Abstract
Viral infection induces numerous tripartite motif (TRIM) proteins to control antiviral immune signaling and viral replication. Particularly, SPRY-containing TRIM proteins are found only in vertebrates and they control target protein degradation by their RING-finger and SPRY domains, and proper cytoplasmic localization. To understand TRIM30 function, we analyzed its localization pattern and putative roles of its RING-finger and SPRY domains. We found that TRIM30 is located in actin-mediated cytoplasmic bodies and produces colocalized ubiquitin chains in SPRY domain- and RING-finger domain-dependent ways that are degraded by autophagy and the proteasome. These results suggest a TRIM protein-dependent degradation mechanism by cytoplasmic body formation with actin networks.
Similar content being viewed by others
Introduction
Many tripartite motif (TRIM) proteins are highly induced upon viral infection and are known to be involved in several pathogen recognition and signaling events during the antiviral host immune response. TRIM proteins share key common structural features at the N terminus: a RING-finger domain, B-box zinc-finger domain(s) and a coiled-coil domain. The RING-finger domain of some TRIM family members was shown to have E3 ubiquitin ligase activity,1 whereas the B-box domain and coiled-coil domain were suggested to mediate protein–protein interactions, probably being required for the enzymatic activities of the protein.2 Contrary to the well-conserved structural organization of TRIM proteins at the N terminus, TRIM proteins possess various C-terminal domains, reflecting their versatile functions.3, 4, 5, 6 In addition, various C-terminal domains of individual TRIM proteins may elicit the distinct function of each TRIM protein in unique subcellular locations. For example, TRIM25 translocates to stress granules upon virus infection to interact with antiviral proteins;7 however, TRIM27 negatively regulates CD4+ T cells by ubiquitinating PI3KC2β in recycling endosomes.8
Among the diverse C-terminal domains found in TRIM proteins, the SPRY domain is found in about half of the TRIM proteins. The SPRY domain is present only in vertebrates and is suggested to act as a protein-interaction domain. The SPRY domain of human TRIM5 was shown to interact with the surface lattice of viral capsids,9 and its amino-acid sequence appears to dictate the susceptibility to HIV-1 infection.10, 11 Therefore, the SPRY domain appears to have an important role in the restriction of retrovirus by regulating viral capsid generation, but the precise mechanism underlying the TRIM5-dependent antiviral activity is not well understood.
Mouse TRIM30, which has a SPRY domain, is homologous to primate TRIM5. TRIM30 was suggested to negatively regulate Toll-like receptor and Nalp3 inflammasome signaling by targeting TAB1- and TAB2-signaling molecules for degradation. However, both in vivo and in vitro studies with TRIM30 null mice showed slight defects in T-cell proliferation, but no obvious defects in Toll-like receptor-signaling pathways in the absence of TRIM30.12, 13, 14 To resolve the contradicting observations, we aimed to examine the subcellular localization pattern of TRIM30 along with other cellular components and the functions associated with each domain of the protein. Using green fluorescent protein (GFP)-tagged TRIM30, we found that TRIM30 formed cytoplasmic bodies that overlap with the unanchored ubiquitin chains produced by TRIM30, and with an autophagy marker, LC3, but failed to colocalize with other cellular components involved in cellular signaling. We confirmed that the inhibition of autophagy or the proteasome stabilized TRIM30. Moreover, assembly into the cytoplasmic body of TRIM30 was dependent on the SPRY domain through interaction with actin and myosin. Therefore, TRIM30-dependent ubiquitination can induce the degradation of target proteins in cytoplasmic bodies similar to the degradation of retroviral capsid proteins by TRIM5.
Materials and methods
Cell lines and culture
The NIH-3T3 cell line and Atg7+/+ and Atg7−/− mouse embryonic fibroblast (MEF) cells were used for transfection studies. Cells were maintained in Dulbecco’s modified Eagle’s medium (Gibco, Grand Island, NY, USA) supplemented with heat-inactivated 10% fetal bovine serum (Gibco), 50 Units ml−1 penicillin and 50 μg ml−1 streptomycin (Pen Strep; Gibco). The cell line was cultured in 5% CO2 in a 37 °C cell incubator. Atg7 MEF cells were provided by Noboru Mizushima (University of Tokyo).
Transfection of plasmids
All of the plasmids were transfected into cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. A day before transfection, the cell line was cultured in 24-well plates with a microscope cover glass on the bottom of the well for 24 h. For 24-well plates, 0.5 μg of DNA was mixed with 50 μl of Opti-MEM (Gibco), and 1 μl of Lipofectamine was added. This mixture was incubated for 5 min at room temperature before being added to the cells.
Plasmid construction
Open reading frame full-length of TRIM30 complementary DNA was obtained from bone marrow-derived macrophages by PCR amplification using appropriate primers. The TRIM30 mutant form (deletion of RING domain: ΔRING; deletion of SPRY domain: ΔSPRY) was amplified from the full-length of the TRIM30-coding region using the following primers: Trim30 forward primer 5′-CCCGAATTCTATGGCCTCATCAGTC-3′ and reverse primer 5′-ATAGGATCCTTAGGAGGGTGGCCCG-3′; Trim30 ΔRING forward primer 5′-CCCGAATTCAGTTCCTTACCCATTTG-3′ and Trim30 ΔSPRY reverse primer 5′-CCGGGATCCCCTTGCATGTAGAGT-3′; and Trim30 C35A forward primer 5′-GCTGATTGTAACCACAGCTTCGCTAGAGCCTGCATCACA-3′ and Trim30 C35A reverse primer 5′-TGTGATGCAGGCTCTAGCGAAGCTGTGGTTACAATCAGC-3′. The C35A mutant was constructed using a site-directed mutagenesis kit (Stratagene, Palo Alto, CA, USA). All of these inserts were cloned into the pEGFP-C1 and pCS4-3 × Flag plasmids by the available restriction enzyme sites.
Antibodies and reagents
The following antibodies and reagents were used in the immunofluorescence or immunoblotting experiments: monoclonal anti-multi ubiquitin (FK2; D058-3; MBL, Nagoya, Japan), monoclonal anti-K48 (05–1307; Millipore, Billerica, MA, USA), monoclonal anti-K63 (05–1308; Millipore), monoclonal anti-Flag (F7425; Sigma-Aldrich, St Louis, MO, USA), anti-β-Actin (#4967; Cell signaling, Danvers, MA, USA), Alexa Fluor 488 Phalloidin (A12379; Molecular Probes, Eugene, OR, USA) and cytochalasin D (Sigma-Aldrich).
Immunofluorescence confocal microscopy imaging
NIH-3T3 cells were seeded on microscope cover glasses in 24-well plates and were transfected for immunofluorescence. After 24 h of transfection, the microscope cover glasses were washed with cold phosphate-buffered saline (PBS), and cells were fixed with 4% (w/v) paraformaldehyde in PBS for 10 min. After fixation, cells were permeabilized using 10 × BD Perm/Wash (BD Bioscience, San Jose, CA, USA) diluted in distilled H2O. To block non-specific binding of antibody, cells were treated with blocking solution (TBST (0.1% (v/v) Tween 20 in PBS) with 10% (v/v) goat serum) for 1 h. Next, cells were incubated with the primary antibody at 4 °C overnight. Cells were then washed with PBS and incubated with tetramethylrhodamine-conjugated anti-rabbit secondary antibody (1:100; Sigma-Aldrich) or anti-mouse secondary antibody (1:100; Sigma-Aldrich) diluted in blocking solution. The nucleus was stained with 4′,6-diamidino-2-phenylindole (Sigma-Aldrich). Microscope cover glasses were mounted on the slide glasses with mounting medium (Dako, Glostrup, Denmark). Finally, the samples were analyzed using a confocal microscope (LSM700; Carl Zeiss, New York, NY, USA).
Immunoblotting
Whole cells were lysed in radioimmunoprecipitation solution (100 mM Tris, 5 mM EDTA, 50 mM NaCl, 50 mM beta-glycerol phosphate, 50 mM NaF, 0.1 mM sodium orthovanadate, 1 mM phenylmethanesulfonylfluoride, 0.5% NP-40, 1% Triton X-100 and 0.5% sodium deoxycholate) with freshly added protease inhibitor cocktail (Roche, Indianapolis, IN, USA). To remove cell debris, lysates were centrifuged at 14 000 g for 10 min at 4 °C. The supernatants were quantified using a BCA protein assay kit (Pierce, Rockford, IL, USA), electrophoresed on an SDS-polyacrylamide gel and then transferred to nitrocellulose membranes using a wet transfer system (Bio-Rad, Hercules, CA, USA). The blots were blocked with 5% w/v skim milk in PBS with 0.1% Tween-20 for 1 h at room temperature or overnight at 4 °C. After blocking, membranes were probed with specific antibodies diluted in blocking buffer, followed by incubation with horseradish peroxidase-conjugated secondary antibody at 1:5000 dilution. Blots were developed using the ECL prime kit (GE Healthcare, Little Chalfont, UK), and images were detected using the LAS 4000 system (GE Healthcare).
Immunoprecipitation and mass spectrometric analysis
NIH-3T3 cells transiently expressing Trim30 and ÄSPRY with a Flag and SBP tag at its N terminus were used for immunoprecipitation analysis. First, transfected cells were lysed and subjected to Anti-Flag M2 affinity gel (Sigma-Aldrich). Elution fractions were obtained using Flag peptide. The first eluate was immunoprecipitated with Streptavidin Sepharose (GE Healthcare) in the second purification. Immunocomplexes were subjected to SDS-polyacrylamide gel electrophoresis, stained with Coomassie blue and the protein bands were digested with trypsin and subjected to mass spectrometric analysis at Yonsei Proteome Research Center.
Results
TRIM30 is localized in cytoplasmic bodies with ubiquitin chains and autophagy markers
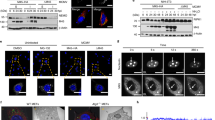
TRIM family proteins displayed various subcellular localizations that are related to their biological functions. To identify the subcellular localization of TRIM30, GFP-TRIM30 was transfected into NIH-3T3 cells. Immunofluorescence staining showed that GFP-TRIM30 formed several large cytoplasmic body structures in the cytosol. Double staining with appropriate antibodies against various cellular organelle markers (KDEL, RCAS2, Lamp1 and PMP70) or with Mitotracker revealed that the TRIM30 cytoplasmic bodies are distinct from the endoplasmic reticulum (ER), Golgi, lysosomes, peroxisomes and mitochondria (Figures 1a–e). Most of the TRIM30 cytoplasmic bodies appeared in the periphery away from the nucleus and ER area. However, TRIM30 colocalized with an autophagy marker, LC3 (Figure 1f), indicating a putative function of TRIM30 in protein degradation. To test the possibility, we examined the colocalization of TRIM30 with polyubiquitin chains by FK2 antibody staining of enhanced GFP–TRIM30-expressing cells. The FK2 antibody, which recognizes polyubiquitinated proteins, labeled several cytoplasmic bodies throughout the cytoplasm, including the ER-associated areas. TRIM30 appeared to colocalize with polyubiquitin chains at most of the cytoplasmic bodies (Figure 1g). To test the types of lysine linkage in the polyubiquitin chains, the ubiquitin chains were stained with antibodies that recognize K48- or K63-linked polyubiquitins. K48-linked ubiquitination is usually associated with proteasomal degradation, whereas K63-linked ubiquitination is associated with both protein degradation and signaling pathways. Intriguingly, TRIM30 colocalized with both K48 and K63 ubiquitin chains (Figures 1h and i). These results suggested that TRIM30 is associated with a ubiquitin-related protein degradation response.
Subcellular localization of TRIM30. NIH-3T3 cells expressing GFP-TRIM30 were stained with the antibodies against (a) KDEL, (b) RCAS1, (c) Mitotracker, (d) LAMP1, (e) PMP70, (f) LC3, (g) FK2, (h) K48-linked ubiquitin and (i) K63-linked ubiquitin (red), as well as 4′,6-diamidino-2-phenylindole staining (blue). Bars, 10 μm.
The RING-finger domain is required for the production of colocalized ubiquitin chains
Given that TRIM30 localizes with endogenous ubiquitin chains, we next investigated whether the RING-finger domain of TRIM30 is responsible for the synthesis of polyubiquitin chains colocalized with TRIM30 at the cytoplasmic bodies. To this end, we generated enhanced GFP-tagged TRIM30 with the RING-finger domain deletion (Trim30ΔRING; Figure 2a), and examined their localization along with those of K48- and K63-linked polyubiquitin chains in the NIH-3T3 cells transfected with enhanced GFP-tagged Trim30ΔRING. As shown in Figure 2b, TRIM30ΔRING showed large cytoplasmic body formations that were similar to those of TRIM30 wild type, but the cytoplasmic bodies were larger than those of TRIM30 wild type (Figure 2b). However, the number of ubiquitin-containing cytoplasmic bodies diminished greatly in the TRIM30ΔRING-expressing cells, and even the small number of areas showing polyubiquitin signals did not colocalize with the TRIM30ΔRING-containing cytoplasmic bodies (Figure 2b). Therefore, the E3 ligase activity of the TRIM30 RING domain appears responsible for the production of the ubiquitin chains. To confirm the idea, we examined the localization patterns of TRIM30 protein and ubiquitin chains using a mutant version of TRIM30 protein that harbors an inactivating point mutation (substitution of the 35th cysteine with alanine) at the RING-finger domain of TRIM30 (Trim30C35A). Confocal microscopy analysis revealed that TRIM30C35A formed cytoplasmic bodies that are devoid of polyubiquitin chains similar to those observed in TRIM30ΔRING-transfected cells (Supplementary Figure 1). From these findings, we confirmed that the TRIM30 RING-finger domain has potential E3 ligase activity required for polyubiquitin chain production at the cytoplasmic bodies.
Trim30 is degraded by the proteasome and autophagy. (a) The construct diagram of wild-type TRIM30 and mutant forms (Δ RING: deletion of RING domain, Δ SPRY: deletion of SPRY domain and C35A: substitution 35th cysteine for alanine). (b) NIH-3T3 cells were transfected with the TRIM30 Δ RING construct. After 24 h, cells were stained with antibodies to specific ubiquitin markers (K48 and K63 ubiquitin). Bars, 10 μm. (c) After transfection of TRIM30- and TRIM30ΔRING-encoding plasmids into NIH-3T3 cells and treatment with MG132 (10 μM) for 6 h. Cells were harvested at 96 h post transfection and at analysis. Each experiment was repeated at least three times. (d) After transfection of TRIM30- and TRIM30ΔRING-encoding plasmid into Atg7+/+ and Atg7−/− MEF cells, followed by treatment with MG132 (10 μM) for 6 h. Cells were harvested at 72 h post transfection and at analysis. Each experiment was repeated at least three times.
TRIM30 is degraded by proteasome and autophagy in an ubiquitin-dependent way
The E3 ligase activity of TRIM30 and its colocalization with polyubiquitin chains indicate that TRIM30 may be degraded rapidly by auto-ubiquitination similar to most other TRIM proteins.15 To test the idea, we observed the TRIM30 protein levels after blocking proteasome activity with MG132. We found that the expression level of the mutant version of TRIM30 was several fold higher than that of wild type, which was increased up to the mutant level upon MG132 treatment. However, the level of TRIM30ΔRING protein was not affected by the addition of MG132, indicating that proteasomal degradation of TRIM30 is dependent on its own E3 ligase activity (Figure 2c). In addition to proteasomal degradation, autophagy also promotes the removal of ubiquitinated protein aggregates.16 Indeed, as indicated from the colocalization of TRIM30 with LC3 (Figure 1f), the total amount of TRIM30 derivatives were also increased in Atg7−/− MEF cells, which lack functional autophagy (Figure 2d). Although we cannot exclude the possibility of different levels of Trim30 derivatives as a result of different expression levels, the fact that the level of TRIM30 protein was further increased by treatment with MG132 (Supplementary Figures 2a and b) indicates otherwise. These results suggest that TRIM30 protein interacts with ubiquitin chains and is degraded by both ubiquitin–proteasome and ubiquitin-autophagy pathways.
SPRY domain is required for TRIM30 cytoplasmic body formation
Generally, the SPRY domain is implicated in protein–protein interactions, and that of TRIM5 is shown to bind viral capsid protein as the target substrate of its E3 ligase activity. However, the exact role of the SPRY domain in most TRIM protein remains unclear.17 To study the function of the SPRY domain of TRIM30, we made a SPRY domain deletion TRIM30 construct (Trim30ΔSPRY; Figure 2a), and examined defects in the localization pattern and production of polyubiquitin chains. Intriguingly, TRIM30ΔSPRY showed a localization pattern markedly different from that of wild type. TRIM30ΔSPRY did not localize to cytoplasmic bodies but formed a ribbon-like structure in the cytosol (Figure 3). Although cytoplasmic body formation was destroyed, most of TRIM30ΔSPRY maintained colocalization with ubiquitin chains. Both K48- and K63-linked ubiquitination were presented along with the ribbon-like structure of TRIM30ΔSPRY. From these results, we assumed that the SPRY domain is required for cytoplasmic body formation of TRIM30, probably through the interaction with unknown cytosolic proteins.
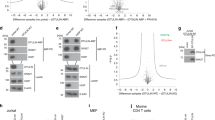
To identify the role of the SPRY domain in the formation of cytoplasmic body complexes through the identification of its binding partner, we tried tandem affinity purification of TRIM30- and TRIM30ΔSPRY-associated proteins. Flag- and SBP-double-tagged TRIM30-derivative-expressing cell extracts were subjected to immunoprecipitation with anti-Flag followed by streptavidin sepharose affinity purification; next, the purified eluates were separated by SDS-polyacrylamide gel electrophoresis for mass spectrometer analysis of differentially purified peptides (Supplementary Figures 3a and b). Wild-type TRIM30 was co-purified with two major additional proteins of ~48 kD and larger than 100 kD in molecular-weight (Supplementary Figures 3a and b)—actin and myosin, respectively (Figures 4a and b). The band intensities corresponding to these peptides were significantly decreased in the affinity-purified fractions of TRIM30ΔSPRY. To examine whether TRIM30 interacts with endogenous actin, we performed co-immunoprecipitation experiments with TRIM30 wild-type and SPRY deletion mutant proteins and confirmed that β-actin indeed binds only to wild-type TRIM30 (Figure 4c). These data indicate that TRIM30 associates with myosin and the actin cytoskeleton through the SPRY domain, which induces distinct cytoplasmic body formation. Taken together, these results suggest that the cytoplasmic body of TRIM30 contributes to the ubiquitin-dependent degradation of target proteins, such as viral capsids, using the proteasome and autophagy.
Actin and myosin are associated with the cytoplasmic body of Trim30. The amino-acid sequence of beta-actin (a) and myosin (b). Red and underlining show peptides identified by mass spectrometry. (c) NIH-3T3 cells transfected with Flag_SBP-TRIM30 or Flag_SBP-TRIM30ΔSPRY were used for immunoprecipitation and immunoblotting. Left panel: input lysate, right panel: eluted fraction.
Discussion
Primate TRIM5 is a restriction factor that inhibits retrovirus infection via capsid recognition by the SPRY domain.18, 19 Species-specific differences in the SPRY domain regulates the efficiency of retrovirus restriction.18 Recent studies have shown that TRIM5 synthesizes K63-linked ubiquitin chains to promote immune signaling,20 and E3 ubiquitin ligase activity is actually required for the restriction of virus infection.21 TRIM30, a close homolog of human TRIM5, has a SPRY domain in the C terminus along with active E3 ligase activity, indicating a TRIM5-related function in mice. In addition, cytoplasmic body formation was also observed in HIV-1-infected cells with primate TRIM5, extending the functional similarities between these homologs.22, 23 The relationship between TRIM30 and retrovirus has not been revealed yet, but another TRIM30 homolog was shown to facilitate lysosome-mediated degradation of viral RNA polymerase and restricts tick-borne encephalitis virus.
Although TRIM5 is a well-known restriction factor that prevents retrovirus infection, the exact mechanism by which TRIM5 limits retroviral infection after capsid recognition is not identified.24 Particularly, the requirement of proteasomal activity for blocking retroviral infection remains controversial.25, 26, 27 Although we could not present the substrate of TRIM30 in this study, identification of a TRIM30 degradation mechanism suggests that TRIM5-mediated destruction of capsid protein may require proteasome and autophagy activities. Because MG132 also inhibits various cysteine proteases and cathepsins, we cannot rule out the involvement of proteases in Trim30 degradation completely. In addition, the binding of SPRY to actin and myosin indicates the involvement of intracellular trafficking in the TRIM30-mediated antiviral function.
References
Deshaies RJ, Joazeiro CA . RING domain E3 ubiquitin ligases. Annu Rev Biochem 2009; 78: 399–434.
Meroni G, Diez-Roux G . TRIM/RBCC, a novel class of 'single protein RING finger' E3 ubiquitin ligases. Bioessays 2005; 27: 1147–1157.
Balastik M, Ferraguti F, Pires-da Silva A, Lee TH, Alvarez-Bolado G, Lu KP et al. Deficiency in ubiquitin ligase TRIM2 causes accumulation of neurofilament light chain and neurodegeneration. Proc Natl Acad Sci USA 2008; 105: 12016–12021.
Ozato K, Shin D-M, Chang T-H, Morse HC . TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol 2008; 8: 849–860.
Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L et al. The tripartite motif family identifies cell compartments. Embo J 2001; 20: 2140–2151.
Schwamborn JC, Berezikov E, Knoblich JA . The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 2009; 136: 913–925.
Yoo JS, Takahasi K, Ng CS, Ouda R, Onomoto K, Yoneyama M et al. DHX36 enhances RIG-I signaling by facilitating PKR-mediated antiviral stress granule formation. PLoS Pathog 2014; 10: e1004012.
Cai XJ, Srivastava S, Sun Y, Li Z, Wu HY, Zuvela-Jelaska L et al. Tripartite motif containing protein 27 negatively regulates CD4 T cells by ubiquitinating and inhibiting the class II PI3K-C2 beta. Proc Natl Acad Sci USA 2011; 108: 20072–20077.
Sebastian S, Luban J . TRIM5a selectively binds a restriction-sensitive retroviral capsid. Retrovirology 2005; 2: 40.
Nisole S, Stoye JP, Saib A . Trim family proteins: retroviral restriction and antiviral defence. Nat Rev Microbiol 2005; 3: 799–808.
Sawyer SL, Wu LI, Emerman M, Malik HS . Positive selection of primate TRIM5 alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci USA 2005; 102: 2832–2837.
Choi UY, Hur JY, Lee MS, Zhang QR, Choi WY, Kim LK et al. Tripartite motif-containing protein 30 modulates TCR-activated proliferation and effector functions in CD4+ T cells. PLoS One 2014 22; 9: e95805.
Hu Y, Mao K, Zeng Y, Chen S, Tao Z, Yang C et al. Tripartite-motif protein 30 negatively regulates NLRP3 inflammasome activation by modulating reactive oxygen species production. J Immunol 2010; 185: 7699–7705.
Shi M, Deng W, Bi E, Mao K, Ji Y, Lin G et al. TRIM30 alpha negatively regulates TLR-mediated NF-kappa B activation by targeting TAB2 and TAB3 for degradation. Nat Immunol 2008; 9: 369–377.
Versteeg GA, Rajsbaum R, Sanchez-Aparicio MT, Maestre AM, Valdiviezo J, Shi M et al. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity 2013; 38: 384–398.
Kirkin V, McEwan DG, Novak I, Dikic I . A role for ubiquitin in selective autophagy. Mol Cell 2009; 34: 259–269.
D'Cruz AA, Babon JJ, Norton RS, Nicola NA, Nicholson SE . Structure and function of the SPRY/B30.2 domain proteins involved in innate immunity. Protein Sci 2013; 22: 1–10.
Stremlau M, Perron M, Welikala S, Sodroski J . Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J Virol 2005; 79: 3139–3145.
Yap MW, Nisole S, Stoye JP . A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr Biol 2005; 15: 73–78.
Pertel T, Hausmann S, Morger D, Zuger S, Guerra J, Lascano J et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 2011; 472: 361–365.
Li X, Kim J, Song B, Finzi A, Pacheco B, Sodroski J . Virus-specific effects of TRIM5alpha(rh) RING domain functions on restriction of retroviruses. J Virol 2013; 87: 7234–7245.
Sakuma R, Ohmine S, Ikeda Y . Determinants for the rhesus monkey TRIM5 alpha-mediated block of the late phase of HIV-1 replication. J Biol Chem 2010; 285: 3784–3793.
Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J . The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 2004; 427: 848–853.
Grutter MG, Luban J . TRIM5 structure, HIV-1 capsid recognition, and innate immune signaling. Curr Opin Virol 2012; 2: 142–150.
Campbell EM, Perez O, Anderson JL, Hope TJ . Visualization of a proteasome-independent intermediate during restriction of HIV-1 by rhesus TRIM5 alpha. J Cell Biol 2008; 180: 549–561.
Perez-Caballero D, Hatziioannou T, Zhang FW, Cowan S, Bieniasz PD . Restriction of human immunodeficiency virus type 1 by TRIM-CypA occurs with rapid kinetics and independently of cytoplasmic bodies, ubiquitin, and proteasome activity. J Virol 2005; 79: 15567–15572.
Wu XL, Anderson JL, Campbell EM, Joseph AM, Hope TJ . Proteasome inhibitors uncouple rhesus TRIM5 alpha restriction of HIV-1 reverse transcription and infection. Proc Natl Acad Sci USA 2006; 103: 7465–7470.
Acknowledgements
This research was supported by the Global Research Laboratory Program of the National Research Foundation (NRF) funded by the Ministry of Science, ICT and Future Planning (MEST; K20705000006-12A0500-00610 to Y-JK), the Bio and Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science, ICT and Future Planning (MEST; 2012028272 to Y-JK), the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare, Republic of Korea (grant number: HI13C08470200), the Introduced Innovative R&D Team Leadership of Guangdong Province, People’s Republic of China.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website
Rights and permissions
This work is licensed under a Creative Commons Attribution 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/3.0/
About this article
Cite this article
Choi, U., Choi, W., Hur, J. et al. Polyubiquitin chain-dependent protein degradation in TRIM30 cytoplasmic bodies. Exp Mol Med 47, e159 (2015). https://doi.org/10.1038/emm.2015.12
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/emm.2015.12