Mann and Whitney two-tailed analysis. Values were considered to be statistically significant if P<0.05.
References
Mendell JR, Rodino-Klapac LR, Rosales-Quintero X, Kota J, Coley BD, Galloway G et al. Limb–girdle muscular dystrophy type 2D gene therapy restores alpha-sarcoglycan and associated proteins. Ann Neurol 2009; 66: 290–297.
Wierzbicki AS, Viljoen A . Alipogene tiparvovec: gene therapy for lipoprotein lipase deficiency. Expert Opin Biol Ther 2013; 13: 7–10.
Mays LE, Wilson JM . The complex and evolving story of T cell activation to AAV vector-encoded transgene products. Mol Ther 2011; 19: 16–27.
Wang L, Dobrzynski E, Schlachterman A, Cao O, Herzog RW . Systemic protein delivery by muscle-gene transfer is limited by a local immune response. Blood 2005; 105: 4226–4234.
Wang Z, Tapscott SJ, Chamberlain JS, Storb R . Immunity and AAV-mediated gene therapy for muscular dystrophies in large animal models and human trials. Front Microbiol 2011; 2: 201.
Li X, Cao H, Wang Q, Di B, Wang M, Lu J et al. Novel AAV-based genetic vaccines encoding truncated dengue virus envelope proteins elicit humoral immune responses in mice. Microbes Infect 2012; 14: 1000–1007.
Herzog RW, Mount JD, Arruda VR, High KA, Lothrop Jr CD . Muscle-directed gene transfer and transient immune suppression result in sustained partial correction of canine hemophilia B caused by a null mutation. Mol Ther 2001; 4: 192–200.
Hartigan-O'Connor D, Kirk CJ, Crawford R, Mule JJ, Chamberlain JS . Immune evasion by muscle-specific gene expression in dystrophic muscle. Mol Ther 2001; 4: 525–533.
Jooss K, Yang Y, Fisher KJ, Wilson JM . Transduction of dendritic cells by DNA viral vectors directs the immune response to transgene products in muscle fibers. J Virol 1998; 72: 4212–4223.
Sarukhan A, Soudais C, Danos O, Jooss K . Factors influencing cross-presentation of non-self antigens expressed from recombinant adeno-associated virus vectors. J Gene Med 2001; 3: 260–270.
Xu D, Walker CM . Continuous CD8(+) T-cell priming by dendritic cell cross-presentation of persistent antigen following adeno-associated virus-mediated gene delivery. J Virol 2011; 85: 12083–12086.
Zhang Y, Chirmule N, Gao G, Wilson J . CD40 ligand-dependent activation of cytotoxic T lymphocytes by adeno-associated virus vectors in vivo: role of immature dendritic cells. J Virol 2000; 74: 8003–8010.
Ribas A . Genetically modified dendritic cells for cancer immunotherapy. Curr Gene Ther 2005; 5: 619–628.
Boisgerault F, Gross DA, Ferrand M, Poupiot J, Darocha S, Richard I et al. Prolonged gene expression in muscle is achieved without active immune tolerance using microRNA 142.3p-regulated rAAV gene transfer. Hum Gene Ther 2013; 24: 393–405.
Majowicz A, Maczuga P, Kwikkers KL, van der Marel S, van Logtenstein R, Petry H et al. Mir-142-3p target sequences reduce transgene-directed immunogenicity following intramuscular adeno-associated virus 1 vector-mediated gene delivery. J Gene Med 2013; 15: 219–232.
Wehling M, Spencer MJ, Tidball JG . A nitric oxide synthase transgene ameliorates muscular dystrophy in mdx mice. J Cell Biol 2001; 155: 123–131.
McDouall RM, Dunn MJ, Dubowitz V . Nature of the mononuclear infiltrate and the mechanism of muscle damage in juvenile dermatomyositis and Duchenne muscular dystrophy. J Neurol Sci 1990; 99: 199–217.
Fougerousse F, Bartoli M, Poupiot J, Arandel L, Durand M, Guerchet N et al. Phenotypic correction of alpha-sarcoglycan deficiency by intra-arterial injection of a muscle-specific serotype 1 rAAV vector. Mol Ther 2007; 15: 53–61.
Darin N, Kroksmark AK, Ahlander AC, Moslemi AR, Oldfors A, Tulinius M . Inflammation and response to steroid treatment in limb-girdle muscular dystrophy 2I. Eur J Paediatr Neurol 2007; 11: 353–357.
Tidball JG, Villalta SA . Regulatory interactions between muscle and the immune system during muscle regeneration. Am J Physiol Regul Integr Comp Physiol 2010; 298: R1173–R1187.
Velazquez VM, Bowen DG, Walker CM . Silencing of T lymphocytes by antigen-driven programmed death in recombinant adeno-associated virus vector-mediated gene therapy. Blood 2009; 113: 538–545.
Riella LV, Paterson AM, Sharpe AH, Chandraker A . Role of the PD-1 pathway in the immune response. Am J Transplant 2012; 12: 2575–2587.
Liao H, Franck E, Freret M, Adriouch S, Baba-Amer Y, Authier FJ et al. Myoinjury transiently activates muscle antigen-specific CD8+ T cells in lymph nodes in a mouse model. Arthritis Rheum 2012; 64: 3441–3451.
Corthay A . CD4+ T cells cooperate with macrophages for specific elimination of MHC class II-negative cancer cells. Adv Exp Med Biol 2007; 590: 195–208.
Villalta SA, Deng B, Rinaldi C, Wehling-Henricks M, Tidball JG . IFN-gamma promotes muscle damage in the mdx mouse model of Duchenne muscular dystrophy by suppressing M2 macrophage activation and inhibiting muscle cell proliferation. J Immunol 2011; 187: 5419–5428.
Sugiura T, Murakawa Y, Nagai A, Kondo M, Kobayashi S . Fas and Fas ligand interaction induces apoptosis in inflammatory myopathies: CD4+ T cells cause muscle cell injury directly in polymyositis. Arthritis Rheum 1999; 42: 291–298.
Adriouch S, Franck E, Drouot L, Bonneau C, Jolinon N, Salvetti A et al. Improved immunological tolerance following combination therapy with CTLA-4/Ig and AAV-mediated PD-L1/2 muscle gene transfer. Front Microbiol 2011; 2: 199.
Lantz O, Grandjean I, Matzinger P, Di Santo JP . Gamma chain required for naive CD4+ T cell survival but not for antigen proliferation. Nat Immunol 2000; 1: 54–58.
Duclos F, Straub V, Moore SA, Venzke DP, Hrstka RF, Crosbie RH et al. Progressive muscular dystrophy in alpha-sarcoglycan-deficient mice. J Cell Biol 1998; 142: 1461–1471.
Turk R, Sterrenburg E, van der Wees CG, de Meijer EJ, de Menezes RX, Groh S et al. Common pathological mechanisms in mouse models for muscular dystrophies. FASEB J 2006; 20: 127–129.
Riviere C, Danos O, Douar AM . Long-term expression and repeated administration of AAV type 1, 2 and 5 vectors in skeletal muscle of immunocompetent adult mice. Gene Therapy 2006; 13: 1300–1308.
Acknowledgements
We are grateful for the help from the Genethon ‘In vivo Evaluation’ and ‘Product Development’ groups for help with animal care, animal experiments, histology and rAAV vector production. We thank Bernard Gjata and Christophe Georger for the involvement in our project. We are indebted to O Lantz, K Campbell and I Richard for providing mice and also obliged to K Campbell for his support in crossing Sgca-null mice. We acknowledge the support from AFM (French Muscular Dystrophy Association) and from the PERSIST EC FP7 large-scale integrating project (GA no. 222878).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Gene Therapy website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Ferrand, M., Galy, A. & Boisgerault, F. A dystrophic muscle broadens the contribution and activation of immune cells reacting to rAAV gene transfer. Gene Ther 21, 828–839 (2014). https://doi.org/10.1038/gt.2014.61
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gt.2014.61