Abstract
Fe65 has been characterized as an adaptor protein, originally identified as an expressed sequence tag (EST) corresponding to an mRNA expressed at high levels in the rat brain. It contains one WW domain and two phosphotyrosine interaction/phosphotyrosine binding domains (PID1/PID2). As the neuronal precursor cell expressed developmentally down regulated 4-2 (Nedd4-2) has a putative WW domain binding motif (72PPLP75) in the N-terminal domain, we hypothesized that Fe65 associates with Nedd4-2 through a WW domain interaction, which has the characteristics of E3 ubiquitin-protein ligase. In this paper, we present evidence for the interaction between Fe65 WW domain and Nedd4-2 through its specific motif, using a pull down approach and co-immunoprecipitation. Additionally, the co-localization of Fe65 and Nedd4-2 were observed via confocal microscopy. Co-localization of Fe65 and Nedd4-2 was disrupted by either the mutation of Fe65 WW domain or its putative binding motif of Nedd4-2. When the ubiquitin assay was performed, the interaction of Nedd4-2 (wt) with Fe65 is required for the cell apoptosis and the ubiquitylation of Fe65. We also observed that the ubiquitylation of Fe65 (wt) was augmented depending on Nedd4-2 expression levels, whereas the Fe65 WW domain mutant (W243KP245K) or the Nedd4-2 AL mutant (72PPLP75 was changed to 72APLA75) was under-ubiquitinated significantly. Thus, our observations implicated that the protein-protein interaction between the WW domain of Fe65 and the putative binding motif of Nedd4-2 down-regulates Fe65 protein stability and subcellular localization through its ubiquitylation, to contribute cell apoptosis.
Similar content being viewed by others
Introduction
Fe65 was originally identified as an expressed sequence tag (EST) corresponding to an mRNA expressed at high levels in the rat brain (Duilio et al., 1991). The sequence of the full length Fe65 cDNA revealed that it encodes a protein of 711 amino acids. An initial first comparison of this sequence with other sequenced proteins revealed that a region of Fe65 can be aligned with a region of the retroviral integrases, DNA binding proteins which catalyze the integration of the proviral DNA into the host genome (McLoughlin et al., 1998; Russo et al., 1998). It was also demonstrated that a short segment of Fe65, flanking at the N-terminal side with integrase homology, is a strong transcription activator (Chen and Sudol, 1995). These two observations led to the hypothesis that Fe65 could function as a transcription factor (Nakaya and Suzuki, 2006). In rats and humans, there are at least three genes in this family; Fe65, Fe65L1, and Fe65L2 (Duilio et al., 1998; Tanahashi and Tabira, 2002). Fe65 is a highly conserved protein: its sequences in humans, mice and rats show greater than 95% similarity, and it belongs to a multigene family (King and Scott, 2004; Guenette et al., 2006). Following the discovery of two novel protein-protein interaction domains, one WW domain and two phosphotyrosine interaction/phosphotyrosine binding domains (PID1/PID2), it was suggested that Fe65 has the characteristics of an adaptor protein (Chen and Sudol, 1995; Fiore et al., 1995; Zambrano et al., 1998). All three Fe65 variants have similar structures, in which the three protein-protein interaction domains are significantly conserved, whereas the majority of the remaining portions of the proteins are not (Russo et al., 1998). They are functionally very similar, and are able to interact with β-amyloid precursor protein (APP) through the PID2 of Fe65, as well as with other members of the APP protein family (Ermekova et al., 1998; Nakaya and Suzuki, 2006). The C-terminal PID domain of Fe65 (PID2) binds the YENPTY sequence in the cytoplasmic domain of APP. This interaction has been demonstrated to act as a cytoplasmic docking site for Fe65 (Fiore et al., 1995; Borg et al., 1996). Until now, few proteins have been identified as ligand proteins to the PID1 domain of Fe65. Fe65 interacts with LSF, Tip60 and low-density lipoprotein receptor-related protein through its PID1 domain (Ermekova et al., 1997; Sudol et al., 2001). Thus, because Fe65 functions as an adaptor protein in the cell, the identification of ligand proteins for the PID1/PID2 domain or the WW domain may provide a clue in understanding its biological roles (McLoughlin and Miller, 2008). Recently, we identified that both GSK 3β and Nek6 are ligand proteins of the Fe65 WW domain (Lee et al., 2007, 2008).
Ubiquitin is a 76 amino acid long protein that can be covalently linked to target proteins, a process referred to as ubiquitylation (Petroski, 2008; Tai and Schuman, 2008). Protein ubiquitylation is achieved by specific ubiquitin-protein ligase enzymes (E3s) after ubiquitin has been carried by E1 and E2 enzymes in cascade. The role of this posttranslational modification is to mark target proteins either for degradation or transport toward other membrane compartments (Boehmer et al., 2003). Several membrane proteins have been found to be either mono- or polyubiquitinylated (Dupré et al., 2004; Oo et al., 2007).
Neuronal precursor cell expressed developmentally down regulated 4-2 (Nedd4-2) is a protein that belongs to the Nedd4/Rsp5 family of E3-type ubiquitin ligase, which are characterized by the presence of a C-terminal HECT (homologous to E6-AP protein C-terminal) N-terminal domain (Kamynina et al., 2001; Konstas et al., 2002). These proteins are found in a wide range of species, from yeast to human, are widely expressed in mammalian tissues and cells, and are involved in a wide variety of cellular functions (Snyder, 2005; Malik et al., 2006). These functions include regulation of the cell cycle, intracellular trafficking, and surface expression of many receptors and transporters (Rotin and Schild, 2008). All members of this family share the same WW and HECT domains (Chen et al., 2001; Kamynina et al., 2001). The catalytic HECT domain in the COOH-terminus moves the ubiquitin to lysine residues on target proteins. HECT domains are followed by a number of WW domains, which are 40 amino acids in length and contain two conserved tryptophan (W) residues (Kuratomi et al., 2005) and are well-characterized binding partners for PY motifs (usually consisting of PPXY) (Rotin and Schild, 2008). Nedd4-2 and related E3 ubiquitin-protein ligases contain three to four WW domains that bind to PY motifs (Bruce et al., 2008).
A number of new Nedd4-2 targets have been recently described, including the cardiac voltage-gated Na+ channel, the glucose transporter GLUT1, the neuronal voltage-gated Na+ channel intestinal phosphate cotransporter (NaPi IIb), the voltage gated K+ channel (Kv1.3), and the glutamate transporter EAAT1 (Snrder et al., 2002; Boehmer et al., 2003; Henke et al., 2004; Palmada, 2004). They showed that the PY motif has a high affinity for WW4 of Nedd4-2. However, even though many of the other Nedd4-2-regulated transporters do not have PY motifs, the effect of Nedd4-2 may be mediated by PY-containing intermediaries (Lang et al., 2006).
Inspection of the amino-acid sequence of Nedd4-2 reveals the presence of a conserved PPLP motif which is also found in Nek6 (Lee et al., 2008), while Fe65 does not contain PY motif. Therefore, we hypothesized that Nedd4-2 (with its PPLP motif) may be involved in the regulation of Fe65 through the WW domain protein-protein interaction. To investigate this, we have examined the biochemical interaction between Fe65 and Nedd4-2, the ubiquitylation of Fe65 by Nedd4-2, and the functional consequences of Nedd4-2 activity. Consequently, we identified that Nedd4-2 which contains a WW domain binding motif (72PPLP75), is one of ubiquitin ligases for Fe65 through the association with Fe65 WW domain. Thus, the present study provides evidences that Fe65 can be a main ubiquitylation substrate by Nedd4-2 through the WW domain protein-protein interaction, and that its stability is likely regulated through the interaction between WW domain of Fe65 and its consensus binding 72PPLP75 motif in Nedd4-2.
Results
Interaction between neuronal precursor cell expressed developmentally down regulated 4-2 (Nedd4-2) and Fe65
We noticed that the WW binding motif (72PPLP75) was present in Nedd4-2 (an E3 ubiquitin ligase) (Figure 1A), which suggests that Nedd4-2 may be one of the Fe65 binding proteins (Ermekova et al., 1997; Espanel and Sudol, 1999; Sudol et al., 2001). This information led us to determine whether the WW binding motif (72PPLP75) of Nedd4-2 interacts with WW domain of Fe65. However, the PY motif, which is known as the Nedd4-2 WW domain binding motif, was not found in Fe65 (Sudol et al., 2001; Rotin and Schild, 2008). Therefore, to demonstrate that the WW binding motif (72PPLP75) of Nedd4-2 interacts with WW domain of Fe65, we constructed several Nedd4-2 or Fe65 mutants (Figure 1A) that had EGFP (for eukaryotic cell expression) or GST (for bacterial expression) tagged expression vectors cloned into them.
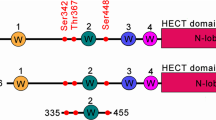
Fe65 and neuronal precursor cell expressed developmentally down regulated 4-2 (Nedd4-2) functional domain and their endogenous protein-protein interaction. The WW domain containing Fe65 functional domain (A) and the functional domain of Nedd4-2 (B) including 72PPLP75 motif are shown. (B) The Nedd4-2 immunoprecipitate contained Fe65 protein. Antibodies directed against Fe65 were also able to capture Nedd4-2 from the same lysates. (C) The Fe65 immunoprecipitate contained Nedd4-2 reciprocally. Antibodies directed against Fe65 were also able to capture Nedd4-2 from the same lysates. (D) Confocal microscopic analysis of endogenous Fe65 (green color) and Nedd4-2 (wt) (red color). Both Nedd4-2 and Fe65 are mainly detected in the nuclear region (yellow color). All figures in this article represent results obtained from five experiments repeated independently.
To confirm whether endogenous Nedd4-2 interacts with Fe65 in the cell, we investigated whether the endogenous Nedd4-2 formed a protein complex with Fe65 in HEK 293 cells via immunoprecipitation. As shown in Figure 1B, the Nedd4-2 immunoprecipitate contained Fe65. Antibodies directed against Fe65 were also able to successfully capture Nedd4-2 from the same lysates (Figure 1B, left lane). Reciprocally, to examine whether the Fe65 immunoprecipitate also contained Nedd4-2, we used antibodies directed against Fe65. As shown in Figure 1C left lane, anti-Fe65 antibodies were also able to successfully capture Nedd4-2 from the same lysates, supporting the hypothesis that the two proteins are physically associated in the cell. For the negative control, an unrelated mouse antibody was used (Figure 1B and 1C, right lane).
Next, we attempted to determine whether Nedd4-2 exists together with Fe65 in cells via confocal microscopy. The results indicated that endogenous Nedd4-2 (red) and Fe65 (green) are co-localized in the nuclear region (yellow) (Figure 1D). These findings strongly suggest that endogenous Nedd4-2 interacts with Fe65 in HEK 293 cells.
The WW domain of Fe65 is required for interaction with Nedd4-2
To determine whether Fe65 interacts with Nedd4-2 through its WW domain, we constructed several deletion mutants of Fe65 (Figure 2A). Each EGFP-Fe65 construct was transfected into a HEK 293 cell. We observed that only the EGFP Fe65 (wt), EGFP Fe65 WW mutant (W243KP245K), EGFP-Fe65 250 (which contains WW domain), or EGFP-Fe65 470 mutant (which contains WW domain) were able to bind the endogenous Nedd4-2. The EGFP-Fe65 170 mutant, which did not contain WW domain, failed to interact with the endogenous Nedd4-2 (Figure 2B, right lane), indicating that WW domain of Fe65 is the crucial factor for Nedd4-2 and Fe65 interaction. To monitor the amount of EGFP fusion protein, a western blot was performed using a Fe65 antibody (Figure 2B). Thus, these results also support that Fe65 interacts with Nedd4-2 through its WW domain, not with Nedd4-2 WW domain.
WW domain of Fe65 interacts with Nedd4-2. (A) WW domain containing Fe65 functional domain and its deletion mutants are indicated below. In contrast to the results obtained with EGFP Fe65 470 or the EGFP Fe65 250, EGFP Fe65 170 mutant failed to interact with Nedd4-2 (left lane). To monitor the EGFP fusion protein amount, western blot was performed with a Fe65 antibody. (B) The purified GST-Fe65 (wt) or GST-Fe65 WW domain mutant (W243KP245K) fusion protein was used as the bait protein to pull down Nedd4-2 from HEK 293 cell lysates as described in the Methods section. Fe65 (wt) brought down Nedd4-2 from HEK 293 cell lysates in high quantities, whereas GST-Fe65 WW domain mutant fusion protein did not. To monitor the GST fusion protein amount, western blot was performed with a GST antibody. (C,D) To determine whether GST-Fe65 (wt) was able to specifically bind to Nedd4-2, EGFP GST-Fe65 (wt) or EGFP Fe65 WW domain mutant was transfected into HEK 293 cells. Immunoprecipitation was performed with GFP antibody and western blot was performed with Fe65 antibody. In contrast to the results obtained with EGFP-Fe65 (wt), the EGFP-Fe65 WW domain mutant (W243KP245K) failed to interact with Nedd4-2 (right lane). To monitor the EGFP fusion protein amount, western blot was performed with a Fe65 antibody.
Next, we constructed Fe65 WW domain mutant (W243KP245K) and performed the pull down assay (Figure 2C) and immunoprecipitation assay (Figure 2D). As shown in Figure 2C and D, we were able to observe the protein-protein interaction between Fe65 and the endogenous Nedd4-2, while we were not able to detect the interaction between Fe65 WW domain mutant (Figure 2C and D). Thus, the protein interaction between Nedd4-2 and Fe65 is mediated by WW domain of Fe65. Furthermore, because the unphosphorylated bacterial Fe65 (wt) protein also brought down Nedd4-2 from HEH293 cell (Figure 2C), it seems that the interaction is not related with the protein phosphorylation. Therefore, the binding between Nedd4-2 and Fe65 seems to be a specific protein interaction, through its WW domain and the motif of Nedd4-2.
The 72PPLP75 motif of Nedd4-2 is required for interaction with Fe65 WW domain
As shown in Figure 1A and 3A, Nedd4-2 also contains WW domains which are able to bind specific motifs, such as PY, PPLP or S*P. Therefore, it is necessary to determine whether Nedd4-2 interacts with Fe65 WW domain through the 72PPLP75 motif of Nedd4-2. To test this hypothesis, we constructed Nedd4-2 deletion mutant, or Nedd4-2 AL mutant (72PPLP75 was changed to 72APLA75) by site directed mutagenesis, shown in Figure 3A (See the Methods section on the creation of the mutants). We transfected Nedd4-2 (wt), 121 or 299aa, mutants and observed in the pull down assay that all three constructs brought down Fe65 from HEK 293 cell lysates (Figure 3B). Because EGFP Nedd4-2 121aa (without its WW domain) also seemed to form a protein complex with Fe65, we speculated that the 72PPLP75 in the N-terminal of Nedd4-2 participated in the binding of the Fe65 WW domain. A Nedd4-2 WW domain binding motif mutant (72APLA75) was constructed (Figure 3A) and a pull down assay (Figure 3C) and an immunoprecipitation assay (Figure 3D) were performed. As shown in Figure 3C, using the purified GST Nedd4-2 (wt) or GST Nedd4-2 AL mutant fusion protein as the bait protein to pull down Fe65 from HEK 293 cell lysates, the Nedd4-2 (wt) brought down Fe65 from HEK 293 cell lysates in high quantities, whereas GST Nedd4-2 AL mutant fusion protein did not. To monitor the GST fusion protein amount, western blot was performed with a GST antibody. Thus, this result also supports our hypothesis that the motif (72PPLP75) in Nedd4-2 N-terminal participates to bind Fe65 WW domain.
PPLP motif of Nedd4-2 interacts with Fe65. (A) Nedd4-2 (wt) or deletion mutant (121 or 299 aa, as indicated above) and AL mutant (72PPLP75 was changed to 72APLA75) which was constructed by site directed mutagenesis are shown. Nedd4-2 deletion mutant or AL mutant were inserted into GST fusion (for bacteria) or EGFP fusion (for cell line) expression vector. (B) After transfection into HEK 293 with Nedd4-2 deletion mutants (121 and 299 aa) or (wt), as EGFP vector alone (for negative control) indicated above, the immunoprecipitation were performed with EGFP antibody and western blot was performed with Fe65 or Nedd4-2 antibody. The results which obtained with EGFP Nedd4-2 (wt), 121 and 299 mutants showed the interaction with Fe65, whereas EGFP vector alone did not (left lane). To monitor the EGFP fusion protein expression level, western blot was performed with a Fe65 antibody. (C) The purified GST Nedd4-2 (wt) or GST Nedd4-2 AL mutant fusion protein was used as the bait protein to pull down Fe65 from HEK 293 cell lysates as described in the Methods section. Nedd4-2 (wt) brought down Fe65 from HEK 293 cell lysates in high quantities, whereas GST Nedd4-2 AL mutant fusion protein did not. To monitor the GST fusion protein amount, western blot was performed with a GST antibody. (D) To determine whether EGFP Nedd4-2 (wt) was able to specifically bind to Fe65, EGFP Nedd4-2 (wt) or EGFP Nedd4-2 AL mutant was transfected into HEK 293 cells. Immunoprecipitation was performed with EGFP antibody and western blot was performed with Fe65 antibody. In contrast to the results obtained with EGFP Nedd4-2 (wt), the EGFP Nedd4-2 AL mutant failed to interact with Fe65 (right lane). To monitor the EGFP fusion protein amount, western blot was performed with a GFP antibody.
To further support that EGFP Nedd4-2 (wt) was specifically bound to Fe65, EGFP Nedd4-2 (wt) or EGFP Nedd4-2 AL mutant was transfected into HEK 293 cells, reciprocally. After transfection, we observed that only the EGFP Nedd4-2 (wt) was able to specifically bind to Fe65 (Figure 3C, left lane), while EGFP Nedd4-2 AL mutant failed to interact with Fe65 (Figure 3C, right lane). Thus, the result also demonstrates that the protein-protein interaction between Nedd4-2 and endogenous Fe65, were observed both in vitro and in the cell, through the motif (72PPLP75) of Nedd4-2 and WW domain of Fe65.
Confocal microscopic analysis of Fe65 and Nedd4-2
Confocal microscopic analysis of transfected EGFP-Fe65 (wt) (Figure 4A), EGFP-Fe65 WW domain mutant (Figure 4B), EGFP Nedd4-2 (wt) (Figure 4C), and EGFP Nedd4-2 AL mutant (Figure 4D) were performed, with all constructs shown as green. The transfected EGFP-Fe65 (wt), detected in both the cytoplasm and the nucleus, was merged (yellow) with Fe65 nuclear speckles around the nuclear pore (Figure 4A). The transfected EGFP-Fe65 WW domain mutant was not detected in the nucleus (Figure 4B). The interaction between Fe65 and Nedd4-2 was detected as nuclear speckles or nuclear pore. The Fe65 WW domain mutant was not merged with Fe65 around nuclear pore (Figure 4B, right lane). In a finding consistent with the endogenous Fe65 results shown in Figure 1D, we observed that exogenous EGFP-Fe65 (green) and Fe65 (red) were localized together, both in the nuclear region (yellow) (Figure 4A).
Confocal microscopic analysis of Nedd4-2 and Fe65. Confocal microscopic analysis of transfected EGFP Nedd4-2 (wt) (A), EGFP Nedd4-2 AL mutant (B) (all constructs were shown as green color). The transfected EGFP-Nedd4-2 (wt) (detected in both the cytoplasm and the nucleus) was merged (yellow) with the endogenous Fe65 nuclear speckles around the nuclear pore (A). The transfected EGFP Nedd4-2 AL mutant was not detected in nuclear speckles around nuclear pore (B). The interaction between Fe65 and Nedd4-2 were detected as nuclear region around nuclear pore. However, Nedd4-2 AL mutant was not merged with Fe65 around nuclear pore (B, right lane). Confocal microscopic analysis of transfected EGFP-Fe65 (wt) and EGFP-Fe65 WW domain mutant (W243KP245K) (C). All constructs were shown as green color. The transfected EGFP-Fe65 (wt) (detected in both the cytoplasm and the nucleus) was merged (yellow) with the endogenous Nedd4-2 nuclear speckles around the nuclear pore (C). The transfected EGFP-Fe65 WW domain mutant was not detected in nuclear speckles around nuclear pore (D). The interaction between Fe65 and Nedd4-2 were detected as nuclear speckles around nuclear pore. EGFP-Fe65 WW domain mutant was not merged with Fe65 around nuclear pore (D, right lane).
The second set of confocal microscopic analysis were performed using the EGFP Nedd4-2 (wt) (Figure 4C) and EGFP Nedd4-2 AL mutant (Figure 4D) transfected into HEK293 cells, with all constructs shown as green. The transfected EGFP-Nedd4-2 (wt), detected in both the cytoplasm and the nucleus, was merged (yellow) with Nedd4-2 nuclear speckles around the nuclear pore (Figure 4C). The transfected EGFP Nedd4-2 AL mutant was not detected in the nuclear speckles around the nuclear pore (Figure 4D). The interaction between Fe65 and Nedd4-2 were detected at the nuclear region. The Nedd4-2 AL mutant was not merged with Nedd4-2 in the nuclear region (Figure 4D, right lane).
The WW domain interaction between Fe65 and Nedd4-2 is necessary for the Fe65 ubiquitylation
In order to characterize the functional consequences of this protein-protein interaction, EGFP Nedd4-2 (wt) or EGFP Nedd4-2 AL mutant were transiently expressed in HEK 293 cells, their ubiquitylation status (Figure 5A) was compared. An anti-GFP, anti-Nedd4-2 antibody was used to monitor the Nedd4-2 protein from the precipitants via immunoblot (Figure 5A, upper lane). In the same samples, an anti-Fe65 antibody was used to determine co-immunoprecipitation of Fe65 with exogenous Nedd4-2 (Figure 5A, middle lane). Consistent with the results shown in Figure 3, we observed Fe65 in EGFP Nedd4-2 immuno-complex, but not in EGFP Nedd4-2 AL mutant (Figure 5A middle lane). To investigate the biological significance of the protein complex, a western blot with an anti-ubiquitin antibody (Ab) was used to determine whether the ubiquitylation of Fe65 was facilitated by the protein-protein interaction (Figure 5A, bottom lane). As shown in Figure 5A, the results of the western blot revealed that there was ubiquitylation of Fe65 by Nedd4-2 (wt) in the cell, while there were none in the EGFP Nedd4-2 AL mutant (Figure 5A).
The ubiquitylation of Fe65 by Nedd4-2 is required for the WW domain interaction. (A) The immunopurified EGFP Nedd4-2 (wt) or its Fe65 mutant protein with EGFP Ab from HEK293 was immunoblotted with anti-Nedd4-2, anti-Fe65 or anti-ubiquitin monoclonal antibody. EGFP Nedd4-2 AL did not form a protein complex with Fe65 nor ubiquitylation of Fe65. (B) The immunopurified EGFP Nedd4-2 (wt) or its Fe65 mutant protein with EGFP Ab from HEK293 was immunoblotted with anti-Nedd4-2, anti-Fe65 anti-ubiquitin monoclonal antibody. EGFP Fe65 WW domain mutant did not form a protein complex with Nedd4-2 nor ubiquitylation of Fe65.
Alternatively, we used the Fe65 WW domain mutant (W243KP245K) to abolish the functionality of the WW domain (Figure 5B). As shown in Figure 5B, the results of the western blot revealed that there was ubiquitylation of Fe65 by Nedd4-2 (wt) in the cell, while the EGFP Fe65 WW domain mutant (not forms a protein complex with Nedd4-2) had none (Figure 5B, middle lane).
The ubiquitylation of Fe65 depending on Nedd4-2 concentration
EGFP Nedd4-2 (wt) was transiently expressed in HEK 293 cells to further characterize the functional implications of the protein-protein interaction by comparing their ubiquitylation status. After transfection with EGFP Fe65 (wt) (2 µg) and Ha Nedd4-2 (0, 1, 2, 3 µg) into HEK 293 respectively, the immunoprecipitation was performed with anti-EGFP antibody (rabbit). Quantification of Ha Nedd4-2 and EGFP Fe65 (wt) DNA were indicated at the top of Figure 6A. The immunopurified Fe65 protein with GFP Ab from HEK 293 cells were immunoblotted with anti-ubiquitin monoclonal antibody (Figure 6A, middle) anti-Fe65 (Figure 6A, bottom).The same cell lysate was immunoblotted with anti-Ha to monitor the amount of Ha Nedd4-2 protein (Figure 6A, upper lane). An anti-Nedd4-2 antibody was used to monitor the quantity of Nedd4-2 protein from the precipitant acquired via anti-GFP (Figure 6A). As expected, the increase of Nedd4-2 augmented the ubiquitylation of Fe65, depending on its concentration.
Ubiquitylation of Fe65 dependent on Nedd4-2 concentration. (A) The immunopurified Fe65 protein with EGFP Ab from HEK 293 was immunoblotted with anti-Fe65 anti-ubiquitin monoclonal antibody. The same cell lysate was immunoblotted with anti-Ha to monitor the Ha Nedd4-2 protein amount. DNA amount of Ha Nedd4-2 (top lane) and that of EGFP Fe65 (wt) were indicated. (B) The immunopurified Nedd4-2 was used as E3 ligase for the in vitro ubiquitinizion assay. The purified GST Fe65 (wt) was ubiqutinized (the right lane), whereas GST-Fe65 W26R mutant was less ubiquitylated, suggesting that K26 residue of Fe65 is one of its major ubiquitylation sites by Nedd4-2. The relative optical density by image analysis with the Fuji Image Quant software is indicated.
Using the putative ubiquitylation site information, we expected that K26 residue of Fe65 is one of the ubiquitylation sites by Nedd4-2. As shown in Figure 6B, there was a dramatic reduction of ubiquitylation with GST Fe65 470 (K26A) protein, suggesting that the K26 residue is one of the ubiquitylation sites used by Nedd4-2 (Figure 6B, top). The relative optical density by image analysis with the Fuji Image Quant software indicated that the ubiquitylation of GST Fe65 470 (K26A) protein is reduced to 1/10 of GST Fe65 470 levels. Immunoblotting with anti-Fe65 was used to monitor the GST Fe65 protein levels (Figure 6B, bottom). The other K/R (K4R, K65R, K160R, and K274R) substitute point mutant ubiquitylation results were not shown.
Protein stability of Fe65 (wt) and its WW domain mutant
To clarify the biological significance of the protein-protein interaction between Fe65 and Nedd4-2, the EGFP-Fe65 or Fe65 WW domain mutant (W243KP245K) were transfected into HEK 293 cells. The transfected cells were treated with cyclohexamide and the protein stability was compared. The Fe65 proteins were chased for the indicated time periods. EGFP Fe65 proteins were immunoprecipitated with a polyclonal anti-GFP antibody and subjected to SDS-PAGE followed by western blotting with a monoclonal Fe65 antibody (Figure 7A). An equal amount of cell lysate was subjected to western blotting with an actin antibody to monitor the protein levels. As shown in Figure 7B, the Fe65 WW domain mutant, was more stable than Fe65 (wt), with the former not associated with Nedd4-2 and less ubiquitylated than the Nedd4-2 associated latter (Figure 5B).
Protein stability of Fe65 (wt) and its WW domain mutant. EGFP-Fe65 or Fe65 WW domain mutant was transfected into HEK293 cells and the cells were treated with cyclohexamide. The Fe65 proteins were chased for the indicated time periods. Western blotting with a monoclonal Fe65 antibody of SDS-PAGE subjected EGFP-Fe65 proteins is shown in (A). To monitor the protein amount, an equal amount of cell lysate was subjected to western blotting with an actin antibody. Results shown are one of five repeated experiments. Quantification of the pulse-chase experiment is shown in (B) by image analysis with the Fuji Image Quant software.
The effect of Fe65 WW domain mutant or Nedd4-2 AL mutant on cell viability
Cell viability was measured using FACS analysis to determine whether the Fe65 WW domain mutant or Nedd4-2 AL mutant altered cell viability. As shown in Table 1, our FACS results indicate that the Fe65 WW domain mutant or Nedd4-2 AL mutant increased the cell survival rate compared to the EGFP Nedd4-2 (wt) or EGFP vector alone. Therefore, the interaction between WW domain of Fe65 and PPLP motif of Nedd4-2 seems to be required for the increase of apoptosis in HEK 293 cell.
Discussion
In this study, we identified that neuronal precursor cell expressed developmentally down regulated 4-2 (Nedd4-2) containing a WW domain binding motifs (72PPLP75) was an ubiquitin ligase of Fe65 via the Fe65 WW domain. Using the pull down method, co-immunoprecipitation, and confocal microscopy analysis, a strong physical association was demonstrated between Nedd4-2 and Fe65 (Figure 1-4). Additionally, clear evidence that this interaction is mediated through Fe65 binding motif (72PPLP75) of Nedd4-2 with the WW domain of Fe65 (Figure 2 and 3) was presented. It was also demonstrated that the interaction between Nedd4-2 and Fe65 regulates/or requires the ubiquitylation for protein stability of Fe65 (Figure 5-7), even though it was unclear that the mutation of PPLP motif itself affects Nedd4-2 activity (Figure 4). Furthermore, the inhibition of Nedd4-2 binding to Fe65 through a mutation in the PPLP motif was sufficient to reduce its apoptotic ability (Figure 5A). Our observation also suggested that Fe65 was one of Nedd4-2 substrates for ubiquitylation (Figure 6), though the ubiquitylation sites in Fe65 remains to be characterized.
The WW domain of Fe65 binds the protein containing the consensus sequence PPLP (Ermekova et al., 1997; Espanel and Sudol, 1999; Sudol et al., 2001). To understand Fe65's function as a cellular scaffolding protein, it is necessary to identify the ligand proteins which interact with its WW, PID1, and PID2 domains (Russo et al., 1998). As the Fe65 WW domains have been shown to bind proteins containing PPLP core motifs (McLoughlin et al., 1998; Russo et al., 1998), it was also determined that the WW domain binding motif (72PPLP75) in Nedd4-2 was the same as the PPLP core motif (Fiore et al., 1995; Borg et al., 1996).
The findings of this paper suggest that the interaction of Nedd4-2 and Fe65 facilitates the ubiquitylation of Fe65 and regulates its subcellular localization (Figure 4 and 6B). However, several questions are also raised regarding the interaction of Nedd4-2 and Fe65. Though the data showed that the Fe65 association with Nedd4-2 was through the ubiquitylation of Fe65 (Figure 6B), it remains to be seen whether the difference was due to cell line differences or to differences in the function of Nedd4-2. Furthermore, though the K26 residue seems to be one of the target sites for Nedd4-2 ubiquitylation, it is still unknown that which lysine residue is the major ubiquitylation site for the regulation of Fe65 biological function. In addition, it remains to be unknown whether the ubiquitin ligase activity of Nedd4-2 to its substrate protein was actually necessary for functional intracellular interaction with Fe65, or Fe65 is one of Nedd4-2 substrate proteins (Figure 5 and 6). Moreover, characterizations of the mechanisms controlling the interactions between Nedd4-2 and Fe65 under different physiological conditions are still necessary. In addition, it remains to be determined whether the Nedd4-2 AL mutant affects the activity of ubiquitin ligase, regardless of protein-protein interactions with Fe65. Consequently, it is necessary to ascertain whether the ubiquitylation of Fe65 is necessary for activation and/or regulation of Nedd4-2.
Because Fe65 also contains PID domains at its C terminal as a scaffolding protein (Ermekova et al., 1997; Espanel and Sudol 1999; Sudol et al., 2001; Nakaya and Suzuki, 2006), the association of Nedd4-2 with Fe65 appears to determine many aspects of its biological functions. Therefore, the identification of adaptor/substrate proteins and signaling properties of Nedd4-2 provide some indications of potential therapeutic strategies for the inhibition of Nedd4-2 activity. Further characterization of the biological impact of the interaction between Nedd4-2 and Fe65 in terms of cell survival, differentiation, Wnt signaling, and cell cycle signaling is also required.
The activated Nedd4-2 mediates ubiquitylation of a variety of intracellular substrates. A number of Nedd4-2 targets have been recently described, including the cardiac voltage-gated Na+ channel, the glucose transporter GLUT1, the neuronal voltage-gated Na+ channel intestinal phosphate cotransporter NaPi IIb, the voltage gated K+ channel Kv1.3, the epithelial Na+ channel, and the Cl- channel CLC-5.9, and the glutamate transporter EAAT1(Boehmer et al. 2003; Palmada et al., 2004; Umadevi et al., 2005; Rotin and Schild, 2008). However, many other authentic substrate proteins of Nedd4-2 remain to be identified. The identification of other substrate proteins may facilitate the characterization of the biological functions of Nedd4-2 in the cell.
It may be amusing to determine that which kinds of cellular signal affect the protein-protein interaction between WW domain of Fe65 and its ligand protein. It also remains to be characterized how the interaction between Fe65 WW domain and its ligand motif (PPLP) is regulated (Ermekova et al., 1997; Nakaya and Suzuki, 2006). In spite of these observations, we can not role out the possibility that a mutation of PPLP motif of Nedd4-2 would affect its ubiquitin ligase activity directly, resulting in the interference of Fe65 binding.
Because Nedd4-2 seems to target Fe65 for degradation (Figure 7), it plays an important role under conditions when it is necessary to minimize APP metabolism (Ermekova et al., 1998; Nakaya and Suzuki et al., 2006). Furthermore, we do not know how the interaction between Fe65 and Nedd4-2 is regulated. We assume that the phosphorylation of Fe65 (which produces the motif, such as S*/T*P, for Nedd4-2 WW domain) seems to be one of the regulation mechanism (Umadevi et al., 2005; Bruce et al., 2008; Rotin and Schild, 2008). However, because the phosphorylation site is not found near the PPLP motif, the possibility that the phosphorylation is involved in the binding regulation between Fe65 and Nedd4-2 is not high. It is possible that the WW domain of Fe65 can be masked by an unknown protein which contains a PPLP motif. In contrast, Fe65 surface expression must be increased to maximize APP expression metabolism (Ermekova et al., 1998; Nakaya and Suzuki et al., 2006). Ubiquitylation of Fe65 seems to inhibit APP metabolism by blocking its binding to Fe65. Thus, presently it is unclear whether the ubiquitylation of Fe65 has any effects on APP metabolism. It seems likely that ubiquitylation might also increase Fe65 and APP endocytosis, as described for some membrane proteins (Fiore et al., 1995; Borg et al., 1996), although this hypothesis remains unproven. Additional work is required to determine whether Fe65 ubiquitylation by Nedd4-2 or other ubiquitin ligases alters the binding to Nedd4-2, and to investigate the physiological relevance of Fe65 ubiquitylation in vivo.
For Fe65, it has been demonstrated that Nedd4-2, via its WW-domains, binds to specific regions of the Fe65 subunits called PPLP motif. This interaction leads to the ubiquitylation of Fe65 subunits from the cell protein pool. Mutations to the PPLP motif of Nedd4-2 is unable to bind to Fe65, resulting in intracellular accumulation of the Fe65. However, the effect of Fe65 accumulation in the cell on the cellular function is presently unknown.
Recently, Minopoli et al. (2007) reported that Fe65 plays essential roles in the cellular response to DNA damage as a signal transduction adaptor protein. The identification of a ligand protein interacting with each Fe65 domain seems to be required to elucidate the mechanism of Fe65 as an adaptor protein in the signal transduction pathway, as Fe65 contains three domains (WW, PID1, and PID2) (McLoughlin et al., 1998; Russo et al., 1998; Espanel and Sudol, 1999; Sudol et al., 2001). In this respect, we speculate that Nedd4-2 (as one ligand protein of Fe65 WW domain) participates in Fe65 biological functions to regulate its ligand protein subcellular localization or cell survival through ubiquitylation. Furthermore, the role of Fe65 for Nedd4-2 regulation seems to be as an adaptor protein, same as Axin protein in Wnt signaling.
Interestingly, Yang et al. (2006) reported that Fe65 itself is one of the real transactivation factors, not as a scaffolding protein for Tip60 or LSF. Therefore, it seems that the interaction between Fe65 and Nedd4-2 in the cytoplasm modulates the transactivation activity of Fe65 and regulates its target gene expressions. Nedd4-2 seems to accelerate the APP ubiquitinylation via Fe65. Thus, the dissociation of Fe65 from APP or other associated proteins (Tip60 or LSF) via the ligand competition to WW domains of Fe65 at the plasma membrane, resulting in their nuclear localization, seems to compose a positive regulation loop to decrease cell survival. However, the biological significances of the direct interaction between Fe65 and Nedd4-2 are presently unknown.
In conclusion, we identified Nedd4-2 as one of Fe65 WW domain binding proteins through its 72PPLP75 motif directly. Further, our data demonstrated that the direct association Fe65 with Nedd4-2 leads to increased Fe65 ubiquitylation and its nuclear localization, leading to a reduction in the protein stability of Fe65. Taken together, our observations might shed light on the molecular mechanism(s) underlying Nedd4-2 and Fe65 regulation, activation, and signaling by the protein-protein interaction with the WW domain of Fe65. These findings also suggest that the interaction between Nedd4-2 and Fe65 may play a role on the regulation of Fe65 function which is related with Alzheimer disease.
Methods
Antibodies
The monoclonal or polyclonal antibody against GFP epitope or GST was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies against Nedd4-2 and actin were purchased from Cell Signaling Co (Boston, MA).
Cell culture
HEK 293 cells were purchased from ATCC (Manassas,VA). Media and supplements were obtained from GIBCO (Grandisland, NY). The cell line was maintained in DMEM containing 10% heat inactivated (for 30 min at 56℃) FBS, 100 U potassium penicillin/ml, 100 µg streptomycin/ml, 2 mM glutamine and 20 mM sodium bicarbonate. The cells were incubated at 5% CO2, 95% humidity, and 37℃ in a chamber. The growth medium was changed every 3 days.
Site-directed mutagenesis of Nedd4-2 and Fe65
To generate Nedd4-2 AL mutants, the sequence (up 5'-GAACTTCCTCCTGCTCCTCTGGCTCCCGGG-3', down 5'-CACCCGGGAGCCAGAGGAGCAGGAGGAAG-3') from EGFP Rat Nedd4-2 or GST Rat Nedd4-2 (purchased from GeneCopia, CA) was used with the "Chameleon" double-stranded site-directed mutagenesis kit (Stratagene, CA), according to the manufacturer's instructions. With the same mutagenesis method, Fe65 K26R mutant with the primers (up 5'-AATGCCAAGTGGTTAAGGGAAGGCCAGAACCAGCTT-3', down 5'-GTTCTGGCCTTCCCTTAACCACTTGGCATTGGCAGG-3'), or Fe65 W243KP245K mutant with the primers (up 5'-GGGACCACCCAGAAGGAAAAACCAGGCCGG-3', down 5'-GGCCCGGCCTGGTTTTTCCTTCTGGGTGGT-3') was generated. EGFP-Fe65 170 deletion mutant with the primers (up 5'-AGCAAGAGTGCTTGACTCCTATTTGGCATG-3', down 5'-AAATAGGAGTCAAGCACTCTTGCTATGTTC-3'), Fe65 250 deletion mutant with the primers (up 5'-CAGGGGAACAGTCCCTAAGAAGAGTCCCAGCTTACC-3', down 5'-CTGGGACTCTTCTTAGGGACTGTTCCCCTGTGATGG-3) or Fe65 470 deletion mutant with the primers (up 5'-GCTCGCTGCTTGGTATAAGGACTCTCCCTGGACCAC-3', down 5'-CAGGGAGAGTCCTTATACCCAGCAGCGAGCATTGCG-3') was generated from Fe65 wild type cDNA (X60469; NCBI Accession Number), which was generously provided by Dr. Margolis as a gift (Lee et al, 2007). After confirming with DNA sequencing, each mutant was inserted into GST fusion (for bacteria) or EGFP fusion (for cell line) expression vector.
Neuronal precursor cell expressed developmentally down regulated 4-2 (Nedd4-2) expression vector transfection and purification
For the mammalian expression, EGFP-Nedd4-2 (wt) or its AL mutant constructs were transfected into HEK 293 cells with the ExGene500 transfection method (Fermantas). Transfected cells (2 × 107) were lysed in RIPA lyses buffer. 1,000 µl of pre-cleaned cell lysate was incubated with anti-GFP polyclonal antibody and precipitated with protein A agarose beads. The beads were washed three times with excess cell lyses buffer. The final pellet was used to detect Fe65 by immunoblotting or to assess the ubiquitylation of Nedd4-2. Western blots also were performed to detect the ubiquitylation of Nedd4-2 using its specific mAb.
Double immunofluorescence microscopy
HEK 293 cells were plated at low confluence (~30%) on two-well Lab-Tek Permanox slides (Nalgene Nunc International, Naperville, IL) and transiently transfected with the EGFP Nedd4-2 (wt) or the EGFP Nedd4-2 AL mutant (72PPLP75 was changed to 72APLA75) plasmid using the ExGene500 procedure (Femantas, CA). Cells were starved for 36 h and subsequently treated with 10% calf serum for 15 h. At no time did cell confluency exceed 60%. Cells were fixed, permeabilized, and processed for indirect double immunofluorescence microscopy as described previously with minor modifications (Chun et al, 2004). Cells were blocked in normal goat and diluted (1:30) in PBS, for 15 min and incubated with affinity-purified, anti-antibodies at 1:150 dilution, in combination with 1:1,000 dilution of murine anti-Nedd4-2 or Fe65 monoclonal antibodies (Santa Cruz Biotech. CA), for 1-2 h at room temperature on a rocking platform. Washed slides were incubated for 1 h at room temperature with 1:150 dilutions anti-rabbit fluorescein isothiocyanate-conjugated secondary antibody (Molecular Probes Inc., Eugene, OR) and Texas red-conjugated goat anti-mouse secondary antibody (Molecular Probes Inc.). Slides were washed and mounted with Vectashield mounting medium (Vector Laboratories Inc) and examined using Zeiss Axiophot optics in The Core Facility of Chungbuk National University, similar with the method described in elsewhere (Park et al, 2007).
Expression and purification of recombinant proteins
Wild type (wt) Nedd4-2, Nedd4-2 deletion or AL mutant, Fe65 (wt), or Fe65 K26R, W243KP245K, deletion mutant protein tagged by GST were expressed in Escherichia coli BL21 and purified with GST-agarose beads, according to the manufacturer's instruction (Amersham Biosciences Co.). Purified proteins were used as a bait protein for the pull down assay.
FACS
EGFP Nedd4-2 (wt), its AL mutant, EGFP-Fe65 (wt), Fe65 WW domain mutant (W243KP245K), or EGFP vector was transfected and the rate of apoptosis was measured by FACS. Trypsinize cells in a 2 ml petri-plate with 70 µl of 1 × Trypsin. Transfected cells were washed twice in cold PBS, and added 2 ml of 70% EtOH (cold) while voltexing gently. The cells were left in EtOH at -20℃ overnight for fixing. The cells were spun at 1,500 rpm for 5 min and resuspended in 2 ml PBS, and spun again at 1,500 rpm for 5 min. After adding 500 ml FACS buffer (PBS plus PI 4 mg/ml plus RNase 30 mg/ml), the cells were incubated at 4℃ for 1 h and analyzed immediately in order to prevent clumping of the cells. FACS was performed on a Coulter Epics Elite equipped with a gated amplifier and upgraded with enhanced system performance in The Core Facility of Chungbuk National University, similar with the method described in elsewhere (Park et al, 2007).
Abbreviations
- APP:
-
amyloid precursor protein
- GLUT1:
-
glucose transporter 1
- HECT:
-
homologous to E6-AP protein C-terminal
- Nedd4-2:
-
neuronal precursor cell expressed developmentally down regulated 4-2
- PID:
-
phosphotyrosine interaction/phosphor-tyrosine binding domain
- PY motif:
-
PPXY motif
- WW domain:
-
domain with two conserved Trp residues
References
Boehmer C, Henke G, Schniepp R, Palmada M, Rothstein JD, Bröer S, Lang F . Regulation of the glutamate transporter EAAT1 by the ubiquitin ligase Nedd4-2 and the serum and glucocorticoid-inducible kinase isoforms SGK1/3 and protein kinase . J Neurochem 2003 ; 86 : 1181 - 1188
Borg JP, Ooi J, Levy E, Margolis B . The phosphotyrosine interaction domains of X11 and FE65 bind to distinct sites on the YENPTY motif of amyloid precursor protein . Mol Cell Biol 1996 ; 16 : 6229 - 6241
Bruce MC, Kanelis V, Fouladkou F, Debonneville A, Staub O, Rotin D . Regulation of Nedd4-2 self-ubiquitination and stability by a PY motif located within its HECT-domain . Biochem J 2008 ; 415 : 155 - 163
Chen H, Ross CA, Wang N, Huo Y, MacKinnon DF, Potash JB, Simposon SG, McMahon FJ, DePaulo JR, McInnis MG . NEDD4L on human chromosome 18q21 has multiple forms of transcripts and is a homologue of the mouse Nedd4-2 gene . Eur J Hum Genet 2001 ; 9 : 922 - 930
Chen HI, Sudol M . The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules . Proc Natl Acad Sci USA 1995 ; 92 : 7819 - 7823
Chun J, Kwon T, Lee EJ, Kim CH, Han YS, Hong SK, Hyun S, Kang SS . 14-3-3-Protein mediates phosphorylation ofmicrituble-associated protein tau by serum- and glucocorticoid- induced protein kinase 1 . Mol Cells 2004 ; 18 : 360 - 368
Duilio A, Zambrano N, Mogavero AR, Ammendola R, Cimino F, Russo T . A rat brain mRNA encoding a transcriptional activator homologous to the DNA binding domain of retroviral integrases . Nucleic Acids Res 1991 ; 19 : 5269 - 5274
Duilio A, Faraonio R, Minopoli G, Zambrano N, Russo T . FE65L2: a new member of the FE65 protein family interacting with the intracellular domain of the Alzheimer's beta-amyloid precursor protein . Biochem J 1998 ; 330 : 513 - 519
Dupré S, Urban-Grimal D, Haguenauer-Tsapis R . Ubiquitin and endocytic internalization in yeast and animal cells . Biochim Biophys Acta 2004 ; 1695 : 89 - 111
Ermekova KS, Zambrano N, Linn H, Minopoli G, Gertler F, Russo T, Sudol M . The WW domain of neural protein FE65 interacts with proline-rich motifs in Mena, the mammalian homolog of Drosophila enabled . J Biol Chem 1997 ; 272 : 32869 - 32877
Ermekova KS, Chang A, Zambrano N, de Candia P, Russo T, Sudol M . Proteins implicated in Alzheimer disease. The role of FE65, a new adapter which binds to beta-amyloid precursor protein . Adv Exp Med Biol 1998 ; 446 : 161 - 180
Espanel X, Sudol M . A single point mutation in a group I WW domain shifts its specificity to that of groupII WW domains . J Biol Chem 1999 ; 274 : 17284 - 17289
Fiore F, Zambrano N, Minopoli G, Donini V, Duilio A, Russo T . The regions of the FE65 protein homologous to the phosphotyrosine interaction/phosphotyrosine binding domain of Shc bind the intracellular domain of the Alzheimer's amyloid precursor protein . J Biol Chem 1995 ; 270 : 30853 - 30856
Guenette S, Chang Y, Hiesberger T, Richardson JA, Eckman CB, Eckman EA, Hammer RE, Herz J . Essential roles for the FE65 amyloid precursor protein-interacting proteins in brain development . EMBO J 2006 ; 25 : 420 - 431
Henke G, Maier G, Wallisch S, Boehmer C, Lang F . Regulation of the voltage gated K+ channel Kvl.3 by the ubiquitin ligase Nedd4-2 and the serum and glucocorticoid inducible kinase SGK1 . J Cell Physiol 2004 ; 199 : 194 - 199
Kamynina E, Debonneville C, Bens M, Vandewalle A, Staub O . A novel mouse Nedd4 protein suppresses the activity of the epithelial Na+ channel . FASEB J 2001 ; 15 : 204 - 214
King GD, Scott Turn R . Adaptor protein interactions: modulators of amyloid precursor protein metabolism and Alzheimer's disease risk ? Exp Neurol 2004 ; 185 : 208 - 219
Konstas AA, Shearwin-Whyatt LM, Fotia AB, Degger B, Riccardi D, Cook DI, Korbmacher C, Kumar S . Regulation of the epithelial sodium channel by N4WBP5A, a novel Nedd4/Nedd4-2-interacting protein . J Biol Chem 2002 ; 277 : 29406 - 29416
Kuratomi G, Komuro A, Goto K, Shinozaki M, Miyazawa K, Miyazono K, Imamura T . NEDD4-2 (neural precursor cell expressed, developmentally down-regulated 4-2) negatively regulates TGF-beta (transforming growth factor-beta) signalling by inducing ubiquitin-mediated degradation of Smad2 and TGF-beta type I receptor . Biochem J 2005 ; 386 : 461 - 470
Lang F, Böhmer C, Palmada M, Seebohm G, Strutz-Seebohm N, Vallon V . (Patho) physiological significance of the serum- and glucocorticoid-inducible kinase isoforms . Physiol Rev 2006 ; 86 : 1151 - 1178
Lee EJ, Hyun SH, Chun J, Kang SS . Human NIMA-related kinase 6 is one of the Fe65 WW domain binding proteins . Biochem Biophys Res Commun 2007 ; 358 : 783 - 788
Lee EJ, Hyun SH, Chun J, Shin SH, Kee KE, Yeon KH, Park TY, Kang SS . The PPLA motif of glycogen synthase kinase 3 beta is required for interaction with Fe65 . Mol Cells 2008 ; 26 : 100 - 105
Malik B, Price SR, Mitch WE, Yue Q, Eaton DC . Regulation of epithelial sodium channels by the ubiquitin-proteasome proteolytic pathway . Am J Physiol Renal Physiol 2006 ; 290 : 1285 - 1294
McLoughlin DM, lrving NG, Miller CC . The Fe65 and X11 families of proteins that interact with the Alzheimer's disease amyloid precursor protein . Biochem Soc Trans 1998 ; 26 : 497 - 500
McLoughlin DM, Miller CC . The FE65 proteins and Alzheimer's disease . J Neurosci Res 2008 ; 86 : 744 - 754
Minopoli G, Stante M, Napolitano F, Telese F, Aloia L, De Felice M, Di Lauro R, Pacelli R, Brunetti A, Zambrano N, Russo T . Essential roles for FE65, Alzheimer amyloid precursor-binding protein, in the cellular response to DNA damage . J Biol Chem 2007 ; 282 : 831 - 835
Nakaya T, Suzuki T . Role of APP phosphorylation in FE65-dependent gene transactivation mediated by AICD . Genes Cells 2006 ; 11 : 633 - 645
Oo ML, Thangada S, Wu MT, Liu CH, Macdonald TL, Lynch KR, Lin CY, Hla T . Immunosuppressive and anti-angiogenic sphingosine 1-phosphate receptor-1 agonists induce ubiquitinylation and proteasomal degradation of the receptor . J Biol Chem 2007 ; 282 : 9082 - 9089
Palmada M, Dieter M, Speil A, Böhmer C, Mack AF, Wagner HJ, Klingel K, Kandolf R, Murer H, Biber J, Closs EI, Lang F . Regulation of intestinal phosphate cotransporter NaPi IIb byubiquitin ligase Nedd4-2 and by serum- and glucocorticoid-dependent kinase 1 . Am J Physiol Gastrointest Liver Physiol 2004 ; 287 : G143 - G150
Park HJ, Seo H, Kim H, Kim JS, Hwang S, Seong Y . The centrosomal localization of KM-HN-1 (MGC33607) dependents on the leucine zipper motif and the C-terminal coiled-coil domain . Exp Mol Med 2007 ; 39 : 828 - 838
Petroski MD . The ubiquitin system, disease, and drug discovery . BMC Biochem 2008 ; 21 : 1 - 7
Rotin D, Schild L . ENaC and its regulatory proteins as drug targets for blood pressure control . Curr Drug Targets 2008 ; 9 : 709 - 716
Russo T, Faraonio R, Minopoli G, De Candia P, De Renzis S, Zambrano N . FE65 and the protein network centered around the cytosolic domain of the Alzheimer's beta-amyloid precursor protein . FEBS Lett 1998 ; 434 : 1 - 7
Snrder PM, Olson DR, Thomas BC . Serum and glucocorticoid-regulated kinase modulates Nedd4-2-mediated inhibition of the epithelial Na+ channel . J Biol Chem 2002 ; 277 : 5 - 8
Sudol M, Sliwa K, Russo T . Functions of PTB1 domains in the nucleus . FEBS Lett 2001 ; 490 : 190 - 195
Synder PM . Minireview: Regulation of epithelial Na+ channel trafficking . Endocrinology 2005 ; 146 : 5079 - 5085
Tai HC, Schuman EM . Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction . Nat Rev Neurosci 2008 ; 9 : 826 - 838
Tanahashi H, Tabira T . Characterization of an amyloid precursor protein-binding protein FE65L2 and its novel isoforms lacking phosphotyrosine-interaction domains . Biochem J 2002 ; 367 : 687 - 895
Umadevi N, Kumar S, Narayana N . Crystallization and preliminary X-ray diffraction studies of the WW4 domain of the Nedd4-2 ubiquitin-protein ligase . Acta Crystallogr Sect F Struct Biol Cryst Commun 2005 ; 61 : 1084 - 1086
Yang Z, Cool BH, Martin GM, Hu Q . A dominant role for FE65 (APBB1) in nuclear signaling . J Biol Chem 2006 ; 281 : 4207 - 4214
Zambrano N, Minopoli G, de Candia P, Russo T . The FE65 adaptor protein interacts through its PID1 domain with the transcription factor CP2/LSF/LBP1 . J Biol Chem 1998 ; 273 : 20128 - 20130
Acknowledgements
This work was supported by KOSEF Grant (R01-2006-000-10167-0) to SSK, and Korea Research Foundation Grant (KRF-2008-331-C00224) to J Chun. We appreciated The Core Facility of Chungbuk National University.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Lee, E., Hyun, S., Chun, J. et al. Ubiquitylation of Fe65 adaptor protein by neuronal precursor cell expressed developmentally down regulated 4-2 (Nedd4-2) via the WW domain interaction with Fe65. Exp Mol Med 41, 555–568 (2009). https://doi.org/10.3858/emm.2009.41.8.061
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3858/emm.2009.41.8.061
Keywords
This article is cited by
-
Computational Challenges in Deciphering Genomic Structures of Bacteria
Journal of Computer Science and Technology (2010)