Abstract
Tumor necrosis factor receptor-related 2 (TR2, HVEM or TNFRSF-14) plays an important role in immune responses, however, the mechanisms regulating its expression are unclear. To understand the control of TR2 gene expression, we studied the upstream region of the gene. Gel supershift assays revealed inducible binding of nuclear factor of activated T cells (NFAT) to a putative NFAT site within the TR2 promoter. Furthermore, cotransfection of a dominant negative NFAT construct, or siRNA for NFAT, resulted in increased expression of a TR2 reporter gene. Our findings demonstrate that NFAT negatively regulates TR2 expression in activated T cells.
Similar content being viewed by others
Introduction
Members of the tumor necrosis factor receptor (TNFR) family interact with a set of ligands forming the TNF family. The resulting signals are involved in diverse cellular immune processes including differentiation, proliferation, and induction of apoptotic cell death (Locksley et al., 2001). Tumor Necrosis Factor Receptor Related 2 (TR2, HVEM or TNFRSF-14) is expressed in various tissues and immune cells, including T cells, B cells and dendritic cells (DC). Signaling through TR2 promotes cell proliferation and the production of cytokines such as interleukin 2 (IL-2), interferon-γ (IFN-γ), IL-4, and TNF-α (Croft, 2003). TR2 interacts with LIGHT (TL5 or TNFSF-14) (Kwon et al., 1999) and B- and T-lymphocyte attenuator (BTLA) (Krieg et al., 2007), and modulates T cell-mediated immune responses in tumors, graft-versus-host-disease (GVHD), and graft rejection (Ye et al., 2002).
Members of the nuclear factor of activated T cells (NFAT) family are widely expressed in cells of the immune system, including T cells. They are involved in T cell regulation (Rengarajan et al., 2002), and the expression of many inducible genes, including not only cytokines such as IL-4, and IFN-γ (Yoshida et al., 1998) but also TNF family members such as TNF-α, and LIGHT (Macian, 2005). In resting T cells, NFAT proteins reside in the cytoplasm, and upon activation translocate to the nucleus (Wisniewska et al., 2007). The nuclear translocation of NFAT proteins is controlled by calcineurin (CaN), a Ca2+-dependent phophatase, and CaN is highly sensitive to cyclosporine A (CsA) (Shaw et al., 1995), which prevents CaN-dependent nuclear translocation of NFAT.
Ye et al. (2002) reported that treatment with CsA, combined with inhibition of the LIGHT-TR2 interaction using either LIGHT knockout mice or blocking antibody against TR2, prevented acute allograft rejection. The combined treatment suppressed the upregulation of many cytokines in the grafts, including IFN-γ, IL-2, and TNF-α. Interestingly, T cells were still activated after transplantation in the presence of CsA, and TR2 was involved in the activation process, although the effects of CsA on TR2 have not been determined. Based on these observations, we hypothesized that TR2 molecules are expressed on T cells in the presence of CsA, and that the expression of TR2 on T cells is increased by CsA.
TR2 is expressed constitutively on the surface of resting T cells and its level of expression decreases after activation (Morel et al., 2000); however, the mechanism by which it is down regulated after activation is not well understood. In this study, we report that NFAT is a negative regulator of TR2 expression in activated T cells.
Results
TR2 expression is increased by treatment with Cs A
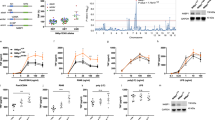
TR2 is expressed constitutively on the surface of resting T cells, and expression decreases after activation (Morel et al., 2000). Also inhibition of the LIGHT-TR2 interaction in the presence of CsA prevents acute allograft rejection (Ye et al., 2002). Therefore, although the effects of CsA on the immune system are complex, we hypothesized that it affects TR2 expression. As shown in Figure 1A, TR2 protein was expressed in resting primary CD4+ T-cells of B57/BL6 mice, and expression was increased by treatment with CsA. Incubation of EL-4 T cells with CsA also resulted in increased expression of TR2 (Figure 1B). To determine whether TR2 expression was affected at the transcriptional level, we analyzed TR2 mRNA levels by RT-PCR. As shown in Figure 1C, TR2 mRNA in EL-4 T cells decreased in response to treatment with PMA/ionomycin (PMA plus ionomycin) and increased when the cells were treated with PMA/ionomycin in the presence of CsA. The effect of CsA on TR2 expression was also assayed by Western blot analysis with anti-TR2 antibody (Figure 1D) at 12 and 24 h after PMA/ionomycin treatment because of the major differences in the RT-PCR results. The protein level of TR2 was also decreased by treatment with PMA/ionomycin and increased by CsA. These experiments point to the presence of negative regulators of TR2 expression.
Effect of CsA on TR2 gene expression in primary murine CD4+ T cells and the EL-4 T cell line. Purified CD4+ T or EL-4 T cells were incubated in the presence or absence of cyclosporin A (400 ng/ml), and TR2 expression was assessed by FACS or RT-PCR. (A) Murine CD4+ T cells or (B) EL-4 cells were stained with anti-TR2 mAb (12H9, rat IgG2b) and FITC-conjugated secondary antibodies, and levels of TR2 expression were determined by FACS analysis. Negative controls with isotypic-matched antibody were indicated in thick line (A) or shaded histogram (B). (C) Total RNA was prepared from EL-4 cells and analyzed by reverse transcript-PCR using mTR2 (27 cycles) and GAPDH (30 cycles) primer pairs in non-saturating conditions. (D) Expression of TR2 protein was determined by Western blot analysis.
An NFAT binding site in the mTR2 promoter is responsible for negative regulation
In a previous study (Sul et al., 2008) we reported that the -273/-3 upstream region of the TR2 gene was important in TR2 gene regulation. Further analysis of this region revealed that deletion of nucleotides -81 to -76 (Figure 2A, D1) resulted in a major increase in the activity of a luciferase reporter gene, pointing to the presence of a negative regulatory element in this interval. Furthermore, changing the TTCCTT sequence to TTAATT (Figure 2B, M1) resulted in comparable activity to that of the D1 deletion. The TTCCTT sequence is identical to the NFAT-binding site generated by the nuclear factor search program TESS (Schug and Overton, 1997). The activity of NFAT proteins is regulated by CaN, which is highly sensitive to the immunosuppressive drug CsA, which prevents CaN-dependent nuclear translocation of NFAT. Since we had found that CsA increases expression of TR2 in T cells, we investigated the effect of CsA on EL-4 cells transfected with TR2 promoter reporter constructs. As demonstrated in Figure 2C, the luciferase activity of the construct pTR2(-276/-3) decreased when cells were activated with PMA/ionomycin, and increased in a dose-dependent manner upon treatment with CsA. On the other hand, exposure of cells harboring the deletion construct D1 to PMA/ionomycin or CsA did not alter promoter activity. These results strongly suggest involvement of NFAT in regulating TR2 expression.
Analysis of the activity of the TR2 promoter and its mutant forms in EL-4 cells. EL-4 cells were transiently transfected with the various constructs, and also with pCMV-β-gal as a control for transfection efficiency. Luciferase activity was measured 24h after transfection. Relative activities were compared with that of the pTR2(-276/-3) construct. This construct was used as the wild type (Wt) construct in subsequent experiments. (A) The deletion mutant construct (D1) was generated from the Wt construct by deleting; the putative NFAT site (from 81 to -76), shown underlined. (B) The substitution mutant construct (M1) was derived from the Wt by replacing the two underlined bases. (C) The effects of PMA/ionomycin and CsA on the expression of the luciferase construct were assayed with the designated mTR2 reporter constructs (Wt or D1). Luciferase assays were repeated at least three times.
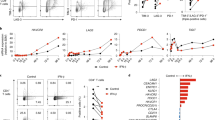
To confirm this role of NFAT, we performed EMSAs with primary CD4+ T cells from C57/BL6 mice. As shown in Figure 3A, nuclear extracts of resting CD4+ T cells formed a complex, C1 (Figure 3A, lane 1) with 32P labeled oligonucleotide P2 containing the wild type NFAT binding site (see Methods), and activation with PMA/ionomycin resulted in the formation of an additional complex, C2 (Figure 3A, lane, 2, 3, and 4). Cold P2 probe (the putative NFAT site) inhibited binding of 32P labeled P2 whereas cold P1 and P3 probes did not (Figure 3B, lanes 1-4). Excess unlabelled commercial NFAT probe (Santa Cruz) also inhibited binding of the P2 probe (Figure 3B, lane 5). Substitution of AA for CC at positions -78 and -79 in the P2 probe (P2M) resulted in loss of competition with P2, indicating that the interaction was sequence specific (Figure 3B, lane 6). When CD4+ T cells were treated with CsA along with PMA/ionomycin the C2 complex was not formed (Figure 3C, lane 2). These results indicate that NFAT binds to the TR2 promoter after activation with PMA/ionomycin. To show that NFAT is recruited to the TR2 promoter, we performed supershift assays. These confirmed that NFATc2 binds to the TR2 promoter of activated CD4+ T cells (Figure 3D, lane 3, arrow C3) since monoclonal antibody against NFATc2 supershifted the C2 complex induced by PMA/ionomycin. To study the effect of the different NFATs, CD4+ T cells were harvested after various times of treatment with PMA/ionomycin. As demonstrated in Figure 3E, binding of NFATc1 and NFATc2 was evident 15 min after activation, and only NFATc1 binding was increased after 20 h of activation whereas no binding of NFATc3 was detected.
NFAT molecules bind to the TR2 gene promoter in CD4+ T cells, and their recruitment is affected by CsA. (A) EMSAs were performed using nuclear extracts from CD4+ T cells which were either in the resting state (lane 1) or activated with PMA/ionomycin for 5 or 30 min and 5.0 h (lane 2-4). Arrows identify retarded complexes putatively containing NFATs (C1, C2) (B) A nuclear extract from activated CD4+ T cells that had been activated with PMA/ionomycin for 30 min, was used in a competition experiment: a 100-fold molar excess of unlabeled P2 competitor oligonucleotide was included in the assays as indicated. P2M is a mutant of P2 with AA substituted for CC (bold, underlined). NFAT STD refers to a commercial NFAT probe (Santa Cruz). (C) The effect of CsA added together with PMA/ionomycin on the formation of the complex with probe P2. (D) Supershift assays were performed using anti-NFATc1, c2, and c3 antibodies. A nuclear extract of PMA/ionomycin-activated CD4+ T cells (for 15 min) was used along with probe P2. Arrows identify the specific supershift complexes corresponding to the NFATs (C3). (E) Recruitment of NFAT components in nuclear extracts of CD4+ T cells activated with PMA/ionomycin for 15 min, 5 h, and 20 h. Supershift assays were performed using antibodies; none for lane 1, 2, 6, and 10; anti-NFATc1 for lane 3, 7 and 11; anti-NFATc2 for lane 4, 8 and 12; anti-NFATc3 for lane 5, 9 and 13.
We also performed EMSA assays with EL-4 T cells. In contrast with the CD4+ T cells, in which the C2 complex appeared after treatment with PMA/ionomycin, nuclear extracts of EL-4 T cells contained the C2 complex in the absence of treatment with PMA/ionomycin (Figure 4A, lane 1), and the C2 complex was supershifted by the anti-NFATc2-Ab to form complex C3 (Figure 4A, lane 3). The supershift patterns of cells exposed to PMA/ionomycin for 20h were almost identical to those of cells not exposed to PMA/ionomycin (Figure 4B).
NFAT molecules bind to the TR2 gene promoter in EL-4 T cells, and TR2 promoter activity is affected by dnNFAT and siRNA. (A) EMSAs were performed using nuclear extracts from EL-4 T cells without treatment with PMA/ionomycin (lane 1), and supershift assays were performed using anti-NFATc1, c2, c3, c4, and c5 (lane 2, 3, 4, 5, and 6). Arrows identify retarded complexes putatively formed by NFATs (C1, C2, C3). (B) Recruitment of NFAT components measured in nuclear extracts of EL-4 T cells activated with PMA/ionomycin for 15 min (lane 2-5), 5 h (lane 6-9), and 20 h (lane 10-13). The letters c1, c2, c3 refer to anti-NFAT c1, c2, and c3 antibodies, respectively. (C) TR2 promoter activity was measured using the luciferase reporter plasmid pTR2(-276/-3) co-transfected into EL-4 T cells with various concentrations of dnNFAT; as a control, dnNFAT was omitted. Cells were harvested 24 h after transfection, and luciferase activity was measured with the dual luciferase assay system (Promega). It was normalized to β-galactosidase activity and is expressed as percentages of the activity with Wt in the absence of dnNFAT. (D) RNA knockdown experiments were performed. EL-4 cells were transfected by electroporation with a siRNA duplex and the TR2 promoter-driven luciferase construct. The transfection efficiency of the siRNA was measured by Western blot analysis using mAb for NFATc2 (Top). TR2 promoter activity was measured using the luciferase reporter plasmid pTR2(-276/-3) cotransfected into EL-4 T cells with NFATc2-specific siRNA; as a control, nonspecific siRNA was used (Bottom). Luciferase activity was measured as in C.
To confirm that TR-2 production is regulated by NFAT, we constructed a dominant negative NFAT mutant (dnNFAT) and measured TR2 expression using the reporter construct. The dnNFAT was derived from NFAT3 and consisted of N-terminal amino acid residues 1 to 160, which, according to Chow et al. (1999), have dominant negative activity against all four known NFATs (NFATc1, c2, c3, and c4). As shown in Figure 4C, co-transfection of EL-4 cells with the dnNFAT construct and the TR-2 reporter gene resulted in increased reporter gene activity. We also demonstrated the effect of NFAT on TR2 regulation using siRNA. The siRNA was constructed for NFATc2 because we found that only NFATc2 interacted with the TR2 promoter in EL-4 T cell (Figure 4A and 4B). As demonstrated in Figure 4D, transfection with NFATc2 siRNA increased reporter gene activity.
Discussion
While the biological importance of TR2 is clear from studies demonstrating its role in T cell activation and regulation, relatively little is known about the factors that affect its expression in T cells. In a previous study (Sul et al., 2008) we reported that the -273/-3 region of the TR2 gene contained binding sites for regulatory factors. In this report, we studied the region involved in negative regulation.
Of the NFAT family members, NFATc1 and NFATc2 are expressed predominantly in lymphocytes, and NFATc3 is also expressed in T cells (Rengarajan et al., 2002; Macian, 2005). NFATc1 is induced by signals such as TCR stimulation, while NFATc2 and NFATc3 are expressed constitutively and their expression is increased by T cell activation (Rengarajan et al., 2002). Using EMSAs, we demonstrated that NFATc2 was recruited to the TR2 promoter as soon as CD4+ T cells were activated by PMA/ionomycin. NFATc1 also became bound to the TR2 promoter as T cell activation proceeded, and binding of these factors were decreased by treatment with CsA. Thus, binding of the NFATs to the TR2 promoter site in activated T cells is associated with decreased TR2 transcription.
NFAT proteins are involved in T cell differentiation and T cell activation. However, their functions are rather controversial, and they also play negative roles. It has been reported that NFATc1 is required for TH2 cell differentiation, whereas loss of NFATc2 diminishes TH1 cell differentiation and causes a mild bias towards TH2 cell differentiation, probably because the T cells of NFATc2 deficient mice maintain IL-4 production for longer (Kiani et al., 1997; Yoshida et al., 1998; Macian, 2005). NFATc2 downregulates cyclin-dependent kinase 4 (CDK4) by directly binding and repressing its transcription through the recruitment of histone deacetylase (Baksh et al., 2002).
NFATs act synergistically with other proteins (Lim et al. 2008) such as AP-1 (Wisniewska et al., 2007), and IRF (Hu et al., 2002). Although we have reported that IRF molecules are involved in TR2 regulation (Sul et al., 2008), the interaction of NFATs with other proteins, and the precise mechanism of regulation of TR2 gene expression, require further investigated.
Although our results demonstrate that NFATs are involved in regulating TR2 expression after T cell activation, the factors regulating TR2 expression in resting T cells are still obscure. In EL-4 T cells, expression of reporter genes was upregulated in mutant forms of the pTR2(-276/-3) construct (Figure 2A and 2B) or by transfection of dnNFAT or siRNA-NFATc2 (Figure 4C and 4D) without PMA/ionomycin treatment. This may be explained by the fact that the C2 complex is formed in EL-4 T cells without PMA/ionomycin treatment, and is supershifted with anti-NFATc2 antibody. However, in resting CD4+ T cells we failed to detect any NFAT isoforms, including NFAT5, in the complexes formed in gel shift assays (data not shown).
As reported previously (Morel et al., 2000), and confirmed by us, TR2 is expressed constitutively in resting T cells and is down regulated after activation with PMA/ionomycin. We have demonstrated above that CsA, by inhibiting the nuclear translocation of NFAT proteins, increases the expression of TR2. Our results indicate that NFATs act as negative regulators of TR2 expression in activated T cells.
Methods
CD4+ T cell purification and stimulation
Cell suspensions were prepared from the spleens and lymph nodes of B57/BL6 mice, and incubated at 37℃ for 1 h in flasks to eliminate adherent cells. CD4+ T cells were then purified using a MACS magnetic separation system according to the manufacturer's instructions (Miltenyl Biotech), and LS columns were used to select the CD4+ T cells. These proved to be routinely > 95% pure by flow cytometry.
Cell culture and transfection experiments
The EL-4 T-cell line was grown in RPMI 1640 medium supplemented with 10% fetal calf serum. For transfection, aliquots of 107 cells were electroporated at 950 microfarads and 300 V using a Bio-Rad Gene Pulser in the presence of 25 µg of pGL2/basic vector or the TR2 promoter-driven luciferase constructs, together with 5 µg of the beta-galactosidase vector. Luciferase activity was determined using the Promega luciferase reporter assay and a Dynex luminometer. Data were normalized to beta-galactosidase activity. Reported values are the averages of three independent transfections, with standard deviations.
Western blot analysis
Cells were lysed and separated on 10% SDS-PAGE gels, transferred to PVDF membranes (Invitrogen), blotted with antibody specific to TR2 (9A3, Immunomics) or NFATc2 (sc-7296x, Santa Cruz), and detected with an ECL detection system (Pierce).
Preparation of reporter gene constructs, expression vectors, and dnNFAT
The genomic sequence upstream of the murine TR2 gene pTR2(-276/-3)-luc (relative to ATG at n1) was prepared as described previously (Sul et al., 2008). The dnNFAT DNA was cloned from EL-4 cells and inserted into pCDNA using primers 5'-GAGCTCATGGGGGCCGCAAGC-3' (sense) and 5'-GTCGACGACGACCTCCAGCCTCTC-'3 (antisense). The cloned domain comprises residues 1 to 160, which include the NFAT transcription activation domain (TAD) (Chow et al., 1999).
Electrophoretic mobility shift assays (EMSAs)
Nuclear extracts of 107 EL-4 T cells were subjected to EMSAs using probes labeled with [γ-32P]ATP using T4 nucleotide kinase. The reaction mixtures were loaded directly onto 6% non-denaturing polyacrylamide gels and electrophoresed at a constant voltage of 200 V at 4℃. The double stranded oligonucleotides used as probes were as follows: wild type NFAT binding site (P2), 5'-TTTCACTTTTCCTTGCCAAG-3'; mutated form of P2 (P2M), 5'-TTTCACTTAACCTTGCCAAG-3'; and control NFAT binding site in the IL-2 promoter (NFAT-STD), 5'-TATGAAACAAATTTTCCTCTTTGGGCG-3' (Santa Cruz). For supershift assays, 1 µg aliquots of monoclonal antibodies against NFATc1 (sc-7294x), NFATc2 (sc-7296x), or NFATc3 (sc-8405x) were added to binding reaction mixtures at room temperature prior to electrophoresis. The reaction mixtures were loaded directly onto a 5% non-denaturing polyacrylamide gel and electrophoresed at room temperature at a constant voltage of 70 V.
Transfection of siRNA into EL-4 cells
RNA knockdown experiments were performed using chemically synthesized and annealed small interfering RNAs (siRNA) specific for NFATc2, and negative control siRNA duplexes; both were purchased from Bioneer (1391490 and SN-1001, respectively). 107 EL-4 cells were transfected by electroporation with 200 nM siRNA duplex, 25 µg of the TR2 promoter-driven luciferase construct and 5 µg of the beta-galactosidase plasmid.
Abbreviations
- CaN:
-
calcineurin
- CsA:
-
cyclosporin A
- HVEM:
-
herpesvirus entry mediator
- LIGHT:
-
lymphotoxin homolog, which exhibits inducible expression and competes with HSV glycoprotein D for HveA and is expressed on T-lymphocytes
- NFAT:
-
nuclear factor of activated T cells
- TNFR:
-
tumor necrosis factor receptor
- TR2:
-
TNFR-related 2
References
Baksh S, Widlund HR, Frazer-Abel A, Du J, Fosmire S, Fisher DE, DeCaprio JA, Modiano JF, Burakoff SJ . NFATc2-mediated repression of cyclin-dependent kanase 4 expression . Mol Cell 2002 ; 10 : 1071 - 1081
Chow CW, Mercedes R, Roger JD . Requirement for Transcription Factor NFAT in Interleukin-2 Expression . Mol Cell Biol 1999 ; 19 : 2300 - 2307
Croft M . Co-stimulatory members of the TNFR family: keys to effective T cell immunity ? Nat Rev Immunol 2003 ; 3 : 609 - 620
Hu CM, Jang SY, Fanzo JC, Pernis AB . Modulation of T cell cytokine production by interferon regulatory factor-4 . J Biol Chem 2002 ; 277 : 49238 - 49246
Kiani A, Viola JP, Lichtman AH, Rao A . Down-regulation of IL-4 gene transcription and control of Th2 cell differentiation by a mechanism involving NFAT1 . Immunity 1997 ; 7 : 849 - 860
Krieg C, Boyman O, Fu YX, Kaye J . B and T lymphocyte attenuator regulates CD8+ T cell-intrinsic homeostasis and memory cell generation . Nat Immunol 2007 ; 8 : 162 - 171
Kwon B, Yu KY, Ni J, Yu GL, Jang IK, Kim YJ, Xing L, Liu D, Wang SX, Kwon BS . Identification of a novel activation-inducible protein of the tumor necrosis factor receptor superfamily and its ligand . J Biol Chem 1999 ; 274 : 6056 - 6061
Lim YS, Lee JS, Huang TQ, Seo JS . Protein kinase Cmu plays an essential role in hypertonicity-induced heat shock protein 70 expression . Exp Mol Med 2008 ; 40 : 596 - 606
Locksley RM, Killeen N, Lenardo MJ . The TNF and TNF receptor superfamilies: integrating mammalian biology . Cell 2001 ; 104 : 487 - 501
Macian F . NFAT proteins: key regulators of T cell development and function . Nat Rev Immunol 2005 ; 5 : 472 - 484
Morel Y, Schiano de Colella JM, Harrop J, Deen KC, Holmes SD, Wattam TA, Khandekar SS, Truneh A, Sweet RW, Gastaut JA, Olive D, Costello RT . Reciprocal expression of the TNF family receptor herpes virus entry mediator and its ligand LIGHT on activated T cells: LIGHT down-regulates its own receptor . J Immunol 2000 ; 165 : 4397 - 4404
Rengarajan J, Tang B, Glimcher LH . NFATc2 and NFATc3 regulate T(H)2 differentiation and modulate TCR-responsiveness of naive T(H) cells . Nat Immunol 2002 ; 3 : 48 - 54
Schug J, Overton GC . TESS: Transcription Element Search Software on the WWW, Technical Report CBIL-TR-1997-1001-v0.0, Computational Biology and Informatics Laboratory, School of Medicine, University of Pennsylvania, 1997 ; URL: http://www.cbil.upenn.edu/tess
Shaw KT, Ho AM, Raghavan A, Kim J, Jain J, Park J, Sharma S, Rao A, Hogan PG . Immunosuppressive drugs prevent a rapid dephosphorylation of transcription factor NFAT1 in stimulated immune cells . Proc Natl Acad Sci USA 1995 ; 92 : 11205 - 11209
Sul OJ, Lee HW, Kim WK, Choi JH, Kwak JS, Choi YJ, Latour AM, Koller BH, Jeong CS, Kwon BS . Regulation of the murine TR2/HVEM gene expression by IRF . Biochem Biophys Res Commun 2008 ; 367 : 277 - 283
Wisniewska MB, Ameyar-Zazoua M, Bakiri L, Kaminska B, Yaniv M, Weitzman JB . Dimer composition and promoter context contribute to functional cooperation between AP-1 and NFAT . J Mol Biol 2007 ; 371 : 569 - 576
Ye Q, Fraser CC, Gao W, Wang L, Busfield SJ, Wang C, Qiu Y, Coyle AJ, Gutierrez-Ramos JC, Hancock WW . Modulation of LIGHT-HVEM costimulation prolongs cardiac allograft survival . J Exp Med 2002 ; 195 : 795 - 800
Yoshida H, Nishina H, Takimoto H, Marengere LE, Wakeham AC, Bouchard D, Kong YY, Ohteki T, Shahinian A, Bachmann M, Ohashi PS, Penninger JM, Crabtree GR, Mak TW . The transcription factor NF-ATc1 regulates lymphocyte proliferation and Th2 cytokine production . Immunity 1998 ; 8 : 115 - 124
Acknowledgements
This work was supported by grants from the SRC Fund to the IRC at the University of Ulsan from the KOSEF and the Ministry of Science and Technology, Korea.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Kim, WK., Sul, OJ., Kwak, JS. et al. Nuclear factor of activated T cells negatively regulates expression of the tumor necrosis factor receptor-related 2 gene in T cells. Exp Mol Med 42, 805–810 (2010). https://doi.org/10.3858/emm.2010.42.12.083
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3858/emm.2010.42.12.083