Abstract
NEDD8 (Neural precursor cell expressed developmentally downregulated protein 8) is an ubiquitin-like protein that is covalently attached to a lysine residue of a protein substrate through a process known as neddylation, catalyzed by the enzyme cascade, namely NEDD8 activating enzyme (E1), NEDD8 conjugating enzyme (E2), and NEDD8 ligase (E3). The substrates of neddylation are categorized into cullins and non-cullin proteins. Neddylation of cullins activates CRLs (cullin RING ligases), the largest family of E3 ligases, whereas neddylation of non-cullin substrates alters their stability and activity, as well as subcellular localization. Significantly, the neddylation pathway and/or many neddylation substrates are abnormally activated or over-expressed in various human diseases, such as metabolic disorders, liver dysfunction, neurodegenerative disorders, and cancers, among others. Thus, targeting neddylation becomes an attractive strategy for the treatment of these diseases. In this review, we first provide a general introduction on the neddylation cascade, its biochemical process and regulation, and the crystal structures of neddylation enzymes in complex with cullin substrates; then discuss how neddylation governs various key biological processes via the modification of cullins and non-cullin substrates. We further review the literature data on dysregulated neddylation in several human diseases, particularly cancer, followed by an outline of current efforts in the discovery of small molecule inhibitors of neddylation as a promising therapeutic approach. Finally, few perspectives were proposed for extensive future investigations.
Similar content being viewed by others
Introduction
The protein post-translational modifications (PDM), such as phosphorylation, acetylation, methylation, glycosylation, nitrosylation, lipidation, ubiquitylation, and neddylation among others, affect many aspects of normal cell biology, and are essential for the maintenance of the homeostasis of a cell.1,2 Specifically, neddylation is an ubiquitylation-like (UBL) modifications.3 NEDD8, one of the UBLs with 59% sequence identity to ubiquitin,4 was synthesized as an 81 amino-acid precursor protein, then converted to the mature form by proteolytic cleavage to expose the C-terminal Gly 76.5,6 The neddylation leads to attachment of NEDD8 to the lysine reside of a substrate protein, catalyzed by a three-step enzymatic cascade of E1 NEDD8-activating enzyme (NAE), NEDD8-conjuagating enzyme E2 and substrate-specific NEDD8-E3 ligases.7,8 Like ubiquitin, the NEDD8 is first adenylated and activated by NAE in the presence of ATP,9 and activated NEDD8 is then transferred to UBE2M (also known as UBC12) or UBE2F, two known E2s via a trans-thiolation reaction.10,11 Finally, an NEDD8 E3 ligase, such as RING-box proteins (RBX1/2), catalyzes the transfer of NEDD8 from E2 to its substrate protein via a covalent bond12 (Fig. 1). Unlike ubiquitylation that normally conjugates ubiquitin to substrate through poly-ubiquitin chains mainly for subsequent degradation, NEDD8 is mainly conjugated to a single lysine residue on substrates leading to mono-neddylation.13 Interestingly, recent proteomics studies provided direct evidence for the existence of poly-neddylation, and the NEDD8 chains are also capable of being linked on K6, K11, K22, K27, K48 and K54 resides on NEDD8.14,15,16 The substrate proteins, upon neddylation modification, are not doomed for degradation, rather have altered properties, such as the changes in structural conformation, stability, activity/binding affinity, or subcellular localization, eventually leading to altered substrate functions.8
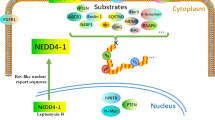
The neddylation pathway. a Three enzymes catalyze the NEDD8 attachment to a substrate: E1 activation enzyme, a heterodimer of catalytic subunit UBA3/NAEβ, and regulatory subunit NAE1/APPBP1; E2 conjugation enzymes, UBE2M/UBC12 and UBE2F; and E3 ligases (e.g., RBX1 and RBX2). One type of enzyme that removes NEDD8 from a substrate: Deneddylase (e.g CNS5). b Neddylation reaction with the UBE2M-RBX1 pair to catalyze neddylation of CULs 1-4, and UBE2M-other E3s pairs for neddylation of non-cullin substrates, and the UBE2F-RBX2 pair to catalyze CUL5 neddylation. c Classification of known non-cullin substrates
In mammalian cells, neddylation substrates are mainly classified into two categories: Cullin family members and non-cullin proteins17 (Fig. 1, Table 1). The cullin family has 8 members, including cullin-1, cullin-2, cullin-3, cullin-4A, cullin-4B, cullin-5, cullin-7, and cullin-9 with last two members less studied. Cullins are the best characterized physiological substrates of neddylation and their neddylation modification causes the conformational change to disrupt inhibitory binding of CAND1 (Cullin-associated and neddylation-dissociated 1), leading to activation of Cullin RING ligases (CRLs). By activating the CRLs, neddylation pathway controls the ubiquitylation of ~20% of cellular proteins doomed degradation via the ubiquitin-proteasome system,18 thus regulating various cellular processes and homeostasis.12,19,20,21,22
The non-cullin substrates can be further classified into seven sub-groups, including oncogenes, tumor suppressor, ribosomal proteins, histone and modifying enzymes, transcription factors and signal transducers, E3 ligases, and quite few others (Fig. 1c and Table 1). The neddylation modifications of these proteins further regulates the biological processes controlled by these substrates. However, most studies on non-cullin substrates involves the use of overexpression system, making it controversy as to whether they are indeed physiological substrates of neddylation. Furthermore, the proteomics-based approaches to identify new neddylation substrates faced major technical challenges due to low abundancy, transient/reversible nature, and contamination of ubiquitin or other ubiquitin-like proteins, among others.15,23
It has been for more than 30 years, since the first discovery of NEDD8 in 1992. Figure 2 summarized the milestone discoveries in the neddylation field, covering from target identification, validation, drug discovery to clinical trials, and clinical approval of a neddylation inhibitor for treatment of human diseases. In this review, we first introduce the components of neddylation cascade, including their crystal structures with substrates, then summarize the roles of neddylation pathway in regulation of normal cell functions and its dysregulation in several human diseases, particularly cancer, if abnormally activated. We then provide an overview on the clinical significances of targeting neddylation pathway. Finally, we summarize the current drug discovery efforts for neddylation inhibitors, and end up with few proposed future perspectives.
The chronicles of neddylation-related research. Timeline showing important discovery events in the field of neddylation research over past 30 years: From the cloning of NEDD8, demonstration of NEDD8 conjugates to cullins, identification of neddylation E1 and E2s, revelation of neddylation E2/E3 crystal structures, to discovery of neddylation E1 inhibitor (MLN4924/Pevonedistat) and its clinical trials, and its FDA approval for the treatment of MDS
Biochemical process of neddylation and deneddylation
The neddylation cascade consists of a single E1 (NAE), two E2s (UBE2F and UBE2M, also known as UBC12), and a few E3s, catalyzing a reaction to add the NEDD8 to its substrates, including cullins or non-cullin proteins to add NEDD8 to a substrate (Fig. 1). The deneddylation is a reverse reaction to remove NEDD8 from a neddylated substrate, catalyzed by an NEDD8 isopeptidase, such as COP9 signalosome complex (CSN).24 Physiologically, both neddylation and deneddylation processes are essential for the viability of most organisms,8 and deregulated neddylation or deneddylation is involved in the development of several human diseases, including neurodegenerative disorders and cancers.12,21
NEDD8
NEDD8 was first cloned in 1992 as a highly expressed gene in mouse neural precursor cells,25 and characterized in 1993 to encode a small new peptide with 81 amino acids, sharing 59% identical and 80% homologous to ubiquitin, and conservatively expressed in most eukaryote and almost all tissues.7,26 It was further defined in 1997 that newly synthesized NEDD8 with 81 amino-acid is actually a precursor, which is cleaved by C-terminal hydrolases, including UCH-L3 and NEDP1/DEN1/SENP8 into mature form of 76 amino acids with glycine 76 at its C-terminus. The mature NEDD8 is then conjugated to other proteins similar to ubiquitylation.7 In 1998, the crystal structure of NEDD8 was resolved with 3D structure similar to ubiquitin.4 Also in 1998, a new NEDD8-ligating system was established for cullin-4A, involving NEDD8 activation by NAE E1 and conjugation by UBC12 E2, eventually the attachment of NEDD8 to cullin-4A.27 And also RUB1, a yeast NEDD8, was identified to conjugate to Cullin-1/CDC53 when coupling with ULA1 and UBA3, a heterodimeric E1, and UBC12 E2 to modulate ubiquitin-proteasome system.27,28,29 While two neddylation E3s RBX1 (also known as ROC1 or Hrt1) and RBX2/SAG were cloned in 1999,30,31,32,33 SAG was first cloned as an antioxidant protein,34 then characterized as RBX1 family member with ligase activity in 2000.35 In 2002, it was found that NEDD8 not only enhances intrinsic ubiquitylation activity of CRLs upon attachment to cullins, but also prevents cullin binding with its inhibitor CAND1 to maintain fully functional CRLs.36,37
NEDD8-activating enzyme E1 (NAE)
NAE consists of catalytic subunit UBA3 (also known as NAEβ) and the regulatory subunit amyloid-beta precursor protein-binding protein 1 (APPBP1, also known as NAE1).38 NAE activates NEDD8 to initiate neddylation process by recognizing the alanine 72 in NEDD8,4,27 and continues the NEDD8 transfer cascade by activating the C-terminus of NEDD8 with diGly.9 There are two distinct active sites resided in NAE, an adenylation site located in UBA3 and a cysteine transthiolation domain located in residues involved in both subunits.9 The holo-enzyme of NAE is an “open” conformation when a large groove separates these two active sites. The third functional domain is the C-terminal ubiquitin fold domain (UFD) in UBA3, which is important for binding to NEDD8 E2 enzymes.9 The catalytic reaction by NAE is highly ordered, involving the conversion of ATP to AMP, and “open” to “close” conformations.39,40 Specifically, NEDD8 is linked to the active site cysteine of NAE via its C-terminus in the “closed” state, whereas the next NEDD8 binding to the adenylation site again will re-adopt the NAE to the “open” conformation.41
NEDD8-specific E2 conjugating enzymes
Mammalian cells have two NEDD8-specific E2 conjugating enzymes, namely, UBE2F and UBE2M (also known as UBC12), which catalyze the transthiolation reaction to transfer NEDD8 from the active cysteine site of NAE onto the active cysteine site on an E2 enzyme.10,11 The C-terminal UFD of UBA3 is responsible for E2 enzyme interaction.10 Specifically, when NAE is double-NEDD8 loaded, the UFD starts to reorient and form a cryptic E2-binding groove together with a portion of the adenylation domain.41 Afterwards, E2 directly binds to the moiety of NEDD8 attached to the NAE.41 In addition, the N-terminus of E2 binding in a groove of the adenylation domain of UBA3 appears to be the third E1-E2 interface, which is unique to the NEDD8 cascade.42 These binding interfaces create an amicable environment for catalysis by positioning the cysteine residue of NAE and E2 enzyme in the close proximity. After transthiolation, two E2 binding sites with doubly NEDD8-loaded NAE are lost, and thus the E1-E2 binding becomes weak, followed by E2 release from E1.17
NEDD8 E3 ligases
More than a dozen of neddylation E3 ligases were identified, including RBX1/2,11,17,30 murine double minute 2 (MDM2),43 ring fingers protein 111 (RNF111),44 tripartite motif (TRIM) 40,45 Smurf1,46 casitas B-lineage lymphoma (c-CBL)47 and yeast Tfb3 (MAT1 in mammal)48 among others12 (Table 2). All these E3s are dual E3s for both neddylation and ubiquitylation which are capable of transferring either NEDD8 or ubiquitin from E2s onto the substrate proteins. RBX1 and RBX2/SAG are neddylation E3s for cullin neddylation. RBX1 couples with UBE2M E2 to promote neddylation of cullins 1-4 (CUL1-4), whereas RBX2/SAG couples with UBE2F E2 for cullin-5 (CUL5) neddylation.11,49 As the dual E3s, RBX1 is the RING component of CRLs 1–4, whereas RBX2/SAG is the RING component of CRL5, required for CRL activity (Fig. 1b). The RING domain of RBX1/2 is structurally organized to bind with two zinc atoms in a cross-braced arrangement, which forms a protein–protein interaction interface for various E2s.50 And such interaction allows the catalyzation and transfer of the NEDD8 onto a lysine residue of the substrate protein. Since the catalytic domain between UBE2M and ubiquitin-specific E2s has similar structure, it is likely that NEDD8 E2 would interact with E3 RING domain similarly as ubiquitin-specific E2s binding to E3s.51,52,53,54 In fact, UBE2M was reported to serve as an ubiquitin E2 for CRL3 E3 under the physiological condition, and Parkin E3 under stressed conditions.55 However, how E3 ligases for non-cullin substrates interact with neddylation E2 (UBE2M or UBE2F) has not been systematically studies which is an interesting area for future investigation.
Neddylation chains formation and its regulation
In the most cases, NEDD8 is conjugated to a single Lys residue of a substrate to form mono-neddylation.13 However, in vitro, NEDD8 is capable of forming multiple NEDD8 chains on their substrates by the linkage via Lys6, Lys11, Lys22, Lys27, Lys48, Lys54, and Lys60.14,15,56,57,58 The poly-NEDD8 chains could be pre-formed on the catalytic cysteine residue of E2, then transferred to substrates.59 Recent proteomics studies also confirmed the existence of polyneddylated chains.14,15,16,60 However, for in vivo substrates, the neddylation occurs mainly in mono-NEDD8 form on a single or few conserved lysine residues, and the physiological significance of poly-NEDD8 chains remains elusive.58 Two recent studies showed possible role of poly-neddylated form. The first study reported that the conversion from poly-NEDD8 chains to mono-NEDD8 could act as a regulatory module for the function of HSP70 (heat shock protein 70), which is required for apoptosis induction upon DNA damage and may also be involved in tumorigenesis.61 The second study showed that the PARP-1 [poly(ADP-ribose) polymerase 1] can be poly-neddylated (tri-NEDD8) upon oxidative stress, and neddylated PARP-1 attenuates its activation to protect cells from death induced by the stress.62 Nevertheless, more extensive studies are needed to precisely characterize the physiological or pathological effect of substrate polyneddylation.
Few mechanisms have been elucidated as to how the neddylation process is being regulated. The yeast defective in cullin neddylation 1 (DCN1) protein, which bind to both NEDD8 E2 enzymes and cullins to increase neddylation efficiency, is considered as the main regulator of neddylation.63,64,65 DCN1 appears to constrain the RBX1–UBE2M ~ NEDD8 complex structurally to direct NEDD8 towards a most optimal way by recognizing the site specifically in the cullin close to its neddylation site and the acetylated N-terminus of the UBE2M.65,66 In budding yeast DCN1 is required for neddylation of CUL1 and CUL3, but not CUL4,48 suggesting that other regulators are also involved in regulating different cullins. Human cells have 5 DCN1-like proteins (DCNL), namely DCNL1–DCNL5. They all share a conserved C-terminal potentiating neddylation (PONY) domain, which is necessary and sufficient for cullin neddylation.64 DCNL1 and DCNL2 are the yeast DCN1 homologs, which interact with cullins and mainly modulate neddylation of CUL1 and CUL3.67 DCNL3 is mainly located at the plasma membrane where it also contributes to CUL3 neddylation,67 while DCNL4 and DCNL5 with a nuclear localization signal are exclusively localized in the nucleus with unknown cullin preference.68 Additionally, the neddylation process can be regulated by a protein synthesis enzyme glycyl-tRNA synthetase, which binds to NAE1 to facilitate capture and protection of activated E2 UBE2M before it reaching to a downstream substrate.69 More recently, we reported that neddylation is subject to self-regulation. Specifically, UBE2M could act as an ubiquitin E2. Under physiological condition, UBE2M couples with Cul-3, whereas under stressed conditions, including exposure to TPA (12-O-tetradecanoylphorbol-13-acetate) or hypoxia, UBE2M couples with Parkin E3 ligase to promote UBE2F ubiquitylation, therefore blocking CUL5 neddylation.55 Finally, in bacteria, NEDD8 can be directly targeted. Specifically, upon bacterial infection, the Cif (Cycle-inhibiting factor) from enteropathogenic Escherichia coli (EPEC) and its homolog from Burkholderia pseudomallei (CHBP) can convert the Gln40 residue to a Glu on NEDD8, therefore abrogating neddylation process.70,71
Deneddylation
Deneddylation is a process by which NEDD8 is removed from a conjugated substrate, catalyzed by deneddylases. The isopeptidases that deconjugate NEDD8 moieties from substrates in vitro, include Constitutive photomorphogenesis 9 (COP9) signalosome (CSN), NEDP1/DEN1, Ataxin-3, USP21, UCH-L1, and UCH-L38,72,73 (Table 2). However, only CSN and DEN1 are NEDD8 specific, the rest are also capable of deubiquitylation.8 The CSN is the major deneddylase comprising a metalloprotease active site that can cleave the NEDD8 from both the cullin and other non-cullin proteins.74,75,76 Given that genetic deletion of any subunits of CSN cause tissue damages or even lethality, CSN is critical and even indispensable for tissue homeostasis and development.77,78,79,80,81,82 CSN5, the catalytic subunit of CSN, is auto-inhibited in the form of substrate-free holoenzyme,83 Upon the interaction between the C-terminus of the cullin and the CSN subunits CSN2/CSN4, CSN5 is allosterically activated with association of CSN6, to form active deneddylase.84,85 Since the CSN tightly binds the deneddylated CRL, the RBX1 is unable to cooperate with activated NEDD8 E2 due to the steric hindrance, while the CRL substrates are capable of competing away CSN, leading to CRL activation by neddylation.86,87 Likewise, the cysteine protease DEN1/NEDP1 also specifically binds to NEDD8 and mainly deconjugate NEDD8 from non-cullin proteins,88 as well as cullin proteins.89 Therefore, DEN1 plays a complementary role in deneddylation to the CSN complex in vivo.88 Notably, DEN1 is more likely to prevent aberrant poly-neddylation via deneddylation, since it tends to deneddylate hyper-neddylated cullins without affecting mono-neddylation.89 Moreover, DEN1 appears to selectively remove the NEDD8 chains with K6-, K11- and K54-linkages, given that depletion of DEN1 causes the accumulation of these linkages of NEDD8 chains.14
Crystal structures of neddylation enzymes in complex with cullin substrates
Cullin neddylation is a complex process requiring the coordinated action of multiple components, including NEDD8, E1, E2, a multifunctional RING E3, and the cullin substrate. Subsequent to neddylation modification, the Cullin-RING complex, in conjunction with ubiquitin E1, E2, and a plethora of substrate recognition subunits and adaptor proteins, promotes the ubiquitylation of substrate proteins. This assemblage constitutes a diverse, intricate, and finely regulated machinery, involving numerous protein-protein interactions and transient or continual conformational changes. The mechanisms by which neddylation achieves timely and accurate regulation remain largely elusive. Structural biology studies offer the most direct and compelling evidence to unravel these mysteries. A comprehensive elucidation of the aforementioned structures would facilitate structure-based drug design and virtual screening for targeting neddylation enzymes, thereby accelerating the discovery process of small molecule inhibitors.90 Table 3 summarizes the published crystal structures of the major components of cullin neddylation. Below we focused on several pivotal structural studies on cullin neddylation.
While early structural biological studies have revealed basic mechanisms of substrate ubiquitylation mediated by CRL, a gap of approximately 50 amino acids was observed between the CRL-bound substrate and the active-site cysteine of a E2.91,92,93 Based on extensive biochemical assays and enzymatic reactions using purified proteins, NEDD8 was proposed to play a crucial role in closing this 50 Å gap.94 In 2008, a crystal structure of the neddylated CUL5CTD-RBX1 was resolved by the Schulman group, revealing that neddylation induces a significant change in conformation, pushing RBX1 and its bound activated E2 enzyme into a close conformation, thus bridging the 50 Å gap to bring the E2 ~ Ub near the substrate.95 Later in 2014, the same group unveiled the first co-crystal structure of a RING E3-E2 ~ NEDD8-CUL1CTD-DCN1P at 3.1 Å, providing intricate insights into mechanisms that govern NEDD8 ligation and specificity.96 This crystal structure of CUL1-RBX1 in complex with NEDD8-conjugated UBE2M revealed how cullin neddylation activates CRLs. Specifically, the UBE2M ~ NEDD8 is anchored to the RBX1 RING domain and forms a compact structure that is stabilized by a “linchpin” arginine residue unique to RBX1 (Arg46). Notably, the C terminus of NEDD8 is covalently linked to a Ser residue, replacing the catalytic Cys111 in the modified UBE2M. The oxyester-bonded UBE2M ~ NEDD8 intermediate binds to a specific linker sequence that bridges the initial beta-strand of the RBX1 protein with its RING domain. This precise interaction aligns the NEDD8 transfer module in an optimal orientation, ensuring that the catalytic site of the NEDD8 E2 enzyme is situated adjacent to the neddylation site Lys720 on CUL1. Additionally, it is positioned near the WHB subdomain, which is detailed in Fig. 3a. This model further highlights the importance of the multifunctional RING E3s with regards to the specificity in neddylation modifications.
The crystal structures of neddylation enzymes in complex with cullin substrates. a The model derived from the co-crystal structure of RBX1-UBE2M ~ NEDD8-CUL1CTD-DCN1P revealed how neddylation induces a conformational change to bring the E2 close to the substrate (modified based on the reported structure96). b The crystal structure of a neddylated CRL1β-TRCP-UBE2D~Ub-substrate intermediate revealed how NEDD8 acts as a nexus between cullin elements and the RING-activated ubiquitin-linked E2 to promote substrate ubiquitylation (PDB ID: 6TTU). c Co-crystal structure of CUL5-ARIH2 E3-E3 ubiquitin ligase unveiled the unique role of NEDD8 in promoting interactions of CUL5-RBX2 with a RING-between-RING (RBR) E3, ARIH2 (PDB ID: 7ONI). d An alignment of the reported structures of CRL1β-TRCP, SAG-CUL5, and CRL7FBXW8 (PDB ID: 6TTU, 6V9I and 7Z8B) shows the different CRL structures with the reported neddylation site. A model of CRL7 in complex with neddylated CRL1 illustrates a unique CRL-CRL interaction that enhances substrate ubiquitylation. e The Electron microscopy structure of CAND1-CUL1-RBX1-SKP1-SKP2-CKS1-CDK2 complex (PDB ID: 8OR0). f The Electron microscopy structure of NEDD8-CUL2-RBX1-ELOB-ELOC-VHL-CSN (PDB ID: 6R7F). All protein structures were illustrated by PyMol software (The PyMol Molecular Graphics System, Version 2.6.0a0 Open-Source, Schrödinger, LLC)
Subsequent studies provided a more comprehensive view of the concerted mechanism of ubiquitylation and neddylation, revealing numerous NEDD8-dependent inter-protein interactions. In 2020, a cryo-electron microscopy structure of a neddylated CRL1β-TRCP-UBE2D~Ub-substrate intermediate showed how NEDD8 acts as a nexus between cullin component and ubiquitin-loaded E2 activated by the RING to promote substrate ubiquitylation.97 Based on structural remodeling, the study showed that numerous interactions among NEDD8, the cullin and RING-bound E2~Ub intermediates for rapid substrate ubiquitylation (Fig. 3b), and the specificity of neddylation RING E3s in complex with specific cullin partners.97
Recent structural and biochemical studies have discerned significant differences between the NEDD8/CUL1 (or CUL1-4, 7, and 9) and NEDD8/CUL5 interactions,95,97,98,99 although the exact mechanism governing the specific arrangement of the two RING E3s, RBX1 and RBX2, in neddylation remains elusive. Nevertheless, a few recent structural biology studies provide some mechanistic insights. For example, co-crystal structure of CUL5-ARIH2 E3-E3 ubiquitin ligase unveiled the unique role of NEDD8 in promoting interactions of CUL5-RBX2 with a RING-between-RING (RBR) E3 ARIH2.100 In that reported structure of NEDD8/CUL5CTD/RBX2/ARIH2 complex (Fig. 3c), the surface of NEDD8 interacts differently with CUL5, as compared to CUL1. The Leu710 and Glu717 on CUL5 bind to Ile44 patch on NEDD8, while the corresponding CUL1 residues Asp and Ala, which are conserved across CUL1-4, are incompatible with the Ile44 patch on NEDD8. This results in a distinct structural arrangement, highlighting the cullin-specific regulation by NEDD8. Furthermore, the study also uncovered an allosteric mechanism as to how the unique CUL5-ARIH2 E3-E3 interaction is induced by NEDD8,100 potentially paving the road for future discovery of allosteric drugs (Fig. 3c).
Earlier structural biology studies, including mutation experiments, have revealed that a specific lysine residue on the WHB domain of cullins (K720 on CUL196 and K724 on CUL5100) serves as the neddylation site (Fig. 3d, top two panels). In a recent study, the cryo-EM and biochemical analyses were used to resolve the structure of CRL7-FBXW8 complex, which shows a distinctive mechanism of forming a specific CUL-RBX partnership.101 Specifically, the CRL7-FBXW8 complex adopts a T-shaped structure, which is characterized by CUL7–RBX1 forming a long bar that intersects the CUL7-FBXW8–SKP1 base at a significant angle. This unique arrangement is indicative of the distinct architecture of complex.101 However, despite the presence of a corresponding lysine residue on CUL7 (K1576), structural differences result in this site being fixed and buried, rendering CUL7 incapable of undergoing neddylation (Fig. 3d, bottom left panel). Moreover, contrary to CUL1-RBX1, CUL7–RBX1 in the CRL7-FBXW8 complex does not exhibit auto-ubiquitylation activity under conditions where CUL1-RBX1 complexes effectively auto-ubiquitinate. These observations align with the structural findings, revealing a restricted orientation of RBX1 RING domain within CRL7-FBXW8. The N-terminal region of RBX1 forms a β-strand within the α/β region of CUL7, creating an intermolecular cullin/RBX domain crucial for the stability of the complex.101 And this unique structure prevents the binding of the RBX1 RING domain to an active E2 ~ NEDD8 or E2~ubiquitin intermediate, leading to the lack of auto-neddylation and ubiquitylation activities in the purified CRL7-FBXW8 complex. More intriguingly, CRL7-FBXW8 could form a complex with neddylated CUL1-RBX1, presenting a special CRL-CRL partnership. This duo-CRL complex facilitates full E3 ligase activity and therefore promotes substrate ubiquitylation, even if the CRL7-FBXW8 is not neddylated101 (Fig. 3d bottom right panel). However, any other involving mechanism or whether RBX1 plays an additional role within this complex remain elusive.
While the co-crystal structure of CUL1/RBX1 and CUL5/RBX2 has both been resolved, the scenario is quite different when comes to the E2-RING partners. Surprisingly, although the co-crystal structure of the RBX1-UBC12 complex was resolved 10 years ago,96 no co-crystal structure of the RBX2-UBE2F complex was reported so far. Additional effort should be made to resolve the structure of this specific neddylation enzyme complex to elucidate the underlying mechanism related to the specificity of CUL5 neddylation, which should pave the road for structure-based design and virtual screening to discover small molecule inhibitors of the RBX2-UBE2F complex. Furthermore, the co-crystal structures of various neddylation E3 with their non-cullin substrates are also lacking, likely due to the lack of solid evidence that the pair of given E3-substrate is physiologically relevant in vivo.
CAND1 and CSN are crucial components in neddylation, either as a negative regulator by selectively binding to unneddylated CUL1-RBX1, thus blocking neddylation, or reversing the neddylation reaction via removal of NEDD8 from neddylated substrates, respectively.102
The early X-ray crystallographic as well as kinetic studies on the effect of CAND1 binding to CUL1/RBX1 or CUL4/RBX1 suggested a model where CAND1 binds to unneddylated cullins, facilitating the removal of substrate receptor (SR) modules like SKP1-SKP2.87,103 However, the detailed mechanism of how CAND1 drives the reaction of SR exchange remained elusive. Most recently, the cryo-EM structure of a substrate-bound CAND1-SCF complex, comprising CAND1-CUL1-RBX1/SKP1/SKP2/CKS1-CDK2, was resolved at 3.1 Å resolution,104 which demonstrates the dynamic interaction of CAND1 with CUL1/RBX1 and SRs (Fig. 3e). Consistently, the Schulman group published a series of cryo-EM structures of the CAND1-SCF complex, illustrating the assembly and disassembly processes of CAND1 and SRs, and these dynamic conformational changes regulate the activity of SCF/CRL1 E3 complexes.105
The CSN deneddylase consists of eight subunits (CSN1-8), with CSN5 serving as the catalytic subunit.83 A series of EM structures of CSN-CRL complexes have been released, including CSN in complex with CRL1, CRL2, CRL3 and CRL4A,85,106,107 and the cryo-EM structures of the NEDD8-CRL2-CSN108 (Fig. 3f). These structures data suggest a relatively conserved mechanism for CSN deneddylation, involving at least three steps: 1) CSN2 and CSN4 initiate the process through conformational clamping of the CRL substrate, 2) the CSN5/CSN6 heterodimer is then released, and 3) CSN5 activates the deneddylation machinery.83,85,106,107,108
Despite substantial progress has been made in understanding the functions of CAND1 and CSN in neddylation, the intricate details of the mechanism remain largely unknown. The structural versatility of CAND1, coupled with the inherent flexibility of CRL structures, suggests a wide range of possible conformations, possibly tailored to specific cullins. Furthermore, the dynamic and complex nature of CSN binding and its deneddylation process remains ambiguous. The future studies are geared toward in-depth structural and kinetic studies to elucidate the underlying mechanisms by which CAND1-mediated exchanges of CRL substrate receptors and the CSN-driven CRL deneddylation.
Neddylation in health: regulation of biochemical reactions and biological processes
By targeting numerous cullin and non-cullin proteins, neddylation pathway is able to directly or indirectly regulate multiple signaling transduction pathways involved in a variety of biological processes, including cell cycle and cell survival, cell metabolism, DNA damage responses and repair, cellular immune-responses, among others.17,109 Thus, precise regulation of neddylation pathway is crucial for the maintenance of the homeostasis of a cell to ensure a healthy operation.
The role in signaling transduction and epigenetic regulation
The NF-κB signal
Nuclear factor-kappa B (NF-κB) is a transcription factor serving as a hub for transactivation of a variety of proteins which are essential for cell survival and immune responses.110 Two pathways known to activate NF-κB are the canonical and the non-canonical pathways. In canonical pathway, NF-κB is activated upon induced ubiquitylation and degradation of the inhibitor of nuclear factor kappa B (IκB).110 Upon several stimuli, such as TNFα or LPS, IκB is phosphorylated by a heterotrimeric kinase composing of IKKα, IKKβ, and IKKγ, followed by targeted ubiquitylation and degradation by SCFβTrCP (CRL1βTrCP) E3 ligase. The removal of IκB triggers p65 nucleus translocation where NF-κB (complex of p50/p65) transactivates the expression of many genes.111 Neddylation of CUL1 activates SCFβTrCP which is required for optimal ubiquitylation and degradation of IκB.112 Another example of neddylation activation of the NFκB signal was observed through CRL5. Upon LPS exposure, NEDD8 is recruited and neddylated CUL5 to activate CRL5, which promotes polyubiquitylation of tumor necrosis factor receptor-associated factor 6 (TRAF6) via the K63 linkage to activate NF-κB and induced expression of proinflammatory cytokines in bone marrow-derived macrophages (BMDMs).113 Interestingly, NF-κB is also subjected to neddylation-mediated negative regulation. It was shown that the IKKγ is neddylated by RING E3 ligase TRIM40, leading to inhibition of kinase activity of the IKK complex, thus preventing IκB from degradation and NF-κB from activation, which appears to be an important mechanism to prevent inflammation-associated carcinogenesis in gastrointestinal epithelium.45
The NF-κB-inducing kinase (NIK) is a central signaling component of the noncanonical NF-κB pathway. In response to some stimuli, such as the ligands of TNFR superfamily members, NIK acts together with the IKKα to trigger phosphorylation-dependent processing of p100 for its proteolytic cleavage, generating the mature p52 form as the non-canonical NF-κB2.114 Importantly, NIK was recently reported as a substrate, subjecting to neddylation, which triggers its degradation.115 Hepatic-specific Nae1 deletion that inhibits its neddylation causes NIK accumulation and aberrant activation, resulting in activation of non-canonical NF-κB pathway, contributing to inflammation and hepatocyte damage.115
The HIF‐1α signal
Hypoxia-inducible factor 1 (HIF-1), a heterodimer consisting of an oxygen inducible factor (HIF-1α) and the constitutive HIF-1β (AhR nuclear translocator, ARNT), is a paramount transcription factor that upregulates a wide range of hypoxia-inducible genes, and acts as a signaling hub.116 Under normoxic conditions, HIF‐1α is hydroxylated by prolyl hydroxylase domain (PHD) proteins, then is recognized by the von Hippel-Lindau (VHL) protein for ubiquitylation and subsequent proteasomal degradation.117 VHL is a substrate-receptor subunit of CRL2, composing of CUL2, elongins B and C, and RBX1.118 Importantly, neddylation of CUL2 activates CRL2VHL to ubiquitylate HIFα.119 Interestingly, the recognition of HIFα by VHL is essential for triggering RBX1-mediated CUL2 neddylation, since HIF-1α mutation on the VHL binding residues reduces CUL2 neddylation.119,120 MLN4924, a small molecule inhibitor of neddylation E1,18 inhibits CUL2 neddylation, leading to stabilization of HIF-1α.121,122 Moreover, VHL itself can be neddylated, possibly mediated by MDM2, which precludes CRL2 complex formation to recruit HIF-1α.123,124 Interestingly, our previous study revealed that UBE2M is subjected to HIF-1 transactivation upon exposure to hypoxia, whereas MLN4924 increases UBE2M expression via inhibiting HIF‐1α degradation.55 Thus, HIF‐1α also regulates the expression of neddylation enzyme, not just as a substrate of CRL2 upon neddylation activation.
Receptor tyrosine kinases and MAPK signals
Receptor tyrosine kinases are cell surface receptors which are activated upon binding to their ligands such as growth factors, cytokines, and hormones to trigger phosphorylation and activation of the downstream proteins important for cell proliferation, differentiation and metabolism.125 Transforming growth factor β type II receptor (TGFβRII) and epidermal growth factor receptor (EGFR) are two examples of RTK signaling subjected to NEDD8 regulation. TGF-βRII has potent anti-proliferative effect in multiple types of cells, and deregulation of TGF-β signaling is highly implicated in tumorigenesis.126 The c-Cbl E3 was identified as a NEDD8 E3 ligase of TGFβRII that promotes conjugation of NEDD8 to TGFβRII at Lys556 and Lys567.47 The neddylated TGFβRII keeps itself from endocytosis to caveolin-positive compartments, leading to a prolonged stability,47 whereas deneddylation of TGFβRII by DEN1/NEDP1/SENP8, one of NEDD8 isopepitidases, results in TGFβRII ubiquitylation and degradation through lipid caveolin-mediated endocytosis.47 Finally, aberrant TβRII neddylation appears to be associated with leukemia development, since the c-Cbl neddylation-dead mutation with potential low levels of TGF-βRII neddylation is a common event in leukemia.47
EGFR receptor tyrosine kinase, upon binding to extracellular growth factors or growth hormones, is activated through dimerization to trigger its intrinsic intracellular protein-tyrosine kinase activity, followed by activation of numerous down signaling cascades in cells.127 The c-Cbl is a well-known ubiquitin E3 ligase to target EGFR for ubiquitylation, which serves as a sorting signal for endocytosis and lysosomal degradation.128 Interestingly, the c-Cbl also acts as an NEDD8 E3 to promote EGFR neddylation, contributing to subsequent EGFR ubiquitylation and endocytic internalization.129 HER2 belongs to the family of EGFR, which is phosphorylated upon binding to growth factors to activate downstream kinases for intracellular signal transduction.130 HER2 was recently reported as a neddylation substrate, and neddylation modification reduces UPS-mediated degradation, leading to its stabilization.131
The neddylation pathway was also found to be required for the activation of the RAS/ERK signal.132 Specifically, the SHC, an adaptor protein bridging the antigen receptor and the RAS/ERK signal, was reported to be a neddylation substrate, and the SHC neddylation at lysine 3 mediated by UBE2M facilitates the formation of a ZAP-SHC-GRB2 signaling complex, which modulate the downstream ERK activation and aggravation of inflammation.132 Furthermore, a recent study reported that KDM2A, a lysine demethylase, promotes the proteasomal degradation of transcription factors TCF/LEF in a manner independent of its demethylase domain. This effect is blocked by neddylation inhibitor MLN4924, suggesting that neddylation also regulates the canonical Wnt signaling.133 Taken together, neddylation regulates a number of important signal transduction pathways, highlighting its importance in cell growth and proliferation.
Epigenetic regulation
Epigenetic regulation is referred to a variety of covalent modifications of nucleic acids and histone proteins, which can heritably and reversibly modify gene expression and chromatin structure without affecting DNA sequence.134 Specifically, epigenetic regulation can occur via DNA methylation, histone modifications, chromatin remodeling, and noncoding RNA modulation,134 which play essential role in various biological processes, and dysregulation of which is associated with few human diseases.135,136,137 Notably, NEDD8-conjugated protein appears to be critical for DNA methyltransferases 3b (DNMT3b)-dependent DNA methylation, since DNMT3b interacts with the neddylated form of CUL4A preferentially, and inactivation of NAE1 inhibits DNMT3b-dependent DNA methylation.138 The neddylation-controlled ubiquitin E3 ligase CUL4DDB1 could bind to WDR5 and RBBP5, and regulate histone H3K4 methylation.139 Moreover, CUL4 is also associated with Clr4, a histone methyltransferase in S. pombe to promote lysine 9 methylation on histone H3, a critical process to heterochromatin formation.140 Histone deacetylases (HDACs) are enzymes that remove acetyl groups from histones, making a more tight wrap of histones around the DNA, thus inhibiting gene transcription.141 Neddylation pathway controls the activity of HDACs, since NEDD8 facilitated the ubiquitylation and clearance of HDAC1 and HDAC2,142,143 and inhibition of NEDDylation by MLN4924 increases protein levels of HDAC4/5,144 and significantly increases HDAC6 activity.145
The role in stress responses
DNA damage response
DNA double-strand breaks are common lesions that cause genomic instability, cell death and tumorigenesis when left unrepaired.146 In response to DNA damage, p53 is phosphorylated within the MDM2-binding motif that disrupts the p53-MDM2 binding and MDM2-mediated degradation. Increased p53 then induces cell cycle arrest to trigger DNA repair process or causes apoptosis, if the damage is too severe.147 Interestingly, p53 was subjected to neddylation modification by MDM2 as a part of cellular response to UV radiation in breast cancer cells.43 Furthermore, MDM2 can be self-neddylated in a way similar to its autoubiquitylation,43 and the neddylated MDM2 maintains its protein stability to keep p53 level in check.148 On the other hand, the NEDD8-specific isopeptidase DEN1 is significantly induced during DNA damage response, leading to MDM2 deneddylation and destabilization, and p53 activation.148
The transcription factor E2F-1 is also induced in analogous to p53 in response to DNA damage.149 While the E2F-1 activity is usually suppressed by neddylation modification, and the neddylated E2F-1 levels appear to decrease by ~80% upon etoposide or doxorubicin treatment, suggesting that E2F-1 neddylation is strictly regulated in response to DNA damage.150 DNA-dependent serine/threonine protein kinase catalytic subunit (DNA-PKcs) is a heterotrimeric protein kinase that initiates the signaling cascades in the non-homologous end-joining (NHEJ) repair upon DNA damage.151 DNA-PKcs was reported to be neddylated by the NAE-UBE2M-HUWE1 cascade, which is required for DNA-PKcs activation, since blockage of DNA-PKcs neddylation by MLN4924 or depletion of HUWE1 inhibits the autophosphorylation of DNA-PKcs and lessens the efficiency of NHEJ.152 Consistently, we previously showed that CRL1/SCFFBXW7 E3 promotes polyubiquitylation of XRCC4 at K296 via the K63 linkage to facilitate NHEJ repair process. Inactivation of SCFFBXW7 by MLN4924 blocks this process, abrogates NHEJ and renders cancer cells much more sensitive to radiation.153
In another interesting study, NEDD8 was found to be accumulated and co-localized at DNA damage sites upon DNA damage stimuli, followed by conjugation of NEDD8 chains to the N-terminal lysine residues of histone H4, catalyzed by the UBE2M-RNF111 cascade. Such polyneddylation chain induced by DNA damage is recognized by the UIM (motif interacting with ubiquitin) domain of another RING E3 ligase, RNF168 to recruit BRCA1 and 53BP1, two homologous recombination repair factors, thus amplifying the DNA damage response cascade.44 Besides, RNF168 mediates both ubiquitylation and neddylation of H2A. Upon DNA damage, H2A neddylation is inhibited, whereas its ubiquitylation is induced, which facilitates DNA damage repair.154 Finally, RNF168 itself is neddylated, which activates its E3 ligase activity.154
Interestingly, there is a balance between mono- and poly-neddylation, which is sensed by Heat Shock Protein 70 (HSP70). Upon DNA damage, NEDP1 is induced to convert unanchored poly-NEDD8 chains to mono-NEDD8 in HSP70, which activates ATPase activity of HSP70, and promotes the formation of APAF1 oligomerization to induce apoptosis.61 Furthermore, non-cullin conjugates poly-NEDD8 chains were detectable in cells, which are stabilized upon oxidative stress or when NEDP1 is depleted. Moreover, an unanchored, trimeric NEDD8 interacts with poly(ADP-ribose) polymerase 1 (PARP-1), and to attenuate its activation, thus protecting cell death induced by oxidative stress.62 Taken together, neddylation pathway appears to play a significant role in modulating DNA damage responses.
Nucleolar stress
Ribosome biogenesis is an active nucleolar process that consumes large amount of the cellular energy, thus is regulated tightly.155 Impaired ribosome assembly upon harmful stimuli could cause cell cycle arrest by activated p53, given several ribosomal proteins are capable of interacting with MDM2 to disrupt the MDM2-p53 binding.156 Interestingly, neddylation-deneddylation of ribosomal protein L11 (RPL11) regulates its subcellular localization and stability. Specifically, MDM2 acts as an NEDD8 E3 to directly neddylate RPL11, leading to its stabilization and nucleolus localization, whereas deneddylation by DEN1 results in its relocalization from nucleolus to the nucleoplasm where RPL11 is degraded by an unknown E3 ligase.157,158 Upon nucleolar stress, the neddylated RPL11 is rapidly located to the promoter sites of p53 target genes through interacting with MDM2, and consequently enhances p53 transactivation activity.159 Likewise, upon nucleolar stress the ribosomal protein S14 (RPS14) is neddylated and stabilized to induce p53 by inhibiting MDM2-mediated p53 degradation.160 In addition, MDM2 and DEN1 also mediate neddylation and de-neddylation of the ribosomal protein S27 and RPS27L, respectively, which regulate their stability and survival of cancer cells.161 More interestingly, MDM2 acts as a dual E3 acting either as ubiquitin E3 to degrade p53 or as neddylation E3 to inhibit transcriptional activity of p53.43 Similarly, the ubiquitin ligase SCFFBXO11 also promotes p53 neddylation, and suppresses its transcriptional activity.162 Taken together, ribosomal stress triggers p53 activation via modulating the MDM-RPs axis and neddylation plays a significant role in the process. Given p53 neddylation suppresses its transcriptional activity, treatment of cancer cells harboring a wt p53 with neddylation inhibitor would, in principle, reactivate p53 to induce growth arrest or apoptosis.
The role in development
NEDD8 was first isolated from the mouse brain as a possible regulator for the differentiation and development of the central nervous system.25 Interestingly, while Nedd8 knockout stain is available (https://www.informatics.jax.org/marker/MGI:97301), no study was reported in PubMed to define the phenotypes of Nedd8 total knockout, although it is expected to be embryonically lethal. In fact, our unpublished data showed that it is indeed the case; the Nedd8-null embryos die before E10.5. Likewise, the mice with total knockout of Uba3 die in utero at the stage of peri-implantation. Furthermore, Uba3-null trophoblastic cells fail to enter the S phase of the endoreduplication cycle with aberrant expression of cyclin E and p57.163
Several studies on tissue-specific knockout of neddylation E1 were conducted. Specifically, the conditional KO of Nae1 in the cortical progenitors causes premature death at the postnatal day (P) 28 with disorganized cerebral cortex, as evidence by thinner cortex and enlarged lateral ventricle, similar to the phenotypes observed in mutant mice with gain-of-function of β-catenin, as confirmed by a rescue experiment via suppressing β-catenin expression, indicating the Wnt/β-catenin signaling is subject to neddylation regulation in vivo.164 For postnatal brain development, neddylation pathway is also critical, given that mice with cKO of Nae1 in forebrain excitatory neurons in adulthood led to synaptic instability, impaired neurotransmission and cognitive deficits.165 On the other hand, a study using primary rat cultured neurons showed that the expression of deneddylating enzyme DEN1/SENP8/NEDP1 reaches the peak around the first postnatal week and gradually diminishes in mature brains and neurons. In fact, DEN1 negatively regulates neurite outgrowth and plays an important role in neuronal development via targeting a number of signaling pathways, including actin dynamics, Wnt/β-catenin, and autophagic processes.166
Neddylation pathway also regulates the heart development. Both neddylation enzymes and neddylated proteins are expressed highly in embryonic hearts, but remarkably downregulated after birth.167 Targeted deletion of Nae1 in mouse cardiomyocytes results in myocardial hypoplasia and ventricular non-compaction, and eventually causes heart failure and perinatal lethality, demonstrating its critical role during the development of cardiac chamber in embryonic hearts.167 For postnatal cardiac development, neddylation is required for proper control of metabolic transition in cardiomyocytes by regulating the conversion between glycolysis and oxidative metabolisms. Postnatal deletion of Nae1 in mice hearts causes disruption of transverse tubule formation and repression of fetal-to-adult isoform switching, leading to immature cardiomyocytes, and eventually cardiomyopathy and heart failure.168
The PubMed search did not yield any publications on the total knockout studies of neddylation E2s Ube2f or Ube2m in mice, although it is again expected to be embryonically lethal. In fact, we have attempted to generate a KI mouse model with enzymatic dead mutant (C116A) of Ube2f, but unable to obtain homozygous KI alleles. Further careful examining the KI construct revealed an error in design that fails to generate desired KI, rather a total KO mouse, suggesting that total KO of Ube2f in mice is embryonic lethal as expected (unpublished data).
For two neddylation E3s, Rbx1 and Sag/Rbx2, required for cullin neddylaiton, our earlier studies showed total knockout either caused the embryonic lethality without mutual compensations, indicating their non-redundant role.169,170 Specifically, Rbx1 total knockout caused embryonic death at the E6.5 with remarkable proliferation defects due to p27 accumulation. Simultaneous p27 total KO extends the embryo life to E9.5, indicating accumulation of other critical substrates upon Rbx1 knockout play the essential role during the late-stage development.169 The Sag/Rbx2 total knockout also causes embryonic death at E11.5–12.5 with severe defectives in vascular and nervous system. Mechanistically, some developing organs in the embryos at the stage E8.5-9.5 undergo hypoxia stress,171 which induces Sag172 to recruit Fbxw7 for ubiquitylation and degradation of neurofibromatosis type I (NF1),170 a natural occurring inhibitor of Ras, leading to the activation of MAPK signals to promote proliferation, vasculogenesis, angiogenesis and inhibit apoptosis. Sag knockout causes Nf-1 accumulation to block the Mapk signals, leading to defective proliferation, vasculogenesis, and massive apoptosis. We found that simultaneous Nf1 knockout improves the defects in vasculogenesis, but without extending the lifespan of the embryoes,170 suggesting additional substrates play essential role for the survival. Finally, Tie-Cre driven endothelial Sag deletion also caused embryonic lethality at E15.5 again with poor vasculogenesis.173
Taken together, the neddylation pathway, including its component Nedd8, and its catalyzing enzymes (neddylation E1/2/3) are all essential for development of mouse embryos as well as mouse organs, emphasizing the functional importance of this modification.
The role of immune system
Accumulated data have revealed that inhibition of neddylation pathway via genetic or pharmacological approaches effectively affects cellular immune responses and the functionality of immune cells.22 Indeed, the expression of several components of neddylation complex are markedly altered upon exposure to immune stimuli. For example, the levels of both mRNA and protein of RBX2 are increased ~10 folds in macrophage J774 cells after stimulation with lipopolysaccharides (LPS) or polyinosinic-polycytidylic acid (polyI:C).174 The neddylation of CUL1 is apparently increased in LPS-stimulated human microvascular endothelial cells.175 Moreover, Nedd8 knockout in zebrafish via CRISPR/Cas9 significantly inhibits the expression of key antiviral genes after polyI:C stimulation or SVCV (Spring Viremia of Carp Virus) infection, leading to remarkable increases in SVCV replication, indicating a critical role of neddylation in the antiviral innate immunity.176 Likewise, mice with Ube2m KO in myeloid lineage showed severe defects in innate immune response, and were notably sensitive to infection with Herpes Simplex Virus-1 (HSV-1),177 and RNA viruses including VSV (vesicular stomatitis virus) and H1N1 virus.178 Thus, neddylation pathway is critical for the immune responses, and its abnormal activation or depletion affects the proliferation and function of diverse immune cells, including neutrophils, macrophages, T cells, and Treg cells as described in more details below.179
Neddylation regulation in neutrophils
Neutrophils is one type of polymorphonuclear leukocytes that are recruited to the inflammatory site to eliminate pathogens, thus acting as the first line of defense in innate immune response.180 Neddylation is critically involved in the regulation of neutrophils. Deficiency of Cul5 attenuated alveolar neutrophil recruitment and reduced lung injury in mice receiving intratracheal instillation of LPS.113 Mice with Sag deletion at the myeloid lineage (via LyzM-Cre) are more susceptible to LPS exposure with high mortality. Mechanistically, in response to LPS, the Sag-deficient CD11b+Gr-1+-neutrophils released more TNFα with significantly decreased expression of myeloperoxidase (Mpo), a neutrophil enzyme, and Elane, a neutrophil expressed elastase.181 Neddylation blockage by MLN4924 led to suppression of neutrophil function by inhibiting the NF-κB signal to reduce the production of TNF-α, IL-6, and IL-1β induced by LPS.182 Another study reported that the in vivo treatment with MLN4924 increased neutrophil counts in blood, which could be explained by the overcompensation for early-stage anti-inflammatory effects of MLN4924 on endothelium.183 Moreover, MLN4924 treatment reduced ischemic brain injury via reducing neutrophil extravasation and maintaining the integrity of blood–brain barrier in mice.184
Neddylation regulation in macrophages
Neddylation pathway appears to regulate the survival of macrophages, since MLN4924 significantly induces the arrest cells at the G2 phase, and apoptosis in RAW264.7 macrophages.185 In addition, neddylation pathway also determines the inflammatory responses of macrophages. Depletion of NEDD8 apparently represses LPS-induced secretion of proinflammatory cytokines (e.g., TNF-α, IL-1β, CXCL1, and IL-6) from macrophages by inhibiting IκB degradation to inactivate NF-κB, thus blocking the transactivation of these cytokine expression.113,186 Similarly, RBX2/SAG knockdown induces apoptosis in macrophages, resulting from the accumulation of SAG/E3-regulated pro-apoptotic proteins (e.g., BAX, SARM) when challenged by pathogen-associated molecular patterns (PAMPs), whereas SAG overexpression in BMDMs facilitates the production of proinflammatory cytokines (IL-1β, IL-6, and TNF-α) and concordantly downregulates anti-inflammatory cytokine (IL-10).174 Moreover, SAG-deficient macrophages have impaired production of proinflammatory cytokines in response to LPS stimulation.181
A recent study revealed that Ube2m is essential to obesity-related inflammation induced by macrophages. Specifically, the mice with macrophage depletion of Ube2m have alleviated levels of insulin resistance, obesity, and hepatic steatosis induced by a high-fat diet. Mechanistically, Ube2m depletion blocks the neddylation of TRIM21, an E3 ligase, leading to reduced degradation of VHL. Increased VHL promotes HIF-1α degradation to reduce IL-1β levels.187 Paradoxically, while elevated neddylation pathway promotes the infiltration of tumor-associated macrophages and forms a tumor-promoting microenvironment by activating the NF-κB-CCL2 signal in lung cancer cells,188 neddylation inhibition also promotes M2 macrophage infiltration and worsens the inflammatory microenvironment by activating the HIF1α-CCL5 signaling in chronic pancreatitis.189 Moreover, neddylation enhancement by CSN5 deletion promotes the production of proinflammatory cytokines in macrophages under inflammatory environment by regulating the NF-κB activity, also highlighting the critical role of neddylation pathway in inflammatory responses of macrophages.183
Neddylation regulation in dendritic cells (DCs)
Neddylation pathway is essential to the survival and activity of DCs, which produce several cytokines and co-stimulatory molecules to strengthen T cell activation.22,190,191 During the Mycobacterium tuberculosis infection, CUL1 neddylation is increased in DCs, whereas NEDD8 knockdown promotes phagolysosome fusion in the infected DCs with increased ROS levels, eventually inducing autophagy and apoptosis.192 Similarly, inactivation of neddylation pathway also suppresses the survival of DCs by triggering apoptosis or necroptosis through the caspase-dependent manner.193,194 Moreover, RBX2 depletion or MLN4924 treatment remarkably inhibits the secretion of proinflammatory cytokines and co-stimulatory molecules, such as TNF-α, IL-6, IL-12p70, thus abrogating the ability of DCs to induce proliferation and activation of T cells.191,193,194 Mechanically, neddylation inhibition causes the DEPTOR accumulation by inactivating the CRL-1,170,193,195 therefore dampening the mTOR signaling to repress the functions of DCs.193,196,197 Furthermore, MLN4924 treatment or SAG knockdown also causes the IκBα accumulation through inactivation of CRL-1, thus inhibiting LPS-induced production of cytokines and blocking the functions of DCs.191
Neddylation regulation in T cells
T cell activation mediated by the engagement of T-cell antigen receptor (TCR) and co-stimulatory molecules will eventually cause proliferation, cytokine production and differentiation into T helper cells, which is essential for immune response, particularly for antitumor immune response.198,199 neddylation blockage by MLN4924 or UBE2M knockdown suppresses proliferation and cytokine production of CD4 + T cells induced by T-cell receptor/CD28 both in vitro and in vivo.132 Furthermore, UBA3 depletion compromises the production of antigen-driven cytokines (e.g., IFN-γ, IL-2 and IL-4) in of CD4 + T cells, and impairs the differentiation of follicular helper T (Tfh) cells.132,200 Although Sag deficiency has less effect on T-cell development, it apparently attenuates T-cell responses upon in vitro allogeneic stimulation and remarkably reduces graft-versus-host disease.201 Mechanically, neddylation pathway appears to directly modulate the adaptor protein Shc to facilitate the formation of a ZAP70-Shc-Grb2 complex, thus inducing the downstream Erk signal,132,202 which is essential for T-cell development.203 In CD4 + T cells, MLN4924 treatment or UBA3 knockdown profoundly impairs Erk1/2 activation, and dampens differentiation and function.132 Moreover, Bcl-2 induction is remarkably inhibited in UBA3-deficient CD4 + T cells upon TCR activation, which causes higher percentage of cell undergoing apoptosis, implying that neddylation pathway regulates the survival of CD4 + T cells through upregulating Bcl-2-mediated apoptosis resistance.200 Moreover, neddylation pathway appears to affect the homeostasis of double-negative (CD3 + CD4-CD8-, DN) T cells in a lupus-prone mouse model. Specifically, conditional knockout Ube2m in DN-T cells, or treatment with MLN4924 caused accumulation of Bim by blocking its ubiquitylation and degradation to induce apoptosis, leading to decreased DN T cell population and reduced development of murine lupus.204 Furthermore, it was reported that during P. yoelii 17XNL infection, neddylation pathway facilitates differentiation of Tfh cells by promoting the neddylation and activation of ubiquitin ligase Itch, which degrades the substrate FoxO1 protein.200,205 Thus, UBA3 knockdown greatly inhibits differentiation of Tfh cells by inactivating the ubiquitin ligase Itch.200,205 Finally, our early collaborative work showed that Sag-deficient mature naïve T cells is linked to reduced cell proliferation, mitigated cytokine secretion, as well as decreased ability to induce graft-versus-host disease in recipient mice with a mechanism involving the accumulation of SOCS1 and SOCS3 (suppressor of cytokine signaling) proteins.201
Regulatory T cells (Treg cells) are a subpopulation of CD4 + T cells which negatively regulate other type of cells in the immune system, thus playing a pivotal role in the maintenance of immunological self-tolerance. It is well-accepted that transcription factor Forkhead box P3 (Foxp3) acts as the master regulator of Treg cells.206 To determine the role of neddylation E2s or E3s in functional regulation of Treg cells, we generated the mice with T cell deficiency of either neddylation E2s, Ube2f (Foxp3Cre;Ube2ffl/fl) or Ube2m (Foxp3Cre;Ube2mfl/fl) or E3s, Rbx1 (Foxp3Cre;Rbx1fl/fl) or Sag (Foxp3Cre;Sagfl/fl).207,208,209 Mice with individual Treg KO of Ube2f or Sag are healthy and fertile, indicating the neddylation Ube2f-Sag axis is dispensable. However, mice with T cell deficiency of either Ube2m or Rbx1 develop fatal inflammatory disorder with severely impaired the immune suppressive functions, indicating that the Ube2m-Rbx1 axis is essential for the maintenance of Treg cell fitness.207,208 Double KO of either E2s or E3s showed that both E2s or E3s have functional redundancy in the functional maintenance of Treg cells.209
The role in stem cell maintenance
The neddylation-activated CRLs play critical regulatory role in the maintenance of stem cell homeostasis, self-renewal and differentiation by targeting degradation of several stemness-governing factors, such as c-MYC,210 SOX2,211 Notch.212 The F-box protein FBXW7 is elevated in embryonic stem cells (ESCs) and its proper expression is essential for proliferation and maintenance of ECSs.213,214 The SAG-CUL1-FBXW7 E3 ligase regulates the NF1-RAS pathway by targeting NF1 for ubiquitylation and degradation, therefore playing an important role in embryogenesis.170 Genetically deletion of Sag in mice causes Nf1 accumulation to suppress the Ras pathway, thus inhibiting endothelial differentiation, teratoma proliferation and angiogenesis in ESCs.170 Sag deletion also impairs the retinoic acids-induced ESCs differentiation.215
FBXW7 acts as a key regulator for the differentiation of neural stem cells by targeting Notch family members.212 Mice with brain-specific Fbxw7 deletion show morphological abnormalities of brain and die shortly after birth.212 Pathologically, the neural stem cells with Fbxw7 deletion lose neuronal differentiation, but skew the differentiation toward astrocytes due to the accumulation of Notch1 and Notch3.212 Similarly, complete deletion of Fbxw7 in the hematopoietic compartment contributes to the development of T cell acute lymphoblastic leukemia (T-ALL) by activating the Notch pathway,216,217 and increases leukemia-initiating cells (LICs) by causing the accumulation of c-MYC.218 In LICs of chronic myeloid leukemia (CML), FBXW7 is required for maintenance of quiescence of LICs by reducing the c-MYC.219 Thus, targeting FBXW7 to purge the LICs with the combination of imatinib provides an attractive therapeutic approach for CML therapy.219 Hepatic deletion of Fbxw7 results in elevated proliferation of the biliary system in mice, and facilitates the differentiation of liver stem cells to cholangiocyte rather than hepatocyte due to Notch1 accumulation in vitro.220 Furthermore, FBXW7 appears to suppress the stemness potential of cholangiocarcinoma cells by targeting mTOR for degradation.221
FBXW2, another F-box protein of CRL1, was recently found to promote ubiquitylation and degradation of MSX2 (muscle segment homeobox 2), a transcription repressor that down-regulates SOX2, leading to de-repression of SOX2 expression, and eventually promote breast cancer stem cell property.211,222,223 β-TrCP, yet another F-box protein of CRL1, promotes ubiquitylation and degradation of REST, leading to inhibition of differentiation of ESCs into neural stem cells by blocking the REST-initiated expression of neuron-specific genes.224,225 Furthermore, CRL2VHL E3 was shown to be critical for the quiescence and long-term maintenance of hematopoietic stem cells (HSCs) by promoting HIF-1α ubiquitylation and degradation.226 Biallelic deletion of VHL, which causes the HIF-1α stabilization, maintains cell cycle quiescence in HSCs, while deletion of HIF-1α otherwise causes a loss of quiescence, specifically in the HSC compartment.226,227 NANOG is an essential transcriptional factor required for the maintenance of ESCs.228 It was found that CRL3SPOP E3 promotes ubiquitylation and degradation of NANOG, thus inhibiting the self-renewal of prostate cancer stem cells.122 Likewise, the CRL4DCAF5 E3 also regulates the self-renewal and pluripotency of ESCs by promoting ubiquitylation and degradation of SOX2.229 Similarly, the CRL4ADET1-COP1 E3 also promotes SOX2 ubiquitylation for proteasomal degradation to enhance neural progenitor cell differentiation.230 The CRL5ASB4 E3 is an oxygen-sensitive ligase that is highly activated during the differentiation of ESCs into endothelial cell lineages.231 By targeting ID2 for ubiquitylation and degradation, CRL5ASB4 E3 promotes differentiation of trophoblast cells into placental vascular lineages.232 All the above pieces of evidence suggest that neddylation pathway, by controlling the activation of CRLs, indirectly regulates stem cell property. Indeed, MLN4924 treatment at nanomolar concentration remarkably stimulates in vitro tumor sphere formation and in vivo wound healing, tumorigenesis and differentiation of human cancer cells and mouse embryonic stem cells, partially through c-MYC accumulation.233 It was further demonstrated that PLGA nanoparticles coated with biomimetic macrophage membranes loaded with MLN4924 enhance the healing of diabetic wounds.234
The role in mitochondria function
Mitochondria, the power house of a cell, are essential hubs for bioenergetics, and malfunction of mitochondria is associated with several human diseases, including metabolic disorders and cancers.235 Structurally, mitochondrial organelles are highly dynamic by undergoing fusion, fission, transport and degradation, which is crucial for maintaining mitochondrial functions and cellular metabolic reprogramming.236 Disrupted mitochondrial dynamics, especially the switch towards fission status, is significantly associated with tumorigenesis236 How neddylation pathway regulates the mitochondrial structure and functions has been extensively reviewed recently.109 Our previous studies revealed that blockage of neddylation pathway by MLN4924 treatment or NAEβ knockdown altered mitochondrial morphology by triggering the fission-to-fusion conversion, resulting in reduced tricarboxylic acid cycle but increased oxidative phosphorylation. Mechanistically, MFN1, a fusion-promoting protein is a substrate of neddylation-activated E3 CRL1β-TrCP. Neddylation blockage inactivates CRL1β-TrCP, leading to accumulation of MFN1 to induce the fission-to-fusion conversion.237 The neddylation process is also highly activated in hepatic mitochondria, and neddylation of the mitochondrial electron transfer flavoproteins (ETFA and ETFB) caused their stabilization by blocking their ubiquitylation and degradation, eventually facilitating fatty acid β-oxidation.238
Reactive oxygen species (ROS) are constantly generated from oxygen during the respiration, and mitochondria are the major place for ROS generation, since it consumes ~80% of oxygen during oxidative phosphorylation.239 The homeostasis of ROS (production and scavenge) is important for proper execution of biological processes, and overproduction of ROS contributes to diverse human diseases, including cancer.240 Nuclear factor erythroid 2-related factor 2 (NRF2), an antioxidant transcription factor, is known to play a major role in maintaining the redox homeostasis by transcriptionally activating downstream genes encoding various antioxidant proteins in cellular defense against ROS.241 NRF2 is also a well characterized substrate of neddylation-activated CRL1 and CRL3 E3s.21 When degraded by CRL1 E3, NRF2 is first phosphorylated by glycogen synthase kinase 3, followed by recognized by β-TrCP for targeted ubiuqitylation and degradation. When degraded by CRL3 E3, NRF2, under the normal conditions, is recognized by Kelch-like ECH-associated protein 1 (Keap1), which recruits CRL3 for targeted ubiquitylation and degradation.21 Under the oxidative stresses, the cysteine residue on Keap1 is oxidized, which causes its conformation change to inactivate CRL3Keap1, resulting in NRF2 accumulation to induce transcriptional expression of phase II antioxidant enzymes for ROS scavenge.242,243
The role in energy metabolism
Energy metabolism is the process of generating ATP from nutrients, including mainly glucose, lipids, and proteins. Neddylation pathway has been implicated in the process of glucose metabolism. Fasting or caloric restriction caused an increased hepatic protein levels of NAE1 and NEDD8, while sugar-fed mice had reduced levels of neddylated cullins. Blockage of hepatic neddylation by MLN4924 or liver-specific knockdown of NAE1 or NEDD8 reduced gluconeogenic capacity and contributed to insulin resistance.244 Mechanistically, neddylation of phosphoenolpyruvate carboxykinase 1 (PCK1) at three lysine residues—K278, K342, and K387, increased its gluconeogenic activity by relocating two loops surrounding the catalytic center into an open configuration.244 The signaling adaptor sequestosome 1 (SQSTM1)/p62 appears to inhibit the interaction of DCNL1 with cullin2 and abolish the cullin2 neddylation to inactivate CRL2VHL E3 ligase activity, leading to HIF-1α accumulation to promote the Warburg effect by enhancing the glucose uptake and lactate production in renal cell carcinoma.245 Likewise, neddylation blockage by MLN4924 also promotes the Warburg effect by activating PKM2 via inducing its tetramerization.237,246
Glutamine metabolism is important for energy supply and redox homeostasis, the pleiotropic glutamine also contributes to the maintenance of tumorigenic state.247 Our recent study revealed that CRL3SPOP targets ASCT2, a major glutamine transporter, for ubiquitylation and degradation, whereas neddylation inhibition by MLN4924 promotes glutamine uptake by inactivating CRL3SPOP to enhance glutamine uptake. Biologically, an ASCT2 inhibitor that blocks glutamine uptake sensitizes breast cancer cells to MLN4924.248
Neddylation pathway is also actively involved in regulation of lipid metabolism. It was observed that NEDD8 is significantly upregulated in white adipose tissues from obese mice.249 The PPARγ (peroxisome proliferator-activated receptor gamma), a nuclear hormone receptor driving adipogenesis,250 is subject to MDM2-mediated neddylation, leading to its stabilization. Blocking neddylation by NEDD8 knockdown or MLN4924 treatment substantially impairs the adipocyte differentiation and fat tissue development due to downregulation of PPARγ.251,252 Sterol regulatory element-binding protein-1c (SREBP1c) is an important regulator for lipid homeostasis by inducing lipogenesis in liver.253 Aberrantly activation of SREBP1c links to progression of hepatic steatosis and eventually development of cirrhosis and liver failure.254 Furthermore, MDM2 was found to promote the conjugation of NEDD8 to SREBP1c, the neddylated SREBP1c is stabilized due to blockage of its ubiquitylation, consequently driving lipid accumulation in the liver.255 Under high glucose conditions, XIAP ligase promotes neddylation of PTEN on Lys197 and Lys402 to induce the nuclear import of PTEN, which dephosphorylated FASN, a critical enzyme in fatty acid synthesis, to inhibit its ubiquitylation and degradation by TRIM21 E3, leading to increase the de novo fatty acid synthesis and tumor progression in breast cancer.256 Taken together, neddylation pathways regulate a variety of biochemical and biological processes directly (via NEDD8 or neddylation enzymes) or indirectly (via activation of CRLs or functional modulation of non-cullin substrates) to ensure a healthy operation of a cell (Fig. 4).
Neddylation regulation of several key biological processes via NEDD8 attachment to various substrates. By attachment of NEDD8 to its cullin and non-cullin substrates, neddylation regulates various key biological processes, including signaling transduction and epigenetic regulation, cellular stress and immune responses, cell growth and organ development, stem cell maintenance, mitochondria function and energy metabolism. Created at BioRender.com
Abnormal neddylation in human diseases
The neddylation pathway is tightly regulated in order to maintain protein homeostasis and proper operation of various cellular processes. Dysregulation of neddylation or de-neddylation pathways is associated with several human diseases, including metabolic disorders,251 liver dysfunction,238 neurodegenerative disorders,165 cardiac diseases,257 immune-related diseases102 (Fig. 5), and most importantly, cancer (Fig. 6).21 Targeting neddylation is, therefore, considered as a potential therapeutic approach for the treatment of these diseases.
Metabolic diseases
Metabolic diseases are a group of medical conditions with the clinical manifestation of high glucose, low levels of HDL cholesterol, high levels of LDL and triglycerides in the blood, high blood pressure, large waist circumference or “apple-shaped” body.258 Metabolic diseases are caused by errors in metabolism, which include type 2 diabetes (T2D), hypertension, hypertriglyceridemia, atherosclerosis, and among others.259 Obesity is one of the signs of metabolic syndrome that significantly increases the risk of related metabolic diseases.260 Importantly, the levels of NEDD8, NAE1 and neddylated cullins are significantly higher in the group of people with obesity and type II diabetes.244,261 Neddylation-mediated PPARγ stabilization is crucial for adipogenesis and lipid accumulation within adipocytes, and neddylation inhibition by MLN4924 therefore effectively inhibits obesity induced by the high-fat diet.251 Our collaborative study recently showed that NEDD8-specific E2 conjugating enzyme, UBE2M, is highly associated with obesity and obesity-related metabolic diseases. Mechanically, UBE2M promotes TRIM21 neddylation to enhance TRIM21-mediated VHL ubiquitylation. As such, HIF-1α, the substrate of CRL2VHL, is accumulated to induce IL-1β production from macrophages, thus aggravating obesity-induced inflammation.187
Moreover, obesity-related inflammation, insulin resistance, and diabetes are closely related.262 Hepatic insulin resistance is highly associated with systemic insulin resistance and dyslipidemia.263,264 The nutrient and stress-sensing kinases, upon abnormal activation, phosphorylate insulin receptor substrate (IRS) for proteasome degradation, which is a key underlying mechanism for hepatic insulin resistance. MLN4924 treatment increases hepatic insulin signaling and decreases hepatic glucose production by inhibiting CRL-mediated IRS degradation, indicating that neddylation pathway indeed regulates the insulin resistance.265
Atherosclerosis is a chronic inflammatory disease characterized by infiltration of blood vessels by lipids and leukocytes, which is mostly caused by metabolic disorders.266 Deneddylase CSN5 was found to be overexpressed in human atherosclerotic lesions,267 and mice lacking myeloid Csn5 developed larger atherosclerotic lesions.183 Finally, the histone deacetylase 6 (HDAC6) is also involved in modulating atherosclerosis, and inhibition of HDAC6 neddylation significantly increases its activity to protect against endothelial dysfunction and atherosclerosis.145
Chronic liver diseases
Chronic liver disease (CLD) is a continuous process of inflammation, destruction, and regeneration of liver parenchyma, leading to development of fibrosis, cirrhosis and eventually carcinoma.268 Dysregulated neddylation pathway was reported in various pathological conditions during the CLD development. For instance, mice with liver-specific knockout of Nedd8 or Uba3 died between postnatal days 1 to 7 or 13 to 18, respectively, with spontaneous fatty liver and senescent hepatic cells.238 Furthermore, both NAE1 and global levels of neddylated proteins are elevated during the liver fibrosis.269 Although the initial triggering events that activates neddylation pathway during CLD remains elusive, diverse stress conditions, including heat shock and oxidative stress, are likely the contributing factors.15,61 Interestingly, it was reported that UBE1, an ubiquitin E1, rather than NAE1, promotes the conjugation of NEDD8 to various proteins under the stress circumstances (e.g., heat shock and oxidative stress).11,15 This E1 mediated ubiquitin-NEDD8 “cross-talk” certainly provides a remarkable amplification of the NEDD8 proteome,15,270 but its physiological and pathological implications remain unknown.
The diversified etiologies of CLD, primarily include chronic infection of hepatitis B or C viruses (HBV or HCV), chronic consumption of alcohol or drugs, chronic aberrant metabolic conditions [e.g., nonalcoholic fatty liver disease (NAFLD)], and abnormal autoimmunity.271 Chronic infection of HBV is a primary cause for CLDs, particularly for Chinese patients. HBV-encoded X protein (HBX) is a small regulatory protein that exhibits pleiotropic activities to stimulate virus gene expression and replication, thereby promoting the development of CLD272 Notably, the damage-specific DNA binding protein 1 (DDB1), an adaptor protein of CRL4 E3 ligase, is a well-characterized HBX binding partner.273,274 HBX redirects CRL4 E3 ligase activity to degrade the negative regulators of HBV transcription, including PRMT1,275 WDR77,276 and SMC5/6,277 thereby promoting HBV gene expression and replication. As such, neddylation pathway activates CRL4 to affect HBV replication indirectly. Moreover, HBX is directly neddylated by HDM2 (human homolog of MDM2), which stabilizes HBX by preventing its ubiquitylation and degradation, thus facilitating the transcription of HBV genes.278 Moreover, HBV infection induces NEDD8 expression in human liver cells, whereas NEDD8 knockdown or MLN4924 treatment inhibits HBV replication.279 Thus, neddylation pathway is actively involved in HBV-induced CLDs.
NAFLD is characterized by excessive accumulation of triglyceride in liver cells, known as steatosis, which frequently progresses to a more severe stage of nonalcoholic steatohepatitis (NASH), consisting of hepatic steatosis, inflammation, and fibrosis.280 Neddylation was reported to regulate this process. Specifically, the mitochondrial electron transfer flavoproteins (ETFA and ETFB) are stabilized by neddylation via reducing their ubiquitylation, eventually preventing fasting-induced liver steatosis.238 On the other hand, neddylation blockade inhibits the accumulation of lipid droplets, in part, via increasing mitochondrial fatty acid oxidation (FAO), which maintains liver function and attenuates pathological remodeling of liver structure, thus delaying the development of obesity and HFD-induced NAFLD.252 Furthermore, the serine-rich splicing factor 3 (SRSF3) is essential to hepatocyte differentiation, and liver glucose and lipid metabolisms.281 Genetic depletion of SRSF3 in hepatocytes causes fibrosis, steatohepatitis.282 Notably, SRSF3 protein is neddylated at lysine 11, which enhances its proteasome degradation via an undefined mechanism, thereby contributing to the progression to NASH and cirrhosis.282,283
Liver fibrosis is a common consequence of most chronic inflammatory diseases, including viral infection, alcohol, and NASH.271 Notably, altered neddylation pathway is associated with liver fibrosis, since the levels of NAE1 and neddylated cullins are increased during liver fibrosis, originating from both hepatitis B infection and alcohol abuse.269 Mechanically, neddylation activation of CRL1βTrCP ubiquitylates IκBα for proteasome degradation, resulting in p65 nuclear localization to activate NF-κB, leading to enhanced expression of inflammatory cytokines to promote the inflammation and fibrogenesis.284 Moreover, neddylation modification stabilizes TGFβ-RII to activates the TGFβ signaling, which plays an important role in activation of hepatic stellate cells (HSCs) and liver fibrosis.269,285 Finally, neddylation of EphB1 (Erythropoietin-producing human hepatocellular receptor tyrosine kinase B1) enhances its kinase activity, and significantly increases the number of HSCs, therefore contributing to liver fibrosis induced by CCl4 in mice.286 Thus, targeting neddylation pathway appears to be a potential and novel therapeutic approach for the treatment of liver fibrosis and CLD.269,287
Neurodegenerative disorders
Neurodegenerative diseases are a group of disorders characterized by progressive loss of selectively vulnerable populations of neurons, including Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease. The homeostasis of neddylation is important for the nerve growth, synapse strength, neurotransmission and synaptic plasticity, and dysregulated neddylation pathway is associated with and may be related causally to the development of neurodegenerative diseases.288 Abnormal deposition of amyloid protein Aβ and the formation of intracellular neurofibrillary tangles by tau hyperphosphorylation are the two major pathological features of AD.289 The UPS plays an essential role in the pathogenesis and progression of AD by controlling the accumulation of insoluble proteins in the brain, implying that neddylation pathway via modifying various E3 ligases, may also be involved in AD pathogenesis.290 More directly, dysregulated neddylation pathway has been found in AD. For examples, while NEDD8 is normally localized in the nucleus of normal hippocampal pyramidal cells and granule cells, it is mostly located in the cytoplasm of AD hippocampal neurons.291,292 Furthermore, APP (β-amyloid precursor protein) is the precursor of Aβ, which is generated from APP by sequential cleavages by β-secretase and γ-secretase complex. Notably, APP-binding protein 1 (APP-BP1), also known as NAE1, acting as a regulatory subunit to complex with catalytic subunit of UBA3, to form an active NEDD8-activating enzyme, which neddylates CUL1 to activate CRL1/SCF for the proper processing of APP.9,291 Finally, presenilin (PS), the constituent of γ-secretase, is the substrate of neddylation-activated CRL1, and PS ubiquitylation could alter cellular levels of unprocessed PS and increase in the production of Aβ.293
PD is another well-known neurodegenerative disease featured by the progressive degeneration of dopaminergic neurons in the substantia nigra complex with Lewy bodies presented in the lesion tissues.294 Several gene mutations, including PARKIN and PINK1, are responsible for the pathogenesis of PD.295 Interestingly, PARKIN and PINK1 are the substrates of neddylation modification, and neddylation potentiates the PARKIN E3 ligase activity and increases the stability of a 55 kDa PINK1 proteolytic fragment, which is an active PINK1 form.296 Dysregulated neddylation of PARKIN and PINK1 likely contributes to PD pathogenesis.297,298
Cardiac diseases
Heart disease is one of the leading causes of global death.299 Recently, substantial pieces of evidence supported the notion that abnormal homeostasis of cellular proteins a key mechanism for the initiation and development of cardiac diseases.257 It was reported that the levels of both neddylation enzymes and neddylated proteins are high in embryonic hearts, but significantly reduced at 1 week after birth.7,167 Transient MLN4924 treatment of neonatal rats inhibits cardiomyocyte proliferation and cardiac hypertrophy, and improves cardiac function, supporting the notion that neddylation play an important role in perinatal cardiac growth.300 Interestingly, mice lacking Nae1 show proliferation arrest of cardiomyocytes at E14.5 and pronounced ventricular non-compaction by E16.5, ultimately leading to heart failure and neonatal lethality.167 On the other hand, the neddylated proteins are significantly accumulated in failing hearts from patients with dilated and ischemic cardiomyopathy, suggesting that dysregulated neddylation is indeed involved in cardiomyopathies.257
Mice with cardiac-specific deletion of Csn8 have increased total neddylated proteins and develop cardiac hypertrophy, dilated cardiomyopathy and eventually died from heart failure by age of 4 weeks.301 Furthermore, tamoxifen-induced Csn8 knockout in the adult heart induces rapid heart failure and mice die within 2 weeks after induction.302 Thus, the CSN8-mediated deneddylation is required in the maintenance of cardiac integrity in both postnatal and adult hearts. Mechanistically, loss of CSN8 is likely to activate the RIPK1-RIPK3 pathway to induce massive cardiomyocyte necroptosis, therefore contributing to heart failure.301,303 As such, treatment of the RIP1K inhibitor or deletion of even one allele of Ripk3 inhibited the death of cardiomyocytes and prolonged the lifespan of Csn8-deficient mice.304 Thus, a precise balance between neddylation and deneddylation is the key to maintain the normal heart physiology.257
Immune-related diseases
Neddylation becomes a critical regulatory mechanism for both innate and adaptive immune system, and their associated signal pathways.305 Dysregulation of neddylation by overactivating CRLs affects the functions of immune cells by enhanced degradation of key signal proteins involved in immunoregulation, thus influencing various immune-related diseases, including viral infection, inflammation, and autoimmune diseases.305,306
By promoting neddylation of proteins involved in the host restriction factors (a group of cellular proteins that inhibit the replication of viruses), neddylation pathway has been shown to modulate the process of viral infections.307 During virus infection, the virus proteins, such as Vif, hijacks CRL5 to promote ubiquitylation and degradation of host restriction factors/anti-viral proteins APOBECs (for review, see308). The blockage of neddylation by MLN4924 represses the degradation of host restriction factors, APOBEC3G and SAMHD1, therefore restoring the restriction on human immunodeficiency virus (HIV).309 At the early phase of infection by herpes simplex virus type I (HSV-1), neddylation is required for production of type I interferon as the host innate immune response. This process was inhibited by MLN4924 or UBA3 knockdown via inactivation of NFκB through induced IκBα accumulation.310 Similarly, etoposide, a neddylation inducer, decreases lung lesions in a mouse model of SARS-CoV-2 infection by activating CRL4BPRPF19 to degrade ORF6, an accessory protein of SARS-CoV-2, which significantly inhibits the production of interferon.311 Other examples include neddylation of the polymerase basic protein 2 (PB2) of influenza A virus (IAV) by HDM2 causes its destabilization, thus blocking the replication of influenza A virus,312 and neddylation of VP2, the structural protein of enterovirus 71 (EV71) on K69 residue promotes its degradation to suppress multiplication of the virus.313
For inflammation responses triggered by a variety of stimuli, neddylation pathway also plays an essential role by regulating the production of proinflammatory cytokines mainly via controlling the CRL1/NFκB signal. Inactivation of neddylation causes accumulation of IκB to inactivate NFκB and inhibits the expression of proinflammatory cytokines (eg, TNFα and IL6), thereby alleviating LPS-induced inflammation.113,314 Similarly, neddylation immunomodulation has also been described in mucosal inflammation, MLN4924 enhances the barrier function of intestinal epithelial cells through a CRL2/HIF-1α dependent mechanism,121 with potential to alleviate inflammatory bowel diseases. Finally, neddylation regulation of other inflammatory diseases as well as several autoimmune diseases have been extensively reviewed most recently,305 highly suggesting that neddylation pathway is a promising therapeutic target for the treatment of these diseases.
Cancers
Accumulated lines of evidence have clearly demonstrated that neddylation pathway is significantly overactivated in a number of human cancers, and in most cases, its overactivation correlates with worse survival of cancer patients315 (Fig. 6).
In lung cancers, including large cell neuroendocrine lung carcinoma (LCNEC), lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC), NEDD8-related proteins are found to be dysregulated and associated with malignant stages.316 Compared to lung tissues, the mRNA expression of UBA3, NAE1 and UBE2M were significantly higher in LUAD, LUSC and LCNEC tissues, and higher UBE2M mRNA is correlated with poor differentiation in the LUAD.316,317,318 Moreover, immuno-histochemical staining and western blot analyses showed that the levels of NAE1, UBA3 UBE2M, UBE2F, RBX1/2 and DCN1 are much higher in tumor tissues than adjacent normal tissues.317,319,320,321 Furthermore, the elevated global neddylation is associated with unfavorable overall survival of lung cancer patients.316,317,322 Mechanically, overactivated neddylation pathway appears to activate CRLs to ubiquitylate and degrade tumor suppressors (e.g., p21 and p27), thus facilitating lung cancer progression.316,317 Furthermore, neddylation pathway promotes recruitment and infiltration of myeloid-derived suppressor cells (MDSCs) to lung tumor sites via the NF-κB-mCXCL5 axis, therefore contributing to tumor progression.323 Our previous study revealed that the UBE2F-SAG pair neddylates CUL5 to activate CRL5 for targeted ubiquitylation and degradation of proapototic protein NOXA, leading to enhanced survival of lung cancer cells.324 UBA3 upregulation in lung cancer cells promotes p53 neddylation to inhibit its transcriptional activity, therefore facilitating cell proliferation and invasion.318 Collectively, activation of neddylation pathway serves as an oncogenic signal.
In breast cancer, the levels of NEDD8, neddylation E1 NAE1 and UBA3, E2 UBE2M, and E3 XIAP are all significantly upregulated, compared to matched adjacent normal tissues, which positively correlated with the worse patient survival.131,256,325,326 The hyperactivated neddylation pathway facilitates HER2 neddylation to inhibit its ubiquitylation and subsequent degradation, therefore promoting the progression of HER2-positive breast cancer.131 Neddylation pathway also regulates estrogen-related receptors (ERRs) by inducing CRL1-mediated ubiquitylation and degradation of ERRs, eventually leading to breast cancer progression.327 Notably, Ran-binding protein 2 (RanBP2) was identified as a neddylation E3, which neddylates MKK7 (mitogen-activated protein kinase kinase 7) to inhibit its basal kinase activity. RanBP2 knockdown reduces proliferation and impairs EMT phenotype, which is rescued by simultaneous MKK7 knockdown in human breast cancer cells.328 The tumor suppressor PTEN is subjected to neddylation, triggered by high levels of glucose. Interestingly, neddylated PTEN accumulates predominantly in the nucleus to promote proliferation, metabolism and progression in breast cancer.256 Finally, we found that neddylation pathway controls glutamine uptake and metabolism. Mechanistically, neddylation inhibition by MLN4924 or UBA3 knockdown inactivates CRL3SPOP E3 ligase to cause accumulation of glutamine transporter ASCT2 for enhanced glutamine transports and metabolism in breast cancer cells.248
In colorectal cancer, the levels of NEDD8, NAE1 and UBE2M are all remarkably elevated in colorectal cancer tissues, as compared to matched adjacent normal tissues.46 Notably, a C2-WW-HECT ligase, Smurf1, is positively correlated with NEDD8, NAE1 and UBE2M. Mechanistic study revealed that Smurf1 acts as a NEDD8 ligase to promote its auto-neddylation when coupling with NEDD8 and UBE2M, which in turn enhances Smurf1-mediated ubiquitylation of Smad4, Smad5, RhoA or ING2, leading to accelerated colorectal cancer progression.46 Furthermore, Smurf1, acting as neddylation E3, promotes RRP9 (ribosomal RNA processing 9, U3 small nucleolar RNA binding protein) neddylation to induce pre-rRNA processing, also accelerating the proliferation of colon cancer cells.329 Oncoprotein Hu antigen R (HuR) is subjected to neddylation by MDM2, leading to its stabilization. High levels of HuR correlates with tumor malignancy and colon cancer metastases.330 Finally, SHP2 (Src homology region 2–containing protein tyrosine phosphatase 2) is also subjected to neddylation, leading to its inactivation through a conformation change, which regulates macrophage-mediated engulfment of opsonized colorectal tumor cells.331
In human intrahepatic cholangiocarcinoma, the levels of NEDD8, NAE1, UBA3, and UBE2M are all elevated and apparently correlated with worse clinical characteristics, while only NAE1 is the independent factor associated with postoperative recurrence.332 In hepatocellular carcinoma (HCC), the entire neddylation pathway, including NEDD8, E1 (NAE1 and UBA3), E2 (UBE2M and UBE2F), E3 NEDD8-ligases (RBX1, RNF7/RBX2/SAG and MDM2), and deneddylation enzymes (COPS5, UCHL1 and USP21) is over-activated, and upregulated NAE1, UBE2M, and UCHL1 is associated with poor survival of HCC patients.325,333,334 Mechanistic studies showed that upon activation of neddylation pathway, AKT and LKB1, hallmarks of proliferative metabolism, are neddylation and stabilized, which is related to the development of liver cancer.333 The tumor suppressor, RhoB (a Rho family member of small GTPases) was identified as the substrate of the neddylation-CRL pathway. Neddylation-activated CRL2 E3 ligase targets RhoB for ubiquitylation and degradation, as a key molecular event that drives liver carcinogenesis.335 The master lipogenic transcription factor SREBP-1 is also subjected to UBE2M-mediated neddylation, and SREBP-1 neddylation leads to its stabilization, which is related to progression of liver cancer.325 Likewise, the oncoprotein Hu antigen R (HuR) is stabilized by Mdm2-mediated neddylation, which contributes to liver cancer metastases.330
In human esophageal squamous cell carcinoma, the levels of NEDD8, NAE1, UBA3, and UBE2M are increased significantly, which is positively correlated with high-grade malignancy and postoperative recurrence as well as worse overall survival of patients.336,337 The hyperactivated neddylation pathway activates CRL/SCFβTrCP E3 to destabilize the activating transcription factor 4 (ATF4), thus protecting cells from apoptosis by inhibiting the extrinsic apoptosis mediated by the ATF4–CHOP–DR5 axis, leading to cancer cell survival.337
In glioblastoma, the levels of NAE1, UBA3 and UBE2M are increased significantly with positively correlation of high-grade malignancy and postoperative recurrence as well as worse overall survival of patients.338 The protein levels of UBE2M, RBX1, and SAG/RBX2 are elevated in pancreatic cancer tissues, and NAE1 and UBE2M are higher in osteosarcoma tissues than adjacent normal tissues.339,340,341 The elevated NEDD8 expression in nasopharyngeal carcinoma is correlated with positivity of lymph node metastasis, shorter disease-free survival and poor overall patient survival.342
Despite the large amount of clinical data have demonstrated that neddylation-associated proteins are clearly upregulated in tumor tissues, it is still not clear whether activated neddylation pathway plays a causal or a consequential role during tumorigenesis. To this end, we generated conditional Sag or Rbx1 KO models in lung tissues, and found that deletion of either Sag or Rbx1 remarkably suppressed lung tumor formation induced by KrasG12D via inactivating CRLs,343,344 indicating that neddylation E3s Sag and Rbx1 are promoting factors in KrasG12D-induced lung tumorigenesis. Nevertheless, more conditional KO mouse models are needed for neddylation-pathway genes to determine their causal role during tumorigenesis, induced by activated oncogenes (e.g., Kras, or Wnt/β-catenin) or deletion of tumor suppressors (e.g., p53 or Pten).
Targeting neddylation for anticancer therapy
Over-activation of neddylation pathway in variety of human cancer, along with its positive correlation with poor survival of cancer patients, highly suggests that this pathway is an promising drug target for pan cancers. Indeed, many preclinical studies, using the approaches including those in molecular and cellular biology, tumor biology, genetically modified mouse models, have validated that is the case,12,21 which leads to the advancement of MLN4924 (pevonedistat), the first-in-class neddylation E1 inhibitors, to several phases of clinical trials.18 The topic of targeting neddylation for anticancer therapy has been extensively reviewed by a number of investigators including ourselves.90,345,346 We will briefly review a few representative well-characterized neddylation inhibitors, which is summarized in Fig. 7.
Small molecular inhibitors targeting neddylation pathway. a E1 inhibitor, represented by MLN4924. b UBE2F E2 inhibitor HA-9104. c Several inhibitors that disrupt the UBE2M-DCN1 binding (PDB ID: 3TDU), represented by NAcM-OPT, compound 40, DI-591 and DI-1895. d Non-specific E3 inhibitor of gossypol targeting CUL1-RBX1 and CUL5-RBX2. Created at BioRender.com
NAE E1 inhibitor
MLN4924, a highly selective and first-in-class small molecule inhibitor of neddylation E1 NAE, was first reported in 2009.18 By binding to the active site of NAE and form a steady-state covalent adduct with NEDD8 to impair the process of enzymatic reaction,347 MLN4924 blocks the entire neddylation reaction. As a result, neddylation of both cullins and non-cullin substrates is inhibited, resulting in inactivation of all CRLs and accumulation of various CRLs substrates. MLN4924 suppresses the growth and survival of many cancer cell lines by inducing cell cycle arrest, apoptosis, autophagy and senescence.18,122,347,348 A large number of preclinical studies demonstrate that MLN4924 has an effective anticancer activity for a variety of human cancer cells with tolerable cytotoxicity in mice.349,350,351,352,353 More significantly, a series of phase I/II clinical trials were conducted to evaluate its antitumor activity and safety in patients with hematologic or solid malignancies.354,355,356,357,358,359 MLN4924, known as Pevonedistat in clinical trials, has shown some efficacy to cause stable disease, partial response, and in few cases, complete response.12,21 More importantly, the FDA has granted a Breakthrough Therapy Designation to the investigational agent MLN4924 for the treatment of patients with higher-risk myelodysplastic syndrome.360
The mechanisms of MLN4924 action
Inducing cell cycle arrest
Cell cycle arrest is one type of mechanism by which many anti-cancer agents inhibit the proliferation of cancer cells.361 MLN4924 has shown to induces arrest in different phases of cell cycle by causing the accumulation of a variety of CRLs substrates in a manner dependent of cell types. Specifically, in mantle cell lymphoma and activated B cell-like (ABC) diffuse large B-cell lymphoma (DLBCL) cells, MLN4924 treatment causes rapid accumulation of IκBα to inhibit NF-κB, which is important for the G0/G1-to-S-phase transition,362 therefore inducing G1 phase arrest and ultimately leading to cell apoptosis.350,363 In germinal-center B cell-like (GCB) DLBCL and colon cancer cells, MLN4924 treatment causes the accumulation of CRLs substrate, CDT1 (Chromatin licensing and DNA replication factor 1) via inactivation of CRL1 or CRL4364 to deregulate the component of prereplication complex, which enhances DNA synthesis and triggers DNA replication stress to arrest cancer cells in S phase, and subsequent DNA damage and cell death.18,350,365 Finally, the most frequent cell cycle arrest induced by MLN4924 is at the G2 or G2/M phase by inactivation of CRL1 to cause the accumulation of Wee1, an inhibitor of G2–M phase transition.366 By doing so, MLN4924 induces cell cycle arrest at the G2 or G2/M phase in many types of cancer cells, including live cancer,348 cholangiocarcinoma,332 glioblastoma,338 nasopharyngeal carcinoma,342 gastric cancer,367 T-cell acute lymphoblastic leukemia,368 pancreatic cancer,369,370 renal cancer,371 urothelial carcinoma,372 and Ewing sarcoma.373 Interestingly, MLN4924 induces the arrest at the different phase of cell cycle in different types of RB (retinoblastoma) tumor cells. For example, MLN4924 mainly causes G1 arrest in RB1021 cells, but S + G2/M arrest in RB3823 cells, while RB247, WERI-RB1 and Y79 cells appears to undergo G2/M arrest upon MLN4924 treatment. The exact underlying mechanism for these distinct effects is unclear, but it is likely associated with differential modulation by MLN4924 of multiple CRL targets and/or other neddylated proteins in a cell line dependent manner.352
Inducing apoptosis
Virtually all mammal cells have the ability to self-destruct and maintain homeostasis by undergoing apoptosis.374 Induction of apoptosis is the most common effect of MLN4924 contributing to its killing of cancer cells. By causing accumulation of various CRL substrates, MLN4924 triggers apoptosis through multiple mechanisms. By inactivation of CRL1SKP2 and CRL4CDT2, MLN4924 causes the accumulation of their substrate CDT1 to induce DNA re-replication and subsequent cell apoptosis in HCT-116 cells.365 By inactivating CRL1β-TrCP, IκBα is accumulated to block NF-κB activation, leading to apoptosis induction in ABC lymphoma and AML cells.349,350,375 Also in AML cells, inactivation of NF-κB transcriptionally downregulates miR-155 via decreasing NF-κB and binding to the miR-155 promoter. As a result, SHIP1 (SH2 domain–containing inositol phosphatase), a miR-155 inhibitory target, is increased to inhibit the PI3K/AKT pathway, eventually resulting in cell apoptosis.376 Moreover, by inactivating SAG-CRL5, MLN4924 directly causes the accumulation of the proapoptotic protein NOXA,324,377 and by inactivating RBX1-CRL1, MLN4924 increases the levels of c-MYC and transcription factor 4 (ATF4) to promote NOXA transcription, both triggering cell apoptosis.337,378 Furthermore, ATF4 accumulation also transactivates the transcription factor CHOP to induce the expression of Death Receptor 5 (DR5), which activates caspase-8 to trigger extrinsic apoptosis pathway in esophageal cancer cells.337
Inducing senescence
Senescence is characterized by enlarged and flattened cellular morphology and positive staining of senescence associated β-galactosidase, which is another important mechanism by which MLN4924 inhibits the growth of cancer cells.379,380 The p53/p21 and p16/pRB initiated pathways are the two major axes contributing to senescence induction in response to various stresses.381 Indeed, MLN4924 inactivation of CRL1SKP2 causes p21 accumulation to induce senescence in a manner independent of the pRB/p16 and p53, thus suppressing the growth of a variety of cancer cells,379,380 including osteosarcoma,340 lymphoma,382 multiple myeloma,383 breast cancer,384 pancreatic cancer,369 and gastric cancer.367 Notably, the low dose of MLN4924 is able to induce the senescence irreversibly, suggesting an effective anti-cancer strategy in specific types of cancer.379,380
Inducing autophagy
Autophagy is a pro-survival process that allows stressed cells to recoup essential building blocks and ATPs for biosynthesis in most cases, which is beneficial to cell survival and chemoresistance.385 Upon MLN4924 exposure, a broad type of cancer cells undergo autophagy as a cellular response to protect themselves from senescence and apoptosis.348,386,387 Structurally, MLN4924 treatment causes the accumulation of LC3 II, formation of acidic vesicular organelle (AVO), and double-membraned autophagosome.348 Mechanistically, MLN4924, on one hand, inhibits mTORC1 activity by inactivating CRL1β-TrCP to cause accumulation of DPETOR,195,388,389 a naturally occurring inhibition of mTORC,390 and on the other hand, activates the HIF1α-REDD1-TSC1 axis by inactivating CRL2VHL-mediated HIF1α degradation to induce autophagy in multiple human cancer cells.386 Moreover, given the ROS stress serves as a potential trigger of autophagy,391 and impaired mitochondrial electron transport chain serves as a major source of ROS generation.392 MLN4924 appears to induce autophagy by enhancing mitochondrial membrane depolarization to induce the generation of ROS,393 which is blocked by N-acetyl cysteine (NAC), a classical ROS scavenger, in Huh-7 and Hep G2 cells.348 While MLN4924 suppresses the generation of ROS in macrophages by inhibiting the NEDD8-Cullin3-Nrf2 axis,394 indicating a cell type-dependent way of ROS induction by MLN4924. Since the autophagy response triggered by MLN4924 is likely a pro-survival signal, the combination of MLN4924 with autophagy inhibitor chloroquine indeed enhances anticancer efficacy by inducing apoptosis,348,386,393,395 suggesting a rational drug combination for effective cancer cell killing.
Inhibiting angiogenesis
Abnormal angiogenesis is characterized as an important feature of malignant tumors, and disrupting angiogenesis serves as an attractive strategy for anti-cancer therapy.396 It was reported that all neddylation enzymes, including NAE1 and UBA3, UBE2M, RBX1, SAG and DCN1 are expressed and functional in HUVECs (human umbilical vein endothelial cells).121,317 Neddylation inhibitor MLN4924 apparently inhibits the formation of capillary-like tube networks of HUVECs, and also markedly suppresses angiogenesis and growth in an orthotopic in vivo model of pancreatic cancer.173,397 Mechanistic studies reveal that RhoA, a substrates of CRL3 that controls the assembly of contractile actin and myosin filaments structure,398 is accumulated upon MLN4924 treatment in HUVECs, and RhoA knockdown rescues MLN4924-induced phenotype, indicating its causal role.397 On the other hand, the prolonged exposure of MLN4924 promotes cell cycle arrest and apoptosis in HUVECs by causing the accumulation of p21, p27 and WEE1 and pro-apoptotic proteins NOXA.173,397,399 Moreover, MLN4924 was reported to reduce the tumor microvascular density in a liver metastasis mouse model of uveal melanoma by decreasing the VEGF-C expression due to the inactivation of the NFκB signal.400 Taken together, it appears that targeting neddylation by MLN4924 or other newly developed small molecules may serve as a novel class of antiangiogenic agents.
Inhibiting the survival of cancer stem cells
Cancer stem cells (CSCs) are a small subpopulation of cancer cells with properties of self-renewal and multi-lineage differentiation that are responsible for tumor progression, drug resistance and recurrence.401 Abnormal activation of neddylation is implicated in the self-renewal and survival of CSCs, and targeting NAE by MLN4924 thus could become an effective approach in the elimination of CSCs.200,233,402,403 Indeed, MLN4924 was shown to have more cytotoxic effect against AML stem and progenitor cells than cytarabine, a standard-of-care agent.403 Likewise, the patient-derived glioblastoma stem cells exhibit highly susceptible to MLN4924 through inhibiting the ERK and AKT signaling, while normal human astrocytes are rather resistant.404 In CML, NAE1 expression is remarkably increased in CSCs relative to counterparts, and MLN4924 apparently reduces self-renewal and survival of leukemia stem cells via causing the accumulation of p27.402 Similarly, MLN4924 suppresses the proliferation and stemness characteristics in nasopharyngeal carcinoma cells.342 In breast cancer cells, MLN4924 causes the accumulation of transcription repressor MSX2 by inactivating CRL1FBXW2 E3 ligase. As such, the stem cell factor SOX2 is transcriptionally inhibited by increased MSX2, leading to suppression of the stem cell property.211 In uveal melanoma (UM), MLN4924 treatment inhibits the self-renewal of spherogenic UM cells in vitro and reduces the frequency of UM CSCs by 98% in vivo through increasing the level of FBXO11 (by an unknown mechanism), which promotes Slug degradation.400 Interestingly, our previously study revealed that MLN4924, when applied at nanomolar concentrations, stimulates unexpectedly cell growth and stem cell self-renewal by inhibiting c-MYC degradation and activating EGFR pathways.233 Taken together, it appears that inhibition of CSCs survival is another important anticancer mechanism of MLN4924 action when applied at appropriate concentrations, while neddylation blockage with low concentrations of MLN4924 may be used for stem cell therapy and tissue regeneration.
Regulating inflammatory responses
Chronic inflammation is a promoting event in the progression and development of some types of human cancer, and targeting inflammatory responses appears to be another promising approach for anti-cancer strategy405 Neddylation pathway is implicated to regulate the survival and activity of several immune cells during inflammatory responses, and blocking neddylation pathway by MLN4924 therefore has the robust anti-inflammatory effect under numerous situations.12,22,185
In case of macrophages, short-term MLN4924 treatment (12 h) significantly suppresses the proinflammatory cytokine production induced by LPS in macrophages via blocking the NF-κB activity, without affecting cell viability, while prolonged treatment (over 48 h) apparently induces G2 arrest and apoptosis.185,186 Consistently, MLN4924 treatment dramatically decreases LPS-induced iNOS expression and NO production, as well as reduces LPS-induced expression of cytokines and chemokines, including IL-1β, TNFα and MCP-1, via inhibiting NF-κB/MAPK-mediated signaling in macrophages.406 Moreover, MLN4924-treated macrophages express lower levels of classical cytokines of M1 type such as TNF-α, IL-12, and IL-6; whereas MLN4924 treatment of BMDMs isolated from Apoe−/− mouse (a model for studying cardiovascular disease, atherosclerosis and fat metabolism), shows upregulation of M2-polarizing cytokine IL-13 and the M2 marker Arginase-1, and downregulation of M1 cytokines after 24 h of LPS exposure, suggesting that MLN4924 is likely to skew the macrophage polarization toward an anti-inflammatory M2 status.183
In case of T cells, MLN4924 treatment significantly decreases the proliferation, activation, and effector cytokine release of T cells after stimulation with α-CD3 and α-CD28.201 Besides, MLN4924 impairs the functions of T cells by inhibiting T-cell receptor/CD28-induced proliferation and cytokine production, through inhibiting the Shc and Erk signaling.132 MLN4924 also impedes downstream TCR signaling via interference with the NF-κB pathway, therefore reducing T-cell activation with reduced IL-2 secretion and cell proliferation.407 By inhibiting the NF-κB activation, MLN4924 treatment in CD4 + T cells also reduces the differentiation of inducible regulatory T-cells (iTregs), and shifts the polarization in favor of the Th1 phenotype, likely due to the downregulation of the IL-2/STAT5 signaling, a crucial pathway in Treg differentiation, and decreased production of TNF-α, an NF-κB-regulated cytokine.407 Similarly, in DC cells, MLN4924 significantly attenuates the LPS-induced TNF-α and IL-6 production by inactivation of NF-κB, thus suppressing the ability of DC to stimulate T-cell response.191 Finally, in human microvascular endothelial cells, MLN4924 significantly abrogates the NF-κB pathway and stabilizes the HIF-1α, thus inhibiting the secretion of TNF-α-elicited pro-inflammatory cytokines during the vascular inflammatory response.175 Collectively, it appears that MLN4924 exerts promising anti-inflammatory effect by inhibiting the functions of multiple immune cells. Future study should be geared to investigate the in vivo role of MLN4924 in modulating the function of immune cells infiltrated into the tumor microenvironment.
Serving as a sensitizer for other anticancer therapies
In addition to acting as a single anticancer agent, MLN4924 has been used in combinations with chemotherapy, radiotherapy and immunotherapy, and demonstrates its sensitizing effect.12,408
For chemotherapy, MLN4924 has been shown to sensitize a number of anti-cancer drugs.409 For example, MLN4924 sensitizes ovarian cancer cells to cisplatin even in cisplatin-resistant ovarian cancer cells by increasing the levels of pro-apoptotic protein, BIK and inducing DNA damage and oxidative stress both in cell culture model and in mice bearing ovarian tumor xenografts.410 Likewise, MLN4924 sensitizing cytotoxicity effect of cisplatin was also seen in human cervical carcinoma411 and urothelial carcinoma.412 In pancreatic cancer, MLN4924 sensitizes the effect of gemcitabine by inhibiting cell proliferation and promoting cell death via inducing the accumulation of ERBIN, an inhibitor of the RAS-MAPK pathway, and NOXA.369 In colorectal cancer, MLN4924 sensitizes topoisomerase I inhibitors via inactivating the DCAF13-CRL4 E3 ligase.413 In multiple myeloma, MLN4924 induces REDD1 (regulated in development and DNA damage responses 1) expression at both RNA and protein levels to suppress the AKT and mTOR signaling, thereby enhancing the bortezomib-induced cytotoxicity.414 By inducing the accumulation of c-Jun and NOXA, MLN4924 also sensitizes leukemia cells to retinoic acid-induced apoptosis.215 In acute myelogenous leukemia (AML)/myelodysplastic syndrome (MDS), MLN4924 combination with Belinostat, a HDAC inhibitor, was shown to trigger more robust DNA damage and apoptosis, resulting in enhanced suppression of tumor growth with longer animal survival in AML xenograft models.415 Finally, MLN4924 blocks the FA pathway by inhibiting FANCD2 monoubiquitylation and CHK1 phosphorylation to increase cellular sensitivity to several DNA interstrand crosslinks (ICLs)-inducing agents, such as cisplatin, mitomycin C and hydroxyureain in various cancer cell lines.416
MLN4924 also sensitizes various cancer cell lines to targeted therapeutic agents. In breast cancer cells, the AKT signaling pathway is activated to protect cells from apoptosis upon MLN4924 treatment. The combination of MLN4924 with AKT inhibitor MK-2206 causes much greater induction of apoptosis to kill.417 Also in breast cancer cells, MLN4924 transcriptionally blocks ER-α expression through SGK1-dependent cytoplasmic localization of FOXO3a. Thus, MLN4924 combination with fulvestrant, an estrogen receptor antagonist, causes maximal growth suppression of ER-positive breast cancer cells in vivo.326 Moreover, our recent study showed that MLN4924 activates PKM2 via inducing its tetramerization to promote glycolysis as a cellular protective mechanism in response to MLN4924 cytotoxicity. The combination of MLN4924 with PKM2 inhibitor shikonin synergistically inhibits growth of breast cancer cells in both in vitro cell culture setting and in vivo xenograft model.237
MLN4924 also act as a radio-sensitizer to sensitize a variety of cancer cells to radiotherapy. In pancreatic cancer cells, MLN4924 selectively sensitizes cancer cells, but not normal cells to radiation therapy by increasing radiation-induced DNA damage and apoptosis via causing the accumulation of CRL substrates, CDT1 and WEE1.370 Knockdown of CDT1 and WEE1 partially rescues MLN4924-induced enhancement of radiation effects, suggesting their causal roles.370 Likewise, MLN4924 also synergizes head and neck squamous carcinoma cells to irradiation in both in vitro cell culture and in vivo nude mice models, which is mainly attributed to the stabilization of CDT1 and induction of replication.418 In breast cancer cells, MLN4924 confers radio-sensitization through a p21-dependent mechanism, which is partially rescued by p21 knockdown.419 Similarly, MLN4924 serves as an effective radio-sensitizer by significantly enhancing irradiation-induced cell-cycle arrest and apoptosis in hormone refractory prostate cancer, through a mechanism of WEE1/p21/p27 accumulation.420 Finally, the activity of CRL1FBXW7 ligase is required for NHEJ repair in response to ionizing radiation via inducing XRCC4 polyubiquitylation via the K63 linkage. Thus, MLN4924 inactivation of CRL1FBXW7 in human cancers could be a potent strategy for increasing the efficacy of radiotherapy.153
Targeting immune checkpoints by blockage of programmed cell death protein 1 (PD-1) and its ligand PD-L1 have been approved by FDA for the treatment of several human cancers.421 The abundance of PD-L1 levels on tumor tissues often predicts the efficacy of PD-L1 inhibitors.422 Notably, the abundance of PD-L1 is regulated by ubiquitin ligases, such as CRL1 β-TrCP and CUL3SPOP, via proteasome-degradation pathway.423,424 MLN4924 upregulates PD-L1 in cancer cells via directly blocking the PD-L1 degradation by CRL E3,423 or indirectly by inactivating CRL1FBXW7 to cause accumulation of c-MYC, which transactivates PD-L1 expression.408 Thus, MLN4924 enhances the efficacy of anti-PD-L1 therapy through PD-L1 induction, and exerts the synergistic tumor suppressive effect in vivo.408,425 The deficient mismatch repair (dMMR) phenotype, also known as microsatellite instability (MSI), is a significant feature of certain types of human cancers, which is associated with chemotherapy resistance.426,427,428 A recent study showed that dMMR-induced destabilizing mutations triggers proteome instability in dMMR tumors, causing the accumulation of large number of misfolded proteins. The dMMR cells, therefore, develop a NEDD8-dependent degradation system to clean these misfolded proteins.429 As a result, blocking the NEDD8-dependent clearance pathway by MLN4924 increases the accumulation of misfolded protein aggregates with severe immunogenic cell death. The combination of MLN4924 and PD-1 inhibition, therefore, leverages this immunogenic cell death, with synergistically improving the efficacy over either treatment alone.429 Finally, MLN4924 was found to sensitize human cancer cells to VSVΔ51 oncolytic virotherapy via blocking the type 1 interferon (IFN-1) response through repression of interferon-stimulated growth factor 3 (ISGF3) in a neddylation-dependent manner, and NF-κB nuclear translocation.430 Taken together, MLN4924 could serve as a potential sensitizer when in combination with chemo, radio or immune-therapy in clinical application.
Other NAE inhibitors
The success of MLN4924 sparked a surge in the discovery of additional NAE inhibitors, and various classes of NAE inhibitors has then been reported or under investigation, including covalent NAE inhibitors, non-covalent NAE inhibitors and even NAE agonizts.345 TAS4464, with structural similarity to MLN4924, is another potent and selective covalent NAE inhibitor with an impressive IC50 value of 0.96 nM.431 Few preclinical studies have shown that TAS4464 has potent growth-inhibitory effects against various cancer cell lines, derived from colorectal cancer, endometrial carcinoma, leukemia, liver cancer, ovarian cancer, pancreatic cancer and renal cell carcinoma in both in vitro and in vivo models.431,432,433,434 Mechanistically, TAS4464 induces sub-G1 arrest and apoptosis in cancer cells.433,434 Biochemically, TAS4464 blocks cullin neddylation and causes accumulation of CRL substrates, leading to the activation of extrinsic and intrinsic apoptotic pathways.431 Like MLN4924, TAS4464 also inactivate NF-κB pathways in cancer cells,433 thereby interfering with the signaling pathways associated with inflammation and cell survival. However, TAS4464 was advanced to Phase I clinical trial, but was terminated due to the liver toxicity.432
In addition to covalent inhibitors like MLN4924 and TAS4464, there are also few non-covalent NAE inhibitors being investigated. These inhibitors target the activity of NAE through non-covalent binding and have shown potential in suppressing tumor growth in vitro and in vivo,435,436,437,438,439,440,441,442,443,444,445 although none of them has been advanced to the clinical trials. Interestingly, a NAE agonist, first reported in 2020, showed promising antiproliferative activity against various cancer cell lines.446 In contrast to NAE inhibitors, this NAE agonist activated neddylation and induced morphology changes, apoptosis, and cell cycle arrest in cancer cells. It also suppresses tumor growth in xenograft tumor models. Thus, it appears that cancer cells are susceptible to alterations in neddylation pathway, whose inhibition or activation could affect the growth of cancer cells. NAE agonist might be a potential direction for future investigation in NAE-targeted drugs for cancer therapy.
Although MLN4924 and TAS4464 achieved many successes in preclinical studies, their blockage of the entire neddylation pathway bears unavoidable intrinsic toxicity to normal cells. In fact, in clinical trials, MLN4924 has shown a limited overall response rate (17%) in patients with AML or MDS (ClinicalTrials.gov Identifier: NCT00911066), and one of TAS4464 clinical trials was terminated due to liver toxicity (ClinicalTrials.gov Identifier: NCT02978235). Current efforts in the discovery of neddylation pathway inhibitors have focused on neddylation enzymes downstream of E1.
Targeting E1–E2 interaction
The pivotal role of E2 conjugating enzymes in the NEDD8 conjugation process is essential, as they are responsible for engaging specific E3 ligases. Positioned centrally, E2 forms a vital connection, linking with both E1 and E3 enzymes, thus maintaining a crucial bridge between them.324 Targeting E2s, therefore, can be done by disrupting the catalytic core of E2 enzymes, or by blocking the interaction between E1 and E2, or E2 and E3. The crystal structures of UBE2M or UBE2F in complex with UBA3 have been resolved,11,42,96 providing the base for structure-based drug design and virtual screen. However, it’s still challenging due to the fast conformational changes and the absence of deep druggable pockets on its catalytic sites.447 Our group recently identified few potential pockets in UBE2F responsible for the UBA3-UBE2F interactions, and conducted a virtual screening which led to the discovery of a small molecule inhibitor HA-1141.448 HA-1141 directly binds to UBA3 in vitro and in vivo and effectively block neddylations of CUL 1-5, suggesting it targets E1. Unexpectedly, HA-1141 also induces non-canonical endoplasmic reticulum (ER) stress and PKR-mediated terminal integrated stress response (ISR), leading to activated ATF4, decreased protein synthesis and inhibited mTORC1 activity. HA-1141 showed a sound inhibitory effect in both cultured lung cancer cells and an in vivo lung tumor xenograft model.448
The same virtual screening also identified a small molecule HA-9104, which selectively targets the UBE2F-CRL5 axis.449 Specifically, HA-9104 binds to UBE2F, and blocks CUL5 neddylation to cause accumulation of CRL5 substrate NOXA, resulting in induction of apoptosis in lung cancer cells both in vitro cell culture setting and in vivo xenograft model.449 Medicinal chemistry-based SAR (Structure and Activity Relationship) study is under the way to optimize anticancer activity of these compounds.
In another recent study, Mamun et al. established a HTRF based drug screen focusing on searching for UBE2M inhibitors.450 The screen and subsequent biochemical and cellular assays discovered Micafungin, an antifungal agent, as a neddylation inhibitor targeting UBE2M. Micafungin interacts with UBE2M via occupying the catalytic cysteine (Cys111) in UBE2M, thus inhibiting neddylation to accumulate CRLs substrates, resulting in suppression of survival and migration of gastric cancer cells.450
Targeting DCN1-UBE2M
DCN1 (defective in cullin neddylation 1) is a co-E3 ligase of RBX1 that forms a complex with UBE2M to enhance neddylation of cullins.64 Importantly, the crystal structure analysis revealed that DCN1–UBE2M interaction is amenable for the design of small-molecule inhibitors.96,451 To this end, a piperidine-based chemical probe, designated as NAcM-HIT, was discovered through a time-resolved fluorescence energy transfer (TR-FRET) based ligand competition assay from a library of 600,000 chemicals. NAcM-HIT specifically disrupts the DCN1-UBE2M binding, and inhibits the DCN1-dependent neddylation reaction.451 After a series of optimization, a NAcM-HIT analog was synthesized, which has 100-fold increase in potency, but still with low stability.452 Through a series of rational structure-based design and empirical chemistry approaches, a more potent and stable compound, NAcM-OPT was identified, which selectively disrupts the DCN1-UBE2M interaction and inhibit neddylation of CUL1 and CUL3, without affecting neddylation of CUL2, CUL4 and CUL5.453 Subsequent studies from the same group disclosed another series of pyrazolo-pyridones inhibitors, represented by compound 2454 and compound 40,455 with improved efficacy and reasonable oral bioavailability.
Simultaneously, the Wang group, through a structure-based virtual screen, identified the high-affinity, cell-permeable, drug-like peptidomimetic small-molecules, DI-591 and DI-404, which selectively inhibit the DCN1-UBE2M interaction.456,457 Unlike NAcM-OPT and MLN4924, DI-591/DI-404 specifically blocks CUL-3 neddylation without affecting, if any, the neddylation of other cullins. As a result, DI-591/DI-404 cause the accumulation of NRF2, a CUL-3 substrate, in a dose-dependent manner in multiple cancer cell lines.456 However, DI-591/DI-404 fail to show any cytotoxicity in both human cancer cells at concentrations up to 20 µM, suggesting the dispensable functions of CRL3 activity in determining cell survival. Based upon DI-591, the Wang group further designed a class of covalent DCN1 inhibitors and discovered two potent, selective and covalent DCN1 inhibitors, designated as DI-1548 and DI-1859.458 Both inhibitors inhibit CUL-3 neddylation in cancer cells at low nanomolar concentrations with 2–3 orders of magnitude more potent than DI-591. Impressively, DI-1859 induces a robust increase of NRF2 protein in mouse liver and effectively protects mice from liver toxicity induced by acetaminophen. Given that activation of NRF2 has been pursued as a promising therapeutic approach for treatment of multiple sclerosis,459 DI-1859 could be a potential therapeutic agent for the treatment of this disease. DI-1859 may also have a potential therapeutic application to protect normal tissues from acute toxicity induced by oxidative stress through upregulation of NRF2 via blocking CRL3 activity.460
Toward the DCN-1 target, the Liu and Zhao group also discovered several small molecule inhibitors with distinct chemical structures. Triazolo[1,5-α] pyrimidine-based inhibitor WS-291 and its successor WS-383 showed inhibitory effect on neddylation of CUL-1 and CUL-3 in gastric cancer cell lines.461 Another pyrimidines-based small molecular inhibitor DC-2 could specifically inhibits CUL-3 neddylation with IC50 around 15 nM,462 and further SAR yielded DN-2 with even more potent efficacy (IC50 = 9.6 nM).463 The Liu group also reported a phenyltriazole thiol-based inhibitor SK-464 with IC50 value of 26 nM, bringing a new class of potent DCN1 inhibitors into this field.464 Although none of the reported DCN1/UBE2M inhibitors has entered a clinical trial, they provided guidance for future drug design and discovery on this target.
Targeting neddylation E3s
From a broader perspective, achieving therapeutic targeting of RING E3s in neddylation holds the promise of greater selectivity and fewer side effects. However, no such specific inhibitors targeting RBX1 or RBX2/SAG has been identified. From a structural perspective, RBX1 or SAG lacks a well-defined catalytic pocket, making it challenging to identify or design small-molecule inhibitors.465 In addition, the ubiquitylation and neddylation processes both involve a dynamic enzymatic cascade, featuring numerous transient and temporary protein-protein interactions, which makes the RING E3s difficult to target.465 Despite these challenges, several research teams have put the efforts to discover small molecule inhibitors for this seemingly “undruggable” target. Indeed, one reference compound, designated as C64, was extensively characterized for selective binding at the RBX1-binding grooves with VLYRLWLN motif on CUL1-7,466 through various approaches, including a series of bioinformatics analysis on the binding patterns between RBX1 and seven cullins, molecular dynamics (MD) simulations to define critical binding sites and to predict key conformations influencing RBX1-Cullin binding stability, and structure-based virtual screening and docking analyses. However, the study did not confirm the inhibitory activity of these small molecules, although the analysis of the RBX1-Cullin binding site and conformation could provide valuable insights for designing small-molecule inhibitors targeting RBX1-induced cullin neddylation.
Our group recently developed an Alpha-Screen-based high throughput screen (HTS) assay, and identified a natural compound derived from cotton seed named gossypol that can effectively block neddylation of CUL1 and CUL5 by directly binding to RBX1-CUL1 or SAG-CUL5 complex, respectively.467 In this way, gossypol inhibits cullins neddylation in multiple cancer cell lines but selectively causes accumulation of the CRL1 substrate MCL1, and CRL5 substrate NOXA.467 By inducing the accumulation of NOXA, gossypol induces apoptosis in few cancer cell lines. Given that MCL1 is a pro-survival Bcl-2-like protein, and gossypol-induced MCL1 links to the less efficacy of gossypol, we found combination of gossypol with MCL1 inhibitor S63845 indeed achieve a better growth inhibition of cancer cells.467 However, the underlying mechanism for gossypol to selectively cause accumulation of MCL1 and NOXA, but not other substrates of CRL1 and CRL5, is a subject for future investigation.
Conclusion and future perspectives
In this review, we describe how neddylation pathway modulates the biochemical activities of cullin and non-cullin substrates by catalyzing the NEDD8 attachment, leading to activation of CRLs or altered activities, stability or subcellular localization of a variety of non-cullin substrates. We also summarize the essential role of neddylation pathway in the maintenance of healthy cellular homeostasis, and its active involvement, upon abnormal activation or dysregulation, in the development of various human diseases. Finally, we discuss current efforts in the discovery and development of small molecular inhibitors of neddylation pathway and its potential application in the treatment of human diseases, mainly human cancer. However, there are quite a few unsolved puzzles and open areas in the field of neddylation research. Below, we list a number of possible directions for future investigations (Fig. 8).
Future perspectives. A total of nine research directions are proposed for future investigations, covering from basic mechanistic study, target identification and validation, the use of animal disease models to eventually discover specific neddylation inhibitors for clinical trials to treat various human diseases with abnormal activation of the neddylation pathway (see text for details). Created at BioRender.com
What is the underlying mechanism(s) of neddyltion dysregulation in human diseases?
The exact underlying mechanisms by which NEDD8 and neddylation enzymes are altered and in the most cases over-activated in human diseases, such as cancer, remains elusive. The future studies on this aspect should be directed to a systematic elucidation of possible alterations at 1) the transcription levels, including epigenetic modifications of gene promoters and enhancers; 2) the mRNA translational levels including RNA modifications, such as N6-methyladenosine (m6A), 5-methylcytosine (m5C), and 7-methylguanosine (m7G);468 3) the post-translational levels, including phosphorylation, acetylation, methylation, even ubiquitylation, among many others that affect their stability469 as well as the regulators that affect their activities, such as inositol hexakisphosphate (IP6) and other metabolites.470
How many reported non-cullin substrates are physiological substrates?
While cullin family members are well-established as the physiological substrates of neddylation, almost all non-cullin substrates were identified and characterized in the cell culture settings with overexpression or knockdown approaches, which raises a serious consideration as to whether they are indeed physiological substrates of neddylation.17 Furthermore, unlike cullins, none of non-cullin substrates showed in western blotting a fast-migrating (non-neddylated form) and a slow-migrating (neddylated form) bands with conversion of neddylated to non-neddylated form upon treatment of neddylation inhibitor (e.g., MLN4924) or knockdown of neddylation enzymes.12 The big challenge in the field is to validate (for reported non-cullin substrates) and identify novel physiological substrates of neddylation, if any. It is known that under the condition of NEDD8 overexpression, NEDD8 is mistakenly recognized as ubiquitin by UAE, an ubiquitin activating enzyme E1, and conjugated onto its ubiquitin substrates, leading to erroneous identification of neddylation substrates. The possible solution to this scenario is to generate cell lines expressing the CRISPR-Cas9-mediated knock-in of NEDD8 with an N-terminal FLAG- or HA-tag, followed by FLAG-/HA-based pull-down and affinity purification to identify true endogenous substrates. One step further is to generate a knock-in mouse model, in which FLAG- or HA-tagged Nedd8 is expressed in the physiological level to facilitate the identification of spatiotemporal-specific neddylation substrates. Moreover, the combination of deconjugation-resistant form of NEDD8 with a specific antibody against the unique NEDD8 remnant would offer additional options to identify new neddylation substrates.56,471
Are there additional members in the neddylation cascade?
Although the biochemical process of neddylation modification is well established, few questions could be asked: 1) are there any additional E2s for neddylation, beyond UBE2M and UBE2F? 2) can UBE2M or UBE2F act as an E2 for ubiquitylation or other ubiquitin-like modifications? Indeed, we found that UBE2M can act as an ubiquitin E2 for CRL3 and Parkin E3 ubiquitin ligases55; 3) It will not be surprising that additional ubiquitin E3s will be identified to serve as neddylation E3s, and deubiquitylases as deneddylases. 4) While few crystal structures of neddylation E2/E3 complexed with cullin substrates have been resolved,95,96,97,98,100 no such structure was resolved/available for non-cullin substrates.
Does the neddylation pathway function in subcellular organelles?
Although neddylation plays a regulatory role in the morphology and function of mitochondria,109 detailed underlying mechanism remains elusive. For examples, are neddylation enzymes or cullin substrates localized in the mitochondria and if so, in what compartment? And whether and how the neddylation-CRL system modifies mitochondrial proteins, thus affecting the process of ROS generation or energy production? Finally, are neddylation components localized in other subcellular organelles, such as endoplasmic reticulum or lysosome?
Possible role in liquid–liquid phase separation
Liquid–liquid phase separation (LLPS) is ubiquitous phenomenon underlying the formation of membraneless organelles in eukaryotic cells to facilitate local biochemical reaction.472 Several studies have shown that neddylation modulates LLPS. For examples, 1) neddylation inhibitors MLN4924 and TAS4464 promote phase separation of PML/RARα;473 2) DAXX drives LLPS of SPOP, a well-known receptor for CRL3, to promote DAXX ubiquitylation and degradation;474 3) CRL5ASB11 promotes the ubiquitylation of PAICS, leading to the recruitment of UBAP2 to trigger LLPS to form the purinosome.475 The open questions include 1) does LLPS occurs during the neddylation process under specific physiological or pathological/stressed conditions? 2) whether and under what conditions that the substrate receptors of CRL drive their own LLPS to rapidly promote the ubiquitylation of substrates? 3) does LLPS regulation by the neddylation-CRL axis is a common phenomenon to facilitate ubiquitylation of substrates?
Cross-talk between neddylation and other post-translational modifications
The cross-talk between neddylation and ubiquitylation is frequently observed with the same or opposite effect. For example, either neddylation or ubiquitylation of yeast CUL4 orthologue Rtt101 causes its catalytic activation.48 Similarly, in response to DNA damage both NEDD8 and ubiquitin accumulate at the DNA damage sites, and the poly-neddylation and poly-ubiquitylation of histones are functionally redundant.44,476,477 In contrast, neddylation or ubiquitylation of TGFβRII elicit distinct biochemical consequence. While ubiquitylation leads to degradation, neddylation mediates signal transduction through clathrin-mediated endocytosis.478 Furthermore, neddylation on a given protein often inhibits its ubiquitylation and subsequent degradation.47,131,238,251,278,479 An important open question is how a cell coordinates these two types of modifications to gain the growth advantage. Besides ubiquitylation, much less studies were conducted to elucidate the cross-talks between neddylation and other types of ubiquitin-like modifications, such as sumoylation.480 Thorough understanding these cross-talks will deepen our understanding of neddylation regulation of various biological processes.
NEDD8 modifications of itself or other macromolecules
NEDD8 itself may undergo post-translational modifications, since numerous proteomic studies have shown the presence on NEDD8 of ubiquitylation, phosphorylation, acetylation, and succinylation sites.481,482,483,484 However, no follow-up studies, except phosphorylation,485 were conducted to confirm these possible modifications, not to mention their biochemical and functional significance of these modifications.
NEDD8 is also found to incorporate into an existing ubiquitin chain to form mixed NEDD8- and Ub-modified conjugates.15,16,23 It remains to determine whether this process is physiologically relevant or merely an experimental artifact? If the former is true, what is the biochemical consequence or biological significance for this mix conjugation on a protein?
Finally, few recent studies have shown that ubiquitylation can occur on few other macrobiomolecules such as glucosaccharides486 and lipopolysaccharide.487 It is completely unknown but very possible that these macromolecules are also subjected to neddylation modification. It will be an interesting subject for future investigation.
Further validation of neddylation pathway as a therapeutic target for various diseases
The dysregulation of neddylation pathway frequently occurs in several human diseases, particularly in a variety of human cancers, as evidenced by overexpression of NEDD8 and neddylation enzymes.12,21,72 However, these association studies did not reveal any causal or consequential relationship between these alterations in the disease development. Thus, to pinpoint genetically the causal or consequent effects, engineered mouse models, including organ-specific deletion or transgenic expression, should be established alone or in combination with known mouse models for various diseases, including cancer (e.g., Kras activation,488 p53/pTen loss489,490), metabolic disorders (e.g., Leptin-deficient ob/ob model, leptin receptor-deficient db/db model491), neurodegenerative diseases (e.g., APP/PS1 model for Alzheimer’s disease, mutant α-Synuclein or mutant LRRK2 transgenic models for autosomal dominant Parkinson’s disease),492 immune disorders (e.g., SAMP1/YitFc (Samp) mice for Crohn’s disease,493 K/BxN mice for rheumatoid arthritis,494 and cardiovascular diseases (e.g., TAC mice495), among others.
Discovery of neddylation-specific inhibitors
Although neddylation pathway has been validated as a promising anticancer target,12,21 only two inhibitors of neddylation E1 (MLN4924 and TAS4464) were advanced into the clinical trials,354,432 and none of them is granted by FDA for the treatment of human cancer, although preclinical studies showed impressive anti-cancer effect.18,431 The fact that E1 inhibitors block the entire neddylation pathway, the cytotoxicity is inevitable. Furthermore, MLN4924 bears a number of off-target effects.496 Thus, the further efforts should be more focused on the discovery and development of specific inhibitors targeting neddylation E2s, UBE2F or UBE2M or E3s, such as RBX1 or RBX2, leading to indirect inhibition of selective CRLs. Although several such inhibitors were reported,448,449,450,451,452,453,454,455,456,457,458,497 extensive studies with guidance of the crystal structures and AI simulation,498 are needed to discover much potent and selective inhibitors for the advancement to the clinical trials.
References
van der Veen, A. G. & Ploegh, H. L. Ubiquitin-like proteins. Annu. Rev. Biochem. 81, 323–357 (2012).
Ramazi, S. & Zahiri, J. Posttranslational modifications in proteins: resources, tools and prediction methods. Database. 2021, baab012 (2021).
Sun, Y. Introduction. Adv. Exp. Med. Biol. 1217, 1–8 (2020).
Whitby, F. G., Xia, G., Pickart, C. M. & Hill, C. P. Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes. J. Biol. Chem. 273, 34983–34991, (1998).
Gan-Erdene, T. et al. Identification and characterization of DEN1, a deneddylase of the ULP family. J. Biol. Chem. 278, 28892–28900 (2003).
Frickel, E. M. et al. Apicomplexan UCHL3 retains dual specificity for ubiquitin and Nedd8 throughout evolution. Cell Microbiol 9, 1601–1610 (2007).
Kamitani, T., Kito, K., Nguyen, H. P. & Yeh, E. T. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J. Biol. Chem. 272, 28557–28562 (1997).
Rabut, G. & Peter, M. Function and regulation of protein neddylation. EMBO Rep. 9, 969–976 (2008).
Walden, H. et al. The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol. Cell 12, 1427–1437 (2003).
Huang, D. T. et al. Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8’s E1. Mol. Cell 17, 341–350 (2005).
Huang, D. T. et al. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol. Cell 33, 483–495 (2009).
Zhou, L., Zhang, W., Sun, Y. & Jia, L. Protein neddylation and its alterations in human cancers for targeted therapy. Cell Signal 44, 92–102 (2018).
Cappadocia, L. & Lima, C. D. Ubiquitin-like protein conjugation: structures, chemistry, and mechanism. Chem. Rev. 118, 889–918 (2018).
Vogl, A. M. et al. Global site-specific neddylation profiling reveals that NEDDylated cofilin regulates actin dynamics. Nat. Struct. Mol. Biol. 27, 210–220 (2020).
Leidecker, O. et al. The ubiquitin E1 enzyme Ube1 mediates NEDD8 activation under diverse stress conditions. Cell cycle 11, 1142–1150 (2012).
Singh, R. K. et al. Recognition and cleavage of related to ubiquitin 1 (Rub1) and Rub1-ubiquitin chains by components of the ubiquitin-proteasome system. Mol. Cell Proteom. 11, 1595–1611 (2012).
Enchev, R. I., Schulman, B. A. & Peter, M. Protein neddylation: beyond cullin-RING ligases. Nat. Rev. Mol. Cell Biol. 16, 30–44 (2015).
Soucy, T. A. et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 (2009).
Liu, D., Che, X. & Wu, G. Deciphering the role of neddylation in tumor microenvironment modulation: common outcome of multiple signaling pathways. Biomark. Res. 12, 5 (2024).
Zhao, Y. & Sun, Y. Cullin-RING Ligases as attractive anti-cancer targets. Curr. Pharm. Des. 19, 3215–3225 (2013).
Zhao, Y., Morgan, M. A. & Sun, Y. Targeting Neddylation pathways to inactivate cullin-RING ligases for anticancer therapy. Antioxid. Redox Signal 21, 2383–2400 (2014).
Zhou, L. et al. Neddylation: a novel modulator of the tumor microenvironment. Mol. Cancer 18, 77 (2019).
Hjerpe, R. et al. Changes in the ratio of free NEDD8 to ubiquitin triggers NEDDylation by ubiquitin enzymes. Biochem. J. 441, 927–936 (2012).
Du, W., Zhang, R., Muhammad, B. & Pei, D. Targeting the COP9 signalosome for cancer therapy. Cancer Biol. Med. 19, 573–590 (2022).
Kumar, S., Tomooka, Y. & Noda, M. Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem. Biophys. Res. Commun. 185, 1155–1161 (1992).
Kumar, S., Yoshida, Y. & Noda, M. Cloning of a cDNA which encodes a novel ubiquitin-like protein. Biochem. Biophys. Res. Commun. 195, 393–399 (1993).
Osaka, F. et al. A new NEDD8-ligating system for cullin-4A. Genes Dev. 12, 2263–2268 (1998).
Liakopoulos, D., Doenges, G., Matuschewski, K. & Jentsch, S. A novel protein modification pathway related to the ubiquitin system. EMBO J. 17, 2208–2214 (1998).
Lammer, D. et al. Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev. 12, 914–926 (1998).
Kamura, T. et al. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 13, 2928–2933 (1999).
Ohta, T., Michel, J. J., Schottelius, A. J. & Xiong, Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell 3, 535–541 (1999).
Tan, P. et al. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of IkBa. Mol. Cell 3, 527–533 (1999).
Seol, J. H. et al. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 13, 1614–1626 (1999).
Duan, H. et al. SAG, a novel zinc RING finger protein that protects cells from apoptosis induced by redox agents. Mol. Cell Biol. 19, 3145–3155 (1999).
Swaroop, M. et al. Yeast homolog of human SAG/ROC2/Rbx2/Hrt2 is essential for cell growth, but not for germination: chip profiling implicates its role in cell cycle regulation. Oncogene 19, 2855–2866 (2000).
Liu, J., Furukawa, M., Matsumoto, T. & Xiong, Y. NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol. Cell 10, 1511–1518 (2002).
Zheng, J. et al. CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol. Cell 10, 1519–1526 (2002).
Gong, L. & Yeh, E. T. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J. Biol. Chem. 274, 12036–12042 (1999).
Bohnsack, R. N. & Haas, A. L. Conservation in the mechanism of Nedd8 activation by the human AppBp1-Uba3 heterodimer. J. Biol. Chem. 278, 26823–26830 (2003).
Olsen, S. K. et al. Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature 463, 906–912 (2010).
Huang, D. T. et al. Basis for a ubiquitin-like protein thioester switch toggling E1-E2 affinity. Nature 445, 394–398 (2007).
Huang, D. T. et al. A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8. Nat. Struct. Mol. Biol. 11, 927–935 (2004).
Xirodimas, D. P. et al. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell 118, 83–97 (2004).
Ma, T. et al. RNF111-dependent neddylation activates DNA damage-induced ubiquitination. Mol. Cell 49, 897–907 (2013).
Noguchi, K. et al. TRIM40 promotes neddylation of IKKγ and is downregulated in gastrointestinal cancers. Carcinogenesis 32, 995–1004 (2011).
Xie, P. et al. The covalent modifier Nedd8 is critical for the activation of Smurf1 ubiquitin ligase in tumorigenesis. Nat. Commun. 5, 3733 (2014).
Zuo, W. et al. c-Cbl-mediated neddylation antagonizes ubiquitination and degradation of the TGF-beta type II receptor. Mol. Cell 49, 499–510 (2013).
Rabut, G. et al. The TFIIH subunit Tfb3 regulates cullin neddylation. Mol. Cell 43, 488–495 (2011).
Monda, J. K. et al. Structural conservation of distinctive N-terminal acetylation-dependent interactions across a family of mammalian NEDD8 ligation enzymes. Structures 21, 42–53 (2013).
Deshaies, R. J. & Joazeiro, C. A. RING domain E3 ubiquitin ligases. Annu Rev. Biochem 78, 399–434 (2009).
Dou, H. et al. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat. Struct. Mol. Biol. 19, 876–883 (2012).
Plechanovova, A. et al. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 489, 115–120 (2012).
Yunus, A. A. & Lima, C. D. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat. Struct. Mol. Biol. 13, 491–499 (2006).
Berndsen, C. E. et al. A conserved asparagine has a structural role in ubiquitin-conjugating enzymes. Nat. Chem. Biol. 9, 154–156 (2013).
Zhou, W. et al. UBE2M is a stress-inducible dual e2 for neddylation and ubiquitylation that promotes targeted degradation of UBE2F. Mol. Cell 70, 1008–1024 e1006 (2018).
Jeram, S. M. et al. An improved SUMmOn-based methodology for the identification of ubiquitin and ubiquitin-like protein conjugation sites identifies novel ubiquitin-like protein chain linkages. Proteomics 10, 254–265 (2010).
Jones, J. et al. A targeted proteomic analysis of the ubiquitin-like modifier nedd8 and associated proteins. J. Proteome Res. 7, 1274–1287 (2008).
Girdwood, D., Xirodimas, D. P. & Gordon, C. The essential functions of NEDD8 are mediated via distinct surface regions, and not by polyneddylation in Schizosaccharomyces pombe. PloS One 6, e20089 (2011).
Ohki, Y., Funatsu, N., Konishi, N. & Chiba, T. The mechanism of poly-NEDD8 chain formation in vitro. Biochem. Biophys. Res. Commun. 381, 443–447 (2009).
Lobato-Gil, S. et al. Proteome-wide identification of NEDD8 modification sites reveals distinct proteomes for canonical and atypical NEDDylation. Cell Rep. 34, 108635 (2021).
Bailly, A. P. et al. The balance between mono- and NEDD8-chains controlled by NEDP1 upon DNA damage is a regulatory module of the HSP70 ATPase activity. Cell Rep. 29, 212–224.e218 (2019).
Keuss, M. J. et al. Unanchored tri-NEDD8 inhibits PARP-1 to protect from oxidative stress-induced cell death. EMBO J. 38, e100024 (2019).
Kurz, T. et al. The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature 435, 1257–1261 (2005).
Kurz, T. et al. Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol. Cell 29, 23–35 (2008).
Scott, D. C. et al. A dual E3 mechanism for Rub1 ligation to Cdc53. Mol. Cell 39, 784–796 (2010).
Scott, D. C. et al. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science 334, 674–678 (2011).
Meyer-Schaller, N. et al. The human Dcn1-like protein DCNL3 promotes Cul3 neddylation at membranes. Proc. Natl Acad. Sci. USA 106, 12365–12370 (2009).
Keuss, M. J. et al. Characterization of the mammalian family of DCN-type NEDD8 E3 ligases. J. Cell Sci. 129, 1441–1454 (2016).
Mo, Z. et al. Neddylation requires glycyl-tRNA synthetase to protect activated E2. Nat. Struct. Mol. Biol. 23, 730–737 (2016).
Cui, J. et al. Glutamine deamidation and dysfunction of ubiquitin/NEDD8 induced by a bacterial effector family. Science 329, 1215–1218 (2010).
Yao, Q. et al. Structural mechanism of ubiquitin and NEDD8 deamidation catalyzed by bacterial effectors that induce macrophage-specific apoptosis. Proc. Natl Acad. Sci. USA 109, 20395–20400 (2012).
Watson, I. R., Irwin, M. S. & Ohh, M. NEDD8 pathways in cancer, Sine Quibus Non. Cancer Cell 19, 168–176 (2011).
Lyapina, S. et al. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 292, 1382–1385 (2001).
Echalier, A. et al. Insights into the regulation of the human COP9 signalosome catalytic subunit, CSN5/Jab1. Proc. Natl Acad. Sci. USA 110, 1273–1278 (2013).
Cope, G. A. et al. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 298, 608–611 (2002).
Wei, N., Serino, G. & Deng, X. W. The COP9 signalosome: more than a protease. Trends Biochem Sci. 33, 592–600 (2008).
Henning, R. H. & Brundel, B. Proteostasis in cardiac health and disease. Nat. Rev. Cardiol. 14, 637–653 (2017).
Menon, S. et al. COP9 signalosome subunit 8 is essential for peripheral T cell homeostasis and antigen receptor-induced entry into the cell cycle from quiescence. Nat. Immunol. 8, 1236–1245 (2007).
Lykke-Andersen, K. et al. Disruption of the COP9 signalosome Csn2 subunit in mice causes deficient cell proliferation, accumulation of p53 and cyclin E, and early embryonic death. Mol. Cell Biol. 23, 6790–6797 (2003).
Oron, E. et al. COP9 signalosome subunits 4 and 5 regulate multiple pleiotropic pathways in Drosophila melanogaster. Development 129, 4399–4409 (2002).
Yan, J. et al. COP9 signalosome subunit 3 is essential for maintenance of cell proliferation in the mouse embryonic epiblast. Mol. Cell Biol. 23, 6798–6808 (2003).
Zhao, R. et al. Subunit 6 of the COP9 signalosome promotes tumorigenesis in mice through stabilization of MDM2 and is upregulated in human cancers. J. Clin. Investig. 121, 851–865 (2011).
Lingaraju, G. M. et al. Crystal structure of the human COP9 signalosome. Nature 512, 161–165 (2014).
King, R. W. et al. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell 81, 279–288 (1995).
Enchev, R. I. et al. Structural basis for a reciprocal regulation between SCF and CSN. Cell Rep. 2, 616–627 (2012).
Emberley, E. D., Mosadeghi, R. & Deshaies, R. J. Deconjugation of Nedd8 from Cul1 is directly regulated by Skp1-F-box and substrate, and the COP9 signalosome inhibits deneddylated SCF by a noncatalytic mechanism. J. Biol. Chem. 287, 29679–29689 (2012).
Fischer, E. S. et al. The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell 147, 1024–1039 (2011).
Chan, Y. et al. DEN1 deneddylates non-cullin proteins in vivo. J. Cell Sci. 121, 3218–3223 (2008).
Wu, K. et al. DEN1 is a dual function protease capable of processing the C terminus of Nedd8 and deconjugating hyper-neddylated CUL1. J. Biol. Chem. 278, 28882–28891 (2003).
Yu, Q., Jiang, Y. & Sun, Y. Anticancer drug discovery by targeting cullin neddylation. Acta Pharm. Sin. B 10, 746–765 (2020).
Hao, B. et al. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell 26, 131–143 (2007).
Wu, G. et al. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol. Cell 11, 1445–1456 (2003).
Orlicky, S. et al. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112, 243–256 (2003).
Merlet, J., Burger, J., Gomes, J. E. & Pintard, L. Regulation of cullin-RING E3 ubiquitin-ligases by neddylation and dimerization. Cell Mol. Life Sci. 66, 1924–1938 (2009).
Duda, D. M. et al. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134, 995–1006 (2008).
Scott, D. C. et al. Structure of a RING E3 trapped in action reveals ligation mechanism for the ubiquitin-like protein NEDD8. Cell 157, 1671–1684 (2014).
Baek, K. et al. NEDD8 nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation assembly. Nature 578, 461–466 (2020).
Horn-Ghetko, D. et al. Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly. Nature 590, 671–676 (2021).
Henneberg, L. T. et al. Activity-based profiling of cullin-RING E3 networks by conformation-specific probes. Nat. Chem. Biol. 19, 1513–1523 (2023).
Kostrhon, S. et al. CUL5-ARIH2 E3-E3 ubiquitin ligase structure reveals cullin-specific NEDD8 activation. Nat. Chem. Biol. 17, 1075–1083 (2021).
Hopf, L. V. M. et al. Structure of CRL7(FBXW8) reveals coupling with CUL1-RBX1/ROC1 for multi-cullin-RING E3-catalyzed ubiquitin ligation. Nat. Struct. Mol. Biol. 29, 854–862 (2022).
Lydeard, J. R., Schulman, B. A. & Harper, J. W. Building and remodelling Cullin-RING E3 ubiquitin ligases. EMBO Rep. 14, 1050–1061 (2013).
Goldenberg, S. J. et al. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell 119, 517–528 (2004).
Shaaban, M. et al. Structural and mechanistic insights into the CAND1-mediated SCF substrate receptor exchange. Mol. Cell 83, 2332–2346.e2338 (2023).
Baek, K. et al. Systemwide disassembly and assembly of SCF ubiquitin ligase complexes. Cell 186, 1895–1911.e1821 (2023).
Mosadeghi, R. et al. Structural and kinetic analysis of the COP9-Signalosome activation and the cullin-RING ubiquitin ligase deneddylation cycle. Elife. 5, e12102 (2016).
Cavadini, S. et al. Cullin-RING ubiquitin E3 ligase regulation by the COP9 signalosome. Nature 531, 598–603 (2016).
Faull, S. V. et al. Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome. Nat. Commun. 10, 3814 (2019).
Zhou, Q., Zheng, Y. & Sun, Y. Neddylation regulation of mitochondrial structure and functions. Cell Biosci. 11, 55 (2021).
Capece, D. et al. NF-kappaB: blending metabolism, immunity, and inflammation. Trends Immunol. 43, 757–775 (2022).
Amir, R. E., Iwai, K. & Ciechanover, A. The NEDD8 pathway is essential for SCF(beta -TrCP)-mediated ubiquitination and processing of the NF-kappa B precursor p105. J. Biol. Chem. 277, 23253–23259 (2002).
Read, M. A. et al. Nedd8 modification of cul-1 activates SCF(beta(TrCP))-dependent ubiquitination of IkappaBalpha. Mol. Cell Biol. 20, 2326–2333 (2000).
Zhu, Z. et al. Nedd8 modification of Cullin-5 regulates lipopolysaccharide-induced acute lung injury. Am. J. Physiol. Lung Cell Mol. Physiol. 313, L104–l114 (2017).
Sun, S. C. The non-canonical NF-kappaB pathway in immunity and inflammation. Nat. Rev. Immunol. 17, 545–558 (2017).
Xu, C. et al. Hepatic neddylation deficiency triggers fatal liver injury via inducing NF-κB-inducing kinase in mice. Nat. Commun. 13, 7782 (2022).
Taylor, C. T. & Scholz, C. C. The effect of HIF on metabolism and immunity. Nat. Rev. Nephrol. 18, 573–587 (2022).
Semenza, G. L. Oxygen sensing, homeostasis, and disease. N. Engl. J. Med. 365, 537–547 (2011).
Ohh, M. et al. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat. Cell Biol. 2, 423–427 (2000).
Sufan, R. I. & Ohh, M. Role of the NEDD8 modification of Cul2 in the sequential activation of ECV complex. Neoplasia 8, 956–963 (2006).
Chew, E. H. & Hagen, T. Substrate-mediated regulation of cullin neddylation. J. Biol. Chem. 282, 17032–17040 (2007).
Curtis, V. F. et al. Stabilization of HIF through inhibition of Cullin-2 neddylation is protective in mucosal inflammatory responses. Faseb j. 29, 208–215 (2015).
Wang, X. et al. AMPK promotes SPOP-mediated NANOG degradation to regulate prostate cancer cell stemness. Dev. Cell. 48, 345–360.e347 (2019).
Russell, R. C. & Ohh, M. NEDD8 acts as a ‘molecular switch’ defining the functional selectivity of VHL. EMBO Rep. 9, 486–491 (2008).
Wolf, E. R., Mabry, A. R., Damania, B. & Mayo, L. D. Mdm2-mediated neddylation of pVHL blocks the induction of antiangiogenic factors. Oncogene 39, 5228–5239 (2020).
Lemmon, M. A. & Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 (2010).
Derynck, R., Turley, S. J. & Akhurst, R. J. TGFbeta biology in cancer progression and immunotherapy. Nat. Rev. Clin. Oncol. 18, 9–34 (2021).
Oda, K., Matsuoka, Y., Funahashi, A. & Kitano, H. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol. Syst. Biol. 1, 2005.0010 (2005).
von Zastrow, M. & Sorkin, A. Mechanisms for regulating and organizing receptor signaling by endocytosis. Annu. Rev. Biochem 90, 709–737 (2021).
Oved, S. et al. Conjugation to Nedd8 instigates ubiquitylation and down-regulation of activated receptor tyrosine kinases. J. Biol. Chem. 281, 21640–21651 (2006).
Swain, S. M., Shastry, M. & Hamilton, E. Targeting HER2-positive breast cancer: advances and future directions. Nat. Rev. Drug Discov. 22, 101–126 (2023).
Xia, X. et al. Neddylation of HER2 inhibits its protein degradation and promotes breast cancer progression. Int J. Biol. Sci. 19, 377–392 (2023).
Jin, H. S., Liao, L., Park, Y. & Liu, Y. C. Neddylation pathway regulates T-cell function by targeting an adaptor protein Shc and a protein kinase Erk signaling. Proc. Natl Acad. Sci. USA 110, 624–629 (2013).
Šopin, T. et al. Lysine demethylase KDM2A promotes proteasomal degradation of TCF/LEF transcription factors in a neddylation-dependent manner. Cells. 12, 2620 (2023).
Davalos, V. & Esteller, M. Cancer epigenetics in clinical practice. CA Cancer J. Clin. 73, 376–424 (2023).
Phillips, R. E., Soshnev, A. A. & Allis, C. D. Epigenomic reprogramming as a driver of malignant glioma. Cancer Cell 38, 647–660 (2020).
Guo, L., Lee, Y. T., Zhou, Y. & Huang, Y. Targeting epigenetic regulatory machinery to overcome cancer therapy resistance. Semin. Cancer Biol. 83, 487–502 (2022).
Zhang, L., Lu, Q. & Chang, C. Epigenetics in health and disease. Adv. Exp. Med. Biol. 1253, 3–55 (2020).
Shamay, M. et al. De novo DNA methyltransferase DNMT3b interacts with NEDD8-modified proteins. J. Biol. Chem. 285, 36377–36386 (2010).
Higa, L. A. et al. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat. Cell Biol. 8, 1277–1283 (2006).
Jia, S., Kobayashi, R. & Grewal, S. I. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat. Cell Biol. 7, 1007–1013 (2005).
Marks, P. et al. Histone deacetylases and cancer: causes and therapies. Nat. Rev. Cancer 1, 194–202 (2001).
Lai, Q. Y. et al. Histone deacetylase 1 induced by neddylation inhibition contributes to drug resistance in acute myelogenous leukemia. Cell Commun. Signal 17, 86 (2019).
Pandey, D. et al. NEDDylation promotes endothelial dysfunction: a role for HDAC2. J. Mol. Cell Cardiol. 81, 18–22 (2015).
Zhou, H., Su, H. & Chen, W. Neddylation regulates class IIa and III histone deacetylases to mediate myoblast differentiation. Int. J. Mol. Sci. 22, 9590 (2021).
Nomura, Y. et al. Inhibition of HDAC6 activity protects against endothelial dysfunction and atherogenesis in vivo: a role for HDAC6 neddylation. Front. Physiol. 12, 675724 (2021).
Groelly, F. J. et al. Targeting DNA damage response pathways in cancer. Nat. Rev. Cancer 23, 78–94 (2023).
Bode, A. M. & Dong, Z. Post-translational modification of p53 in tumorigenesis. Nat. Rev. Cancer 4, 793–805 (2004).
Watson, I. R. et al. Chemotherapy induces NEDP1-mediated destabilization of MDM2. Oncogene 29, 297–304 (2010).
Blattner, C., Sparks, A. & Lane, D. Transcription factor E2F-1 is upregulated in response to DNA damage in a manner analogous to that of p53. Mol. Cell Biol. 19, 3704–3713 (1999).
Loftus, S. J. et al. NEDDylation regulates E2F-1-dependent transcription. EMBO Rep. 13, 811–818 (2012).
Blackford, A. N. & Jackson, S. P. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol. Cell 66, 801–817 (2017).
Guo, Z. et al. HUWE1-dependent DNA-PKcs neddylation modulates its autophosphorylation in DNA damage response. Cell Death Dis. 11, 400 (2020).
Zhang, Q. et al. FBXW7 facilitates nonhomologous end-joining via K63-linked polyubiquitylation of XRCC4. Mol. Cell 61, 419–433 (2016).
Li, T. et al. RNF168-mediated H2A neddylation antagonizes ubiquitylation of H2A and regulates DNA damage repair. J. Cell Sci. 127, 2238–2248 (2014).
Jiao, L. et al. Ribosome biogenesis in disease: new players and therapeutic targets. Signal Transduct. Target Ther. 8, 15 (2023).
Zhang, Y. & Lu, H. Signaling to p53: ribosomal proteins find their way. Cancer Cell 16, 369–377 (2009).
Sundqvist, A., Liu, G., Mirsaliotis, A. & Xirodimas, D. P. Regulation of nucleolar signalling to p53 through NEDDylation of L11. EMBO Rep. 10, 1132–1139 (2009).
Xirodimas, D. P. et al. Ribosomal proteins are targets for the NEDD8 pathway. EMBO Rep. 9, 280–286 (2008).
Mahata, B., Sundqvist, A. & Xirodimas, D. P. Recruitment of RPL11 at promoter sites of p53-regulated genes upon nucleolar stress through NEDD8 and in an Mdm2-dependent manner. Oncogene 31, 3060–3071 (2012).
Zhang, J. et al. hCINAP is a novel regulator of ribosomal protein-HDM2-p53 pathway by controlling NEDDylation of ribosomal protein S14. Oncogene 33, 246–254 (2014).
Xiong, X. et al. Neddylation modification of ribosomal protein RPS27L or RPS27 by MDM2 or NEDP1 regulates cancer cell survival. FASEB j. 34, 13419–13429 (2020).
Abida, W. M. et al. FBXO11 promotes the Neddylation of p53 and inhibits its transcriptional activity. J. Biol. Chem. 282, 1797–1804 (2007).
Tateishi, K., Omata, M., Tanaka, K. & Chiba, T. The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J. Cell Biol. 155, 571–579 (2001).
Zhang, L. et al. Neddylation is critical to cortical development by regulating Wnt/β-catenin signaling. Proc. Natl Acad. Sci. USA 117, 26448–26459 (2020).
Vogl, A. M. et al. Neddylation inhibition impairs spine development, destabilizes synapses and deteriorates cognition. Nat. Neurosci. 18, 239–251 (2015).
Song, J. M. et al. Deneddylating enzyme SENP8 regulates neuronal development. J. Neurochem 165, 348–361 (2023).
Zou, J. et al. Neddylation mediates ventricular chamber maturation through repression of Hippo signaling. Proc. Natl Acad. Sci. USA 115, E4101–E4110 (2018).
Zou, J. et al. Neddylation is required for perinatal cardiac development through stimulation of metabolic maturation. Cell Rep. 42, 112018 (2023).
Tan, M. et al. RBX1/ROC1 disruption results in early embryonic lethality due to proliferation failure, partially rescued by simultaneous loss of p27. Proc. Natl Acad. Sci. USA 106, 6203–6208 (2009).
Tan, M. et al. SAG/RBX2/ROC2 E3 ubiquitin ligase is essential for vascular and neural development by targeting NF1 for degradation. Dev. Cell 21, 1062–1076 (2011).
Dunwoodie, S. L. The role of hypoxia in development of the Mammalian embryo. Dev. Cell 17, 755–773 (2009).
Tan, M. et al. SAG/ROC2/RBX2 is a HIF-1 target gene that promotes HIF-1 alpha ubiquitination and degradation. Oncogene 27, 1404–1411 (2008).
Tan, M., Li, H. & Sun, Y. Endothelial deletion of Sag/Rbx2/Roc2 E3 ubiquitin ligase causes embryonic lethality and blocks tumor angiogenesis. Oncogene 33, 5211–5220 (2014).
Chang, S. C. & Ding, J. L. Ubiquitination by SAG regulates macrophage survival/death and immune response during infection. Cell Death Differ. 21, 1388–1398, (2014).
Ehrentraut, S. F. et al. Central role for endothelial human deneddylase-1/SENP8 in fine-tuning the vascular inflammatory response. J. Immunol. 190, 392–400 (2013).
Yu, G. et al. Neddylation facilitates the antiviral response in zebrafish. Front. Immunol. 10, 1432 (2019).
Li, C. et al. RNF111-facilitated neddylation potentiates cGAS-mediated antiviral innate immune response. PLoS Pathog. 17, e1009401 (2021).
Kong, X. et al. Type I interferon/STAT1 signaling regulates UBE2M-mediated antiviral innate immunity in a negative feedback manner. Cell Rep. 42, 112002 (2023).
Zhu, J. et al. Association between neddylation and immune response. Front. Cell Dev. Biol. 10, 890121 (2022).
Kolaczkowska, E. & Kubes, P. Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 13, 159–175 (2013).
Xiong, X. et al. SAG/RBX2 E3 ubiquitin ligase differentially regulates inflammatory responses of myeloid cell subsets. Front. Immunol. 9, 2882 (2018).
Jin, J. et al. MLN4924 suppresses lipopolysaccharide-induced proinflammatory cytokine production in neutrophils in a dose-dependent manner. Oncol. Lett. 15, 8039–8045 (2018).
Asare, Y. et al. Inhibition of atherogenesis by the COP9 signalosome subunit 5 in vivo. Proc. Natl Acad. Sci. USA 114, E2766–e2775 (2017).
Yu, H. et al. The NEDD8-activating enzyme inhibitor MLN4924 reduces ischemic brain injury in mice. Proc. Natl Acad. Sci. USA 119, e2111896119 (2022).
Li, L. et al. Neddylation pathway regulates the proliferation and survival of macrophages. Biochem. Biophys. Res. Commun. 432, 494–498 (2013).
Chang, F. M. et al. Inhibition of neddylation represses lipopolysaccharide-induced proinflammatory cytokine production in macrophage cells. J. Biol. Chem. 287, 35756–35767 (2012).
Lu, X. et al. UBE2M-mediated neddylation of TRIM21 regulates obesity-induced inflammation and metabolic disorders. Cell Metab. 35, 1390–1405.e1398 (2023).
Zhou, L. et al. Promotion of tumor-associated macrophages infiltration by elevated neddylation pathway via NF-kappaB-CCL2 signaling in lung cancer. Oncogene 38, 5792–5804 (2019).
Lin, Y. et al. Neddylation pathway alleviates chronic pancreatitis by reducing HIF1α-CCL5-dependent macrophage infiltration. Cell Death Dis. 12, 273 (2021).
Banchereau, J. & Steinman, R. M. Dendritic cells and the control of immunity. Nature 392, 245–252 (1998).
Mathewson, N. et al. Neddylation plays an important role in the regulation of murine and human dendritic cell function. Blood 122, 2062–2073 (2013).
Chadha, A. et al. Suppressive role of neddylation in dendritic cells during Mycobacterium tuberculosis infection. Tuberculosis 95, 599–607 (2015).
Cheng, M. et al. Inhibition of neddylation regulates dendritic cell functions via Deptor accumulation driven mTOR inactivation. Oncotarget 7, 35643–35654 (2016).
El-Mesery, M. et al. MLN4924 sensitizes monocytes and maturing dendritic cells for TNF-dependent and -independent necroptosis. Br. J. Pharm. 172, 1222–1236 (2015).
Zhao, Y., Xiong, X. & Sun, Y. DEPTOR, an mTOR inhibitor, is a physiological substrate of SCF(betaTrCP) E3 ubiquitin ligase and regulates survival and autophagy. Mol. Cell 44, 304–316 (2011).
Cao, W. et al. Toll-like receptor-mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6K pathway. Nat. Immunol. 9, 1157–1164 (2008).
Thomson, A. W., Turnquist, H. R. & Raimondi, G. Immunoregulatory functions of mTOR inhibition. Nat. Rev. Immunol. 9, 324–337 (2009).
Smith-Garvin, J. E., Koretzky, G. A. & Jordan, M. S. T cell activation. Annu. Rev. Immunol. 27, 591–619 (2009).
Joyce, J. A. & Fearon, D. T. T cell exclusion, immune privilege, and the tumor microenvironment. Science 348, 74–80 (2015).
Cheng, Q. et al. Neddylation contributes to CD4+ T cell-mediated protective immunity against blood-stage plasmodium infection. PLoS Pathog. 14, e1007440 (2018).
Mathewson, N. D. et al. SAG/Rbx2-dependent neddylation regulates T-cell responses. Am. J. Pathol. 186, 2679–2691 (2016).
Jang, I. K. et al. Grb2 functions at the top of the T-cell antigen receptor-induced tyrosine kinase cascade to control thymic selection. Proc. Natl Acad. Sci. USA 107, 10620–10625 (2010).
Fischer, A. M. et al. The role of erk1 and erk2 in multiple stages of T cell development. Immunity 23, 431–443 (2005).
Zhang, Y. et al. Neddylation is a novel therapeutic target for lupus by regulating double-negative T-cell homeostasis. Signal. Transduct. Target Ther. 9, 18 (2024).
Xiao, N. et al. The E3 ubiquitin ligase Itch is required for the differentiation of follicular helper T cells. Nat. Immunol. 15, 657–666 (2014).
Hori, S., Nomura, T. & Sakaguchi, S. Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003).
Wu, D. et al. The Ube2m-Rbx1 neddylation-Cullin-RING-Ligase proteins are essential for the maintenance of Regulatory T cell fitness. Nat Commun. 13, 3021 (2022).
Wu, D. & Sun, Y. Neddylation-CRLs regulate the functions of Treg immune cells. Bioessays. 45, e2200222 (2023).
Wu, D. & Sun, Y. The Functional Redundancy of Neddylation E2s and E3s in Modulating the Fitness of Regulatory T Cells. Research (Wash D C). 6, 0212 (2023).
Yada, M. et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. Embo j. 23, 2116–2125 (2004).
Yin, Y. et al. The FBXW2-MSX2-SOX2 axis regulates stem cell property and drug resistance of cancer cells. Proc Natl Acad Sci USA 116, 20528–20538 (2019).
Matsumoto, A. et al. Fbxw7-dependent degradation of Notch is required for control of “stemness” and neuronal-glial differentiation in neural stem cells. J. Biol. Chem. 286, 13754–13764 (2011).
Buckley, S. M. et al. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell 11, 783–798 (2012).
Takeishi, S. & Nakayama, K. I. Role of Fbxw7 in the maintenance of normal stem cells and cancer-initiating cells. Br. J. Cancer 111, 1054–1059 (2014).
Tan, M. et al. Inactivation of SAG E3 ubiquitin ligase blocks embryonic stem cell differentiation and sensitizes leukemia cells to retinoid acid. PloS One 6, e27726 (2011).
Thompson, B. J. et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J. Exp. Med. 204, 1825–1835 (2007).
O’Neil, J. et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J. Exp. Med. 204, 1813–1824 (2007).
King, B. et al. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell 153, 1552–1566 (2013).
Takeishi, S. et al. Ablation of Fbxw7 eliminates leukemia-initiating cells by preventing quiescence. Cancer Cell 23, 347–361 (2013).
Onoyama, I. et al. Fbxw7 regulates lipid metabolism and cell fate decisions in the mouse liver. J. Clin. Investig. 121, 342–354 (2011).
Mao, J. H. et al. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science 321, 1499–1502 (2008).
Zhang, S., Xiong, X. & Sun, Y. Functional characterization of SOX2 as an anticancer target. Signal Transduct Target Ther. 5, 135 (2020).
Zhang, S. & Sun, Y. Targeting oncogenic SOX2 in human cancer cells: therapeutic application. Protein Cell 11, 82–84 (2020).
Westbrook, T. F. et al. SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature 452, 370–374 (2008).
Ballas, N. et al. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 121, 645–657 (2005).
Takubo, K. et al. Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell 7, 391–402 (2010).
Simsek, T. et al. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell 7, 380–390 (2010).
Chambers, I. et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113, 643–655 (2003).
Zhang, C. et al. Proteolysis of methylated SOX2 protein is regulated by L3MBTL3 and CRL4(DCAF5) ubiquitin ligase. J. Biol. Chem. 294, 476–489 (2019).
Cui, C. P. et al. Dynamic ubiquitylation of Sox2 regulates proteostasis and governs neural progenitor cell differentiation. Nat. Commun. 9, 4648 (2018).
Ferguson, J. E. et al. ASB4 is a hydroxylation substrate of FIH and promotes vascular differentiation via an oxygen-dependent mechanism. Mol. Cell Biol. 27, 6407–6419 (2007).
Townley-Tilson, W. H., Wu, Y., Ferguson, J. E. 3rd & Patterson, C. The ubiquitin ligase ASB4 promotes trophoblast differentiation through the degradation of ID2. PloS One 9, e89451 (2014).
Zhou, X. et al. Blockage of neddylation modification stimulates tumor sphere formation in vitro and stem cell differentiation and wound healing in vivo. Proc. Natl Acad. Sci. USA 113, E2935–E2944 (2016).
Zeng, R. et al. Neddylation suppression by a macrophage membrane-coated nanoparticle promotes dual immunomodulatory repair of diabetic wounds. Bioact. Mater. 34, 366–380 (2024).
Porporato, P. E. et al. Mitochondrial metabolism and cancer. Cell Res. 28, 265–280 (2018).
Chen, W., Zhao, H. & Li, Y. Mitochondrial dynamics in health and disease: mechanisms and potential targets. Signal Transduct. Target Ther. 8, 333 (2023).
Zhou, Q. et al. Inhibiting neddylation modification alters mitochondrial morphology and reprograms energy metabolism in cancer cells. JCI Insight. 4, e121582 (2019).
Zhang, X. et al. Hepatic neddylation targets and stabilizes electron transfer flavoproteins to facilitate fatty acid beta-oxidation. Proc. Natl Acad. Sci. USA 117, 2473–2483 (2020).
Zorov, D. B., Juhaszova, M. & Sollott, S. J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 94, 909–950 (2014).
Nakamura, H. & Takada, K. Reactive oxygen species in cancer: current findings and future directions. Cancer Sci. 112, 3945–3952 (2021).
Kensler, T. W., Wakabayashi, N. & Biswal, S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharm. Toxicol. 47, 89–116 (2007).
Adinolfi, S. et al. The KEAP1-NRF2 pathway: targets for therapy and role in cancer. Redox Biol. 63, 102726 (2023).
Baird, L. & Yamamoto, M. The molecular mechanisms regulating the KEAP1-NRF2 pathway. Mol Cell Biol. 40, e00099–20 (2020).
Gonzalez-Rellan, M. J. et al. Neddylation of phosphoenolpyruvate carboxykinase 1 controls glucose metabolism. Cell Metab. 35, 1630–1645.e1635 (2023).
Chen, K. et al. Regulation of glucose metabolism by p62/SQSTM1 through HIF1α. J. Cell Sci. 129, 817–830 (2016).
Dayton, T. L., Jacks, T. & Vander Heiden, M. G. PKM2, cancer metabolism, and the road ahead. EMBO Rep. 17, 1721–1730 (2016).
Altman, B. J., Stine, Z. E. & Dang, C. V. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat. Rev. Cancer 16, 619–634 (2016).
Zhou, Q. et al. Neddylation inhibition induces glutamine uptake and metabolism by targeting CRL3(SPOP) E3 ligase in cancer cells. Nat. Commun. 13, 3034 (2022).
Nadler, S. T. et al. The expression of adipogenic genes is decreased in obesity and diabetes mellitus. Proc. Natl Acad. Sci. USA 97, 11371–11376 (2000).
Montaigne, D., Butruille, L. & Staels, B. PPAR control of metabolism and cardiovascular functions. Nat. Rev. Cardiol. 18, 809–823 (2021).
Park, H. S. et al. PPARgamma neddylation essential for adipogenesis is a potential target for treating obesity. Cell Death Differ. 23, 1296–1311 (2016).
Ge, M. et al. MLN4924 treatment diminishes excessive lipid storage in high-fat diet-induced non-alcoholic fatty liver disease (NAFLD) by stimulating hepatic mitochondrial fatty acid oxidation and lipid metabolites. Pharmaceutics 14, 2460 (2022).
Ferre, P., Phan, F. & Foufelle, F. SREBP-1c and lipogenesis in the liver: an update1. Biochem. J. 478, 3723–3739 (2021).
Bence, K. K. & Birnbaum, M. J. Metabolic drivers of non-alcoholic fatty liver disease. Mol. Metab. 50, 101143 (2021).
Ju, U. I. et al. Neddylation of sterol regulatory element-binding protein 1c is a potential therapeutic target for nonalcoholic fatty liver treatment. Cell Death Dis. 11, 283 (2020).
Xie, P. et al. Neddylation of PTEN regulates its nuclear import and promotes tumor development. Cell Res. 31, 291–311 (2021).
Li, J. et al. Neddylation, an emerging mechanism regulating cardiac development and function. Front. Physiol. 11, 612927 (2020).
Stower, H. Personalizing metabolic disease therapies. Nat. Med. 25, 197 (2019).
Wu, Y. L. et al. Epigenetic regulation in metabolic diseases: mechanisms and advances in clinical study. Signal Transduct. Target Ther. 8, 98 (2023).
Newgard, C. B. Metabolomics and metabolic diseases: where do we stand? Cell Metab. 25, 43–56 (2017).
Yu, M. et al. Emerging role of NEDD8-mediated neddylation in age-related metabolic diseases. Ageing Res. Rev. 94, 102191 (2024).
Rohm, T. V., Meier, D. T., Olefsky, J. M. & Donath, M. Y. Inflammation in obesity, diabetes, and related disorders. Immunity 55, 31–55 (2022).
Biddinger, S. B. et al. Hepatic insulin resistance is sufficient to produce dyslipidemia and susceptibility to atherosclerosis. Cell Metab. 7, 125–134 (2008).
Michael, M. D. et al. Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Mol. Cell 6, 87–97 (2000).
Chen, C. et al. Cullin neddylation inhibitor attenuates hyperglycemia by enhancing hepatic insulin signaling through insulin receptor substrate stabilization. Proc. Natl Acad. Sci. USA. 119, e2111737119 (2022).
Bjorkegren, J. L. M. & Lusis, A. J. Atherosclerosis: recent developments. Cell 185, 1630–1645 (2022).
Asare, Y. et al. Endothelial CSN5 impairs NF-κB activation and monocyte adhesion to endothelial cells and is highly expressed in human atherosclerotic lesions. Thromb. Haemost. 110, 141–152 (2013).
Sharma, A. & Nagalli, S. Chronic liver disease. in StatPearls. (2023).
Zubiete-Franco, I. et al. Deregulated neddylation in liver fibrosis. Hepatology 65, 694–709 (2017).
Schmidt, M. H. H. & Dikic, I. Ubiquitin and NEDD8: brothers in arms. Sci. STKE 2006, pe50 (2006).
Hammerich, L. & Tacke, F. Hepatic inflammatory responses in liver fibrosis. Nat. Rev. Gastroenterol. Hepatol. 20, 633–646 (2023).
Zhao, F. et al. The functions of hepatitis B virus encoding proteins: viral persistence and liver pathogenesis. Front. Immunol. 12, 691766 (2021).
Sekiba, K. et al. Inhibition of HBV transcription from cccDNA with nitazoxanide by targeting the HBx-DDB1 interaction. Cell Mol. Gastroenterol. Hepatol. 7, 297–312 (2019).
Bontron, S., Lin-Marq, N. & Strubin, M. Hepatitis B virus X protein associated with UV-DDB1 induces cell death in the nucleus and is functionally antagonized by UV-DDB2. J. Biol. Chem. 277, 38847–38854 (2002).
Guo, L. et al. HBx affects CUL4-DDB1 function in both positive and negative manners. Biochem. Biophys. Res Commun. 450, 1492–1497 (2014).
Yuan, H. et al. HBx represses WDR77 to enhance HBV replication by DDB1-mediated WDR77 degradation in the liver. Theranostics 11, 8362–8378 (2021).
Murphy, C. M. et al. Hepatitis B Virus X Protein Promotes Degradation of SMC5/6 to Enhance HBV Replication. Cell Rep. 16, 2846–2854 (2016).
Liu, N. et al. HDM2 Promotes NEDDylation of hepatitis B virus HBX to enhance its stability and function. J Virol. 91, e00340–17 (2017).
Xie, M. et al. Neddylation inhibitor MLN4924 has anti-HBV activity via modulating the ERK-HNF1alpha-C/EBPalpha-HNF4alpha axis. J. Cell Mol. Med. 25, 840–854 (2021).
Powell, E. E., Wong, V. W. & Rinella, M. Non-alcoholic fatty liver disease. Lancet 397, 2212–2224 (2021).
Sen, S., Jumaa, H. & Webster, N. J. Splicing factor SRSF3 is crucial for hepatocyte differentiation and metabolic function. Nat. Commun. 4, 1336 (2013).
Sen, S., Langiewicz, M., Jumaa, H. & Webster, N. J. Deletion of serine/arginine-rich splicing factor 3 in hepatocytes predisposes to hepatocellular carcinoma in mice. Hepatology 61, 171–183 (2015).
Kumar, D. et al. Degradation of splicing factor SRSF3 contributes to progressive liver disease. J. Clin. Investig. 129, 4477–4491 (2019).
Luedde, T. & Schwabe, R. F. NF-kappaB in the liver–linking injury, fibrosis and hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 8, 108–118 (2011).
Troeger, J. S. et al. Deactivation of hepatic stellate cells during liver fibrosis resolution in mice. Gastroenterology 143, 1073–1083.e1022 (2012).
Li, R. et al. Neddylation of EphB1 regulates its activity and associates with liver fibrosis. Int. J. Mol. Sci. 24, 3415 (2023).
Yao, J., Liang, X., Liu, Y. & Zheng, M. Neddylation: a versatile pathway takes on chronic liver diseases. Front. Med. 7, 586881 (2020).
He, X., Zhu, A., Feng, J. & Wang, X. Role of neddylation in neurological development and diseases. Biotechnol. Appl. Biochem. 69, 330–341 (2022).
Yamazaki, Y. et al. Apolipoprotein E and Alzheimer disease: pathobiology and targeting strategies. Nat. Rev. Neurol. 15, 501–518 (2019).
Schmidt, M. F., Gan, Z. Y., Komander, D. & Dewson, G. Ubiquitin signalling in neurodegeneration: mechanisms and therapeutic opportunities. Cell Death Differ. 28, 570–590 (2021).
Chen, Y., Neve, R. L. & Liu, H. Neddylation dysfunction in Alzheimer’s disease. J. Cell Mol. Med. 16, 2583–2591 (2012).
Chen, Y. et al. APP-BP1 mediates APP-induced apoptosis and DNA synthesis and is increased in Alzheimer’s disease brain. J. Cell Biol. 163, 27–33 (2003).
Li, J. et al. SEL-10 interacts with presenilin 1, facilitates its ubiquitination, and alters A-beta peptide production. J. Neurochem. 82, 1540–1548 (2002).
Berg, D. et al. Prodromal Parkinson disease subtypes—key to understanding heterogeneity. Nat. Rev. Neurol. 17, 349–361 (2021).
Kitada, T. et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608 (1998).
Xiong, H. et al. Parkin, PINK1, and DJ-1 form a ubiquitin E3 ligase complex promoting unfolded protein degradation. J. Clin. Investig. 119, 650–660 (2009).
Choo, Y. S. et al. Regulation of parkin and PINK1 by neddylation. Hum. Mol. Genet 21, 2514–2523 (2012).
Um, J. W. et al. Neddylation positively regulates the ubiquitin E3 ligase activity of parkin. J. Neurosci. Res. 90, 1030–1042 (2012).
Tsao, C. W. et al. Heart Disease and Stroke Statistics—2022 update: a report from the American Heart Association. Circulation 145, e153–e639 (2022).
Zou, J. et al. Transient inhibition of neddylation at neonatal stage evokes reversible cardiomyopathy and predisposes the heart to isoproterenol-induced heart failure. Am. J. Physiol. Heart Circ. Physiol. 316, H1406–h1416 (2019).
Su, H. et al. Perturbation of cullin deneddylation via conditional Csn8 ablation impairs the ubiquitin-proteasome system and causes cardiomyocyte necrosis and dilated cardiomyopathy in mice. Circ. Res. 108, 40–50 (2011).
Su, H. et al. The COP9 signalosome is required for autophagy, proteasome-mediated proteolysis, and cardiomyocyte survival in adult mice. Circ. Heart Fail. 6, 1049–1057 (2013).
Del Re, D. P. et al. Fundamental mechanisms of regulated cell death and implications for heart disease. Physiol. Rev. 99, 1765–1817 (2019).
Xiao, P. et al. COP9 signalosome suppresses RIPK1-RIPK3-mediated cardiomyocyte necroptosis in mice. Circ. Heart Fail. 13, e006996 (2020).
Mao, H., Lin, X. & Sun, Y. Neddylation regulation of immune responses. Research 6, 0283 (2023).
Zhang, X. et al. Advances in the potential roles of Cullin-RING ligases in regulating autoimmune diseases. Front. Immunol. 14, 1125224 (2023).
Kayesh, M. E. H., Kohara, M. & Tsukiyama-Kohara, K. Effects of neddylation on viral infection: an overview. Arch. Virol. 169, 6 (2023).
Zhao, Y., Xiong, X. & Sun, Y. Cullin-RING Ligase 5: functional characterization and its role in human cancers. Semin. Cancer Biol. 67, 61–79 (2020).
Nekorchuk, M. D. et al. HIV relies on neddylation for ubiquitin ligase-mediated functions. Retrovirology 10, 138 (2013).
Zhang, X. et al. Neddylation is required for herpes simplex virus type I (HSV-1)-induced early phase interferon-beta production. Cell Mol. Immunol. 13, 578–583 (2016).
Zhang, L. et al. CRL4B E3 ligase recruited by PRPF19 inhibits SARS-CoV-2 infection by targeting ORF6 for ubiquitin-dependent degradation. mBio 15, e0307123 (2024).
Zhang, T. et al. NEDDylation of PB2 reduces its stability and blocks the replication of influenza A virus. Sci. Rep. 7, 43691 (2017).
Wang, H. et al. Neddylation of enterovirus 71 VP2 protein reduces its stability and restricts viral replication. J. Virol. 96, e0059822 (2022).
Fu, Z. et al. Inhibition of neddylation plays protective role in lipopolysaccharide-induced kidney damage through CRL-mediated NF-κB pathways. Am. J. Transl. Res. 11, 2830–2842 (2019).
He, Z. X. et al. Targeting cullin neddylation for cancer and fibrotic diseases. Theranostics 13, 5017–5056 (2023).
Li, L. et al. Validation of NEDD8-conjugating enzyme UBC12 as a new therapeutic target in lung cancer. EBioMedicine 45, 81–91 (2019).
Li, L. et al. Overactivated neddylation pathway as a therapeutic target in lung cancer. J. Natl Cancer Inst. 106, dju083 (2014).
Liu, H. et al. UBE1C is upregulated and promotes neddylation of p53 in lung cancer. FASEB J. 37, e23181 (2023).
Zhou, L. et al. NEDD8-conjugating enzyme E2 UBE2F confers radiation resistance by protecting lung cancer cells from apoptosis. J. Zhejiang Univ. Sci. B 22, 959–965 (2021).
Sarkaria, I. et al. Squamous cell carcinoma related oncogene/DCUN1D1 is highly conserved and activated by amplification in squamous cell carcinomas. Cancer Res. 66, 9437–9444 (2006).
Jia, L., Soengas, M. S. & Sun, Y. ROC1/RBX1 E3 ubiquitin ligase silencing suppresses tumor cell growth via sequential induction of G2-M arrest, apoptosis, and senescence. Cancer Res. 69, 4974–4982 (2009).
Jiang, Y. et al. Effective targeting of the ubiquitin-like modifier NEDD8 for lung adenocarcinoma treatment. Cell Biol. Toxicol. 36, 349–364 (2020).
Zhou, L. et al. Neddylation pathway promotes myeloid-derived suppressor cell infiltration via NF-κB-mCXCL5 signaling in lung cancer. Int. Immunopharmacol. 113, 109329 (2022).
Zhou, W. et al. Neddylation E2 UBE2F promotes the survival of lung cancer cells by activating CRL5 to degrade NOXA via the K11 linkage. Clin. Cancer Res. 23, 1104–1116 (2017).
Heo, M. J. et al. UBC12-mediated SREBP-1 neddylation worsens metastatic tumor prognosis. Int. J. Cancer 147, 2550–2563 (2020).
Jia, X. et al. Neddylation inactivation facilitates FOXO3a nuclear export to suppress estrogen receptor transcription and improve fulvestrant sensitivity. Clin. Cancer Res. 25, 3658–3672 (2019).
Naik, S. K. et al. NEDDylation negatively regulates ERRβ expression to promote breast cancer tumorigenesis and progression. Cell Death Dis. 11, 703 (2020).
Zhu, T. et al. Neddylation controls basal MKK7 kinase activity in breast cancer cells. Oncogene 35, 2624–2633 (2016).
Du, M. G. et al. Neddylation modification of the U3 snoRNA-binding protein RRP9 by Smurf1 promotes tumorigenesis. J. Biol. Chem. 297, 101307 (2021).
Embade, N. et al. Murine double minute 2 regulates Hu antigen R stability in human liver and colon cancer through NEDDylation. Hepatology 55, 1237–1248 (2012).
Li, Y. et al. SHP2 deneddylation mediates tumor immunosuppression in colon cancer via the CD47/SIRPα axis. J. Clin. Investig. 133, e162870 (2023).
Gao, Q. et al. Neddylation pathway is up-regulated in human intrahepatic cholangiocarcinoma and serves as a potential therapeutic target. Oncotarget 5, 7820–7832 (2014).
Barbier-Torres, L. et al. Stabilization of LKB1 and Akt by neddylation regulates energy metabolism in liver cancer. Oncotarget 6, 2509–2523 (2015).
Yu, J. et al. Overactivated neddylation pathway in human hepatocellular carcinoma. Cancer Med. 7, 3363–3372 (2018).
Xu, J. et al. The neddylation-cullin 2-RBX1 E3 ligase axis targets tumor suppressor RhoB for degradation in liver cancer. Mol. Cell Proteom. 14, 499–509 (2015).
Xian, J. et al. Overexpressed NEDD8 as a potential therapeutic target in esophageal squamous cell carcinoma. Cancer Biol. Med. 19, 504–517 (2021).
Chen, P. et al. Neddylation inhibition activates the extrinsic apoptosis pathway through ATF4-CHOP-DR5 axis in human esophageal cancer cells. Clin. Cancer Res. 22, 4145–4157 (2016).
Hua, W. et al. Suppression of glioblastoma by targeting the overactivated protein neddylation pathway. Neuro Oncol. 17, 1333–1343 (2015).
Xu, J. et al. Pevonedistat suppresses pancreatic cancer growth via inactivation of the neddylation pathway. Front. Oncol. 12, 822039 (2022).
Zhang, Y. et al. MLN4924 suppresses neddylation and induces cell cycle arrest, senescence, and apoptosis in human osteosarcoma. Oncotarget 7, 45263–45274 (2016).
Zhang, Q. et al. Transgenic expression of Sag/Rbx2 E3 causes early stage tumor promotion, late stage cytogenesis and acinar loss in the Kras-PDAC model. Neoplasia 22, 242–252 (2020).
Xie, P. et al. Promoting tumorigenesis in nasopharyngeal carcinoma, NEDD8 serves as a potential theranostic target. Cell Death Dis. 8, e2834 (2017).
Li, H. et al. Inactivation of SAG/RBX2 E3 ubiquitin ligase suppresses KrasG12D-driven lung tumorigenesis. J. Clin. Investig. 124, 835–846 (2014).
Li, Y., Wu, D., Li, H. & Sun, Y. Inactivation of Rbx1 E3 ligase suppresses Kras(G12D)-driven lung tumorigenesis. MedComm 4, e332 (2023).
Fu, D. J. & Wang, T. Targeting NEDD8-activating enzyme for cancer therapy: developments, clinical trials, challenges and future research directions. J. Hematol. Oncol. 16, 87 (2023).
Zheng, Y. C. et al. Targeting neddylation E2s: a novel therapeutic strategy in cancer. J. Hematol. Oncol. 14, 57 (2021).
Brownell, J. E. et al. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol. Cell 37, 102–111 (2010).
Luo, Z. et al. The Nedd8-activating enzyme inhibitor MLN4924 induces autophagy and apoptosis to suppress liver cancer cell growth. Cancer Res. 72, 3360–3371 (2012).
Swords, R. T. et al. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood 115, 3796–3800 (2010).
Milhollen, M. A. et al. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-kappaB-dependent lymphoma. Blood 116, 1515–1523 (2010).
Zhou, X. et al. Neddylation inactivation represses androgen receptor transcription and inhibits growth, survival and invasion of prostate cancer cells. Neoplasia 22, 192–202 (2020).
Aubry, A., Yu, T. & Bremner, R. Preclinical studies reveal MLN4924 is a promising new retinoblastoma therapy. Cell Death Discov. 6, 2 (2020).
Qin, A. et al. A phase II trial of pevonedistat and docetaxel in patients with previously treated advanced non-small-cell lung cancer. Clin. Lung Cancer 25, 128–134 (2023).
Sarantopoulos, J. et al. Phase I study of the investigational NEDD8-activating enzyme inhibitor pevonedistat (TAK-924/MLN4924) in patients with advanced solid tumors. Clin. Cancer Res. 22, 847–857 (2016).
Shah, J. J. et al. Phase I study of the novel investigational NEDD8-activating enzyme inhibitor pevonedistat (MLN4924) in patients with relapsed/refractory multiple myeloma or lymphoma. Clin. Cancer Res. 22, 34–43 (2016).
Swords, R. T. et al. Pevonedistat (MLN4924), a First-in-Class NEDD8-activating enzyme inhibitor, in patients with acute myeloid leukaemia and myelodysplastic syndromes: a phase 1 study. Br. J. Haematol. 169, 534–543 (2015).
Bhatia, S. et al. A phase I study of the investigational NEDD8-activating enzyme inhibitor pevonedistat (TAK-924/MLN4924) in patients with metastatic melanoma. Investig. N. Drugs 34, 439–449 (2016).
Chen, Y. N. et al. The neddylation inhibitor MLN4924 inhibits proliferation and triggers apoptosis of oral cancer cells but not for normal cells. Environ. Toxicol. 39, 299–313 (2023).
Short, N. J. et al. A phase 1/2 study of azacitidine, venetoclax and pevonedistat in newly diagnosed secondary AML and in MDS or CMML after failure of hypomethylating agents. J. Hematol. Oncol. 16, 73 (2023).
Takeda announces US FDA breakthrough therapy designation granted for pevonedistat for the treatment of patients with higher-risk myelodysplastic syndromes (HR-MDS). Takeda Pharmaceutical Company Limited. News Release, (2020).
Matthews, H. K., Bertoli, C. & de Bruin, R. A. M. Cell cycle control in cancer. Nat. Rev. Mol. Cell Biol. 23, 74–88 (2022).
Hinz, M. et al. NF-kappaB function in growth control: regulation of cyclin D1 expression and G0/G1-to-S-phase transition. Mol. Cell Biol. 19, 2690–2698 (1999).
Czuczman, N. M. et al. Pevonedistat, a NEDD8-activating enzyme inhibitor, is active in mantle cell lymphoma and enhances rituximab activity in vivo. Blood 127, 1128–1137 (2016).
Nishitani, H. et al. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 25, 1126–1136 (2006).
Milhollen, M. A. et al. Inhibition of NEDD8-activating enzyme induces rereplication and apoptosis in human tumor cells consistent with deregulating CDT1 turnover. Cancer Res. 71, 3042–3051 (2011).
Watanabe, N. et al. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFbeta-TrCP. Proc. Natl Acad. Sci. USA 101, 4419–4424 (2004).
Lan, H., Tang, Z., Jin, H. & Sun, Y. Neddylation inhibitor MLN4924 suppresses growth and migration of human gastric cancer cells. Sci. Rep. 6, 24218 (2016).
Han, K. et al. The NEDD8-activating enzyme inhibitor MLN4924 induces G2 arrest and apoptosis in T-cell acute lymphoblastic leukemia. Oncotarget 7, 23812–23824 (2016).
Li, H. et al. Inhibition of Neddylation Modification Sensitizes Pancreatic Cancer Cells to Gemcitabine. Neoplasia 19, 509–518 (2017).
Wei, D. et al. Radiosensitization of human pancreatic cancer cells by MLN4924, an investigational NEDD8-activating enzyme inhibitor. Cancer Res. 72, 282–293 (2012).
Tong, S. et al. MLN4924 (Pevonedistat), a protein neddylation inhibitor, suppresses proliferation and migration of human clear cell renal cell carcinoma. Sci. Rep. 7, 5599 (2017).
Kuo, K. L. et al. MLN4924, a novel protein neddylation inhibitor, suppresses proliferation and migration of human urothelial carcinoma: In vitro and in vivo studies. Cancer Lett. 363, 127–136 (2015).
Mackintosh, C. et al. WEE1 accumulation and deregulation of S-phase proteins mediate MLN4924 potent inhibitory effect on Ewing sarcoma cells. Oncogene 32, 1441–1451 (2013).
Bertheloot, D., Latz, E. & Franklin, B. S. Necroptosis, pyroptosis and apoptosis: an intricate game of cell death. Cell Mol. Immunol. 18, 1106–1121 (2021).
Godbersen, J. C. et al. The Nedd8-activating enzyme inhibitor MLN4924 thwarts microenvironment-driven NF-kappaB activation and induces apoptosis in chronic lymphocytic leukemia B cells. Clin. Cancer Res. 20, 1576–1589 (2014).
Khalife, J. et al. Pharmacological targeting of miR-155 via the NEDD8-activating enzyme inhibitor MLN4924 (Pevonedistat) in FLT3-ITD acute myeloid leukemia. Leukemia 29, 1981–1992 (2015).
Jia, L. et al. Validation of SAG/RBX2/ROC2 E3 ubiquitin ligase as an anticancer and radiosensitizing target. Clin. Cancer Res. 16, 814–824 (2010).
Knorr, K. L. et al. MLN4924 induces Noxa upregulation in acute myelogenous leukemia and synergizes with Bcl-2 inhibitors. Cell Death Differ. 22, 2133–2142 (2015).
Jia, L., Li, H. & Sun, Y. Induction of p21-dependent senescence by an NAE inhibitor, MLN4924, as a mechanism of growth suppression. Neoplasia 13, 561–569 (2011).
Lin, J. J. et al. NEDD8-targeting drug MLN4924 elicits DNA rereplication by stabilizing Cdt1 in S phase, triggering checkpoint activation, apoptosis, and senescence in cancer cells. Cancer Res. 70, 10310–10320 (2010).
Di Micco, R., Krizhanovsky, V., Baker, D. & d’Adda di Fagagna, F. Cellular senescence in ageing: from mechanisms to therapeutic opportunities. Nat. Rev. Mol. Cell Biol. 22, 75–95 (2021).
Wang, Y. et al. Targeting protein neddylation with an NEDD8-activating enzyme inhibitor MLN4924 induced apoptosis or senescence in human lymphoma cells. Cancer Biol. Ther. 16, 420–429 (2015).
Huang, J. et al. NEDD8 inhibition overcomes CKS1B-induced drug resistance by upregulation of p21 in multiple myeloma. Clin. Cancer Res. 21, 5532–5542 (2015).
Misra, S. et al. Both BRCA1-wild type and -mutant triple-negative breast cancers show sensitivity to the NAE inhibitor MLN4924 which is enhanced upon MLN4924 and cisplatin combination treatment. Oncotarget 11, 784–800 (2020).
Qin, Y. et al. Autophagy and cancer drug resistance in dialogue: preclinical and clinical evidence. Cancer Lett. 570, 216307 (2023).
Zhao, Y., Xiong, X., Jia, L. & Sun, Y. Targeting Cullin-RING ligases by MLN4924 induces autophagy via modulating the HIF1-REDD1-TSC1-mTORC1-DEPTOR axis. Cell Death Dis. 3, e386 (2012).
Luo, Z. et al. Inactivation of the Cullin (CUL)-RING E3 ligase by the NEDD8-activating enzyme inhibitor MLN4924 triggers protective autophagy in cancer cells. Autophagy 8, 1677–1679 (2012).
Gao, D. et al. mTOR drives its own activation via SCF(betaTrCP)-dependent degradation of the mTOR inhibitor DEPTOR. Mol. Cell 44, 290–303 (2011).
Duan, S. et al. mTOR generates an auto-amplification loop by triggering the betaTrCP- and CK1alpha-dependent degradation of DEPTOR. Mol. Cell 44, 317–324 (2011).
Peterson, T. R. et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 137, 873–886 (2009).
Zhou, J. et al. Full-coverage regulations of autophagy by ROS: from induction to maturation. Autophagy 18, 1240–1255 (2022).
Palma, F. R. et al. ROS production by mitochondria: function or dysfunction? Oncogene 43, 295–303 (2023).
Chen, P. et al. Synergistic inhibition of autophagy and neddylation pathways as a novel therapeutic approach for targeting liver cancer. Oncotarget 6, 9002–9017 (2015).
Xiu, H. et al. Neddylation alleviates methicillin-resistant staphylococcus aureus infection by inducing macrophage reactive oxygen species production. J. Immunol. 207, 296–307 (2021).
Yang, D. et al. Induction of autophagy and senescence by knockdown of ROC1 E3 ubiquitin ligase to suppress the growth of liver cancer cells. Cell Death Differ. 20, 235–247 (2013).
Lugano, R., Ramachandran, M. & Dimberg, A. Tumor angiogenesis: causes, consequences, challenges and opportunities. Cell Mol. Life Sci. 77, 1745–1770 (2020).
Yao, W. T. et al. Suppression of tumor angiogenesis by targeting the protein neddylation pathway. Cell Death Dis. 5, e1059 (2014).
Chen, Y. et al. Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Mol. Cell 35, 841–855 (2009).
Liu, X. et al. NEDD8-activating enzyme inhibitor, MLN4924 (Pevonedistat) induces NOXA-dependent apoptosis through up-regulation of ATF-4. Biochem. Biophys. Res. Commun. 488, 1–5 (2017).
Jin, Y. et al. Neddylation blockade diminishes hepatic metastasis by dampening cancer stem-like cells and angiogenesis in uveal melanoma. Clin. Cancer Res. 24, 3741–3754 (2018).
Zhou, H. M., Zhang, J. G., Zhang, X. & Li, Q. Targeting cancer stem cells for reversing therapy resistance: mechanism, signaling, and prospective agents. Signal Transduct. Target Ther. 6, 62 (2021).
Liu, C. et al. Antitumor effects of blocking protein neddylation in T315I-BCR-ABL leukemia cells and leukemia stem cells. Cancer Res. 78, 1522–1536 (2018).
Knorr, K. L. et al. Assessment of drug sensitivity in hematopoietic stem and progenitor cells from acute myelogenous leukemia and myelodysplastic syndrome ex vivo. Stem Cells Transl. Med. 6, 840–850 (2017).
Han, S. et al. The protein neddylation inhibitor MLN4924 suppresses patient-derived glioblastoma cells via inhibition of ERK and AKT signaling. Cancers 11, 1849 (2019).
Zhao, H. et al. Inflammation and tumor progression: signaling pathways and targeted intervention. Signal Transduct. Target Ther. 6, 263 (2021).
Deng, Q. et al. MLN4924 protects against bleomycin-induced pulmonary fibrosis by inhibiting the early inflammatory process. Am. J. Transl. Res. 9, 1810–1821 (2017).
Best, S. et al. Immunomodulatory effects of pevonedistat, a NEDD8-activating enzyme inhibitor, in chronic lymphocytic leukemia-derived T cells. Leukemia 35, 156–168 (2021).
Zhou, S. et al. Neddylation inhibition upregulates PD-L1 expression and enhances the efficacy of immune checkpoint blockade in glioblastoma. Int. J. Cancer 145, 763–774 (2019).
Garcia, K. et al. Nedd8-activating enzyme inhibitor MLN4924 provides synergy with mitomycin C through interactions with ATR, BRCA1/BRCA2, and chromatin dynamics pathways. Mol. Cancer Ther. 13, 1625–1635 (2014).
Nawrocki, S. T. et al. Disrupting protein NEDDylation with MLN4924 is a novel strategy to target cisplatin resistance in ovarian cancer. Clin. Cancer Res. 19, 3577–3590 (2013).
Lin, W. C. et al. MLN4924, a Novel NEDD8-activating enzyme inhibitor, exhibits antitumor activity and enhances cisplatin-induced cytotoxicity in human cervical carcinoma: in vitro and in vivo study. Am. J. Cancer Res. 5, 3350–3362 (2015).
Ho, I. L. et al. MLN4924 synergistically enhances cisplatin-induced cytotoxicity via JNK and Bcl-xL pathways in human urothelial carcinoma. Sci. Rep. 5, 16948 (2015).
Sun, Y. et al. Targeting neddylation sensitizes colorectal cancer to topoisomerase I inhibitors by inactivating the DCAF13-CRL4 ubiquitin ligase complex. Nat. Commun. 14, 3762 (2023).
Gu, Y. et al. MLN4924, an NAE inhibitor, suppresses AKT and mTOR signaling via upregulation of REDD1 in human myeloma cells. Blood 123, 3269–3276 (2014).
Zhou, L. et al. The NAE inhibitor pevonedistat interacts with the HDAC inhibitor belinostat to target AML cells by disrupting the DDR. Blood 127, 2219–2230 (2016).
Kee, Y. et al. Inhibition of the Nedd8 system sensitizes cells to DNA interstrand cross-linking agents. Mol. Cancer Res 10, 369–377 (2012).
Chen, X. et al. AKT inhibitor MK-2206 sensitizes breast cancer cells to MLN4924, a first-in-class NEDD8-activating enzyme (NAE) inhibitor. Cell cycle 17, 2069–2079 (2018).
Vanderdys, V. et al. The neddylation inhibitor pevonedistat (MLN4924) suppresses and radiosensitizes head and neck squamous carcinoma cells and tumors. Mol. Cancer Ther. 17, 368–380 (2018).
Yang, D., Tan, M., Wang, G. & Sun, Y. The p21-dependent radiosensitization of human breast cancer cells by MLN4924, an investigational inhibitor of NEDD8 activating enzyme. PloS One 7, e34079 (2012).
Wang, X. et al. Radiosensitization by the investigational NEDD8-activating enzyme inhibitor MLN4924 (pevonedistat) in hormone-resistant prostate cancer cells. Oncotarget 7, 38380–38391 (2016).
Zou, W., Wolchok, J. D. & Chen, L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci. Transl. Med. 8, 328rv324 (2016).
Chen, L. & Han, X. Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J. Clin. Investig. 125, 3384–3391 (2015).
Zhang, J. et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature 553, 91–95 (2018).
Li, C. W. et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat. Commun. 7, 12632 (2016).
Filippova, N. et al. Blocking PD1/PDL1 interactions together with MLN4924 therapy is a potential strategy for glioma treatment. J. Cancer Sci. Ther. 10, 190–197 (2018).
de la Chapelle, A. & Hampel, H. Clinical relevance of microsatellite instability in colorectal cancer. J. Clin. Oncol. 28, 3380–3387, (2010).
Kandoth, C. et al. Integrated genomic characterization of endometrial carcinoma. Nature 497, 67–73 (2013).
Cancer Genome Atlas Research, N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209, (2014).
McGrail, D. J. et al. Proteome instability is a therapeutic vulnerability in mismatch repair-deficient cancer. Cancer Cell 37, 371–386.e312 (2020).
Wong, B. et al. Pevonedistat, a first-in-class NEDD8-activating enzyme inhibitor, sensitizes cancer cells to VSVΔ51 oncolytic virotherapy. Mol. Ther. 31, 3176–3192 (2023).
Yoshimura, C. et al. TAS4464, a highly potent and selective inhibitor of nedd8-activating enzyme, suppresses neddylation and shows antitumor activity in diverse cancer models. Mol. Cancer Ther. 18, 1205–1216 (2019).
Yamamoto, N. et al. A first-in-human, phase 1 study of the NEDD8 activating enzyme E1 inhibitor TAS4464 in patients with advanced solid tumors. Investig. N. Drugs 39, 1036–1046 (2021).
Muraoka, H. et al. Activity of TAS4464, a novel NEDD8 activating enzyme E1 inhibitor, against multiple myeloma via inactivation of nuclear factor kappa B pathways. Cancer Sci. 110, 3802–3810 (2019).
Ochiiwa, H. et al. TAS4464, a NEDD8-activating enzyme inhibitor, activates both intrinsic and extrinsic apoptotic pathways via c-Myc-mediated regulation in acute myeloid leukemia. Oncogene 40, 1217–1230 (2021).
Li, X. et al. Flavokawain B targets protein neddylation for enhancing the anti-prostate cancer effect of Bortezomib via Skp2 degradation. Cell Commun. Signal 17, 25 (2019).
Pham, V. et al. Gartanin is a novel NEDDylation inhibitor for induction of Skp2 degradation, FBXW2 expression, and autophagy. Mol. Carcinog. 59, 193–201 (2020).
Leung, C. H. et al. A natural product-like inhibitor of NEDD8-activating enzyme. Chem. Commun. 47, 2511–2513 (2011).
Zhong, H. J. et al. Discovery of a natural product inhibitor targeting protein neddylation by structure-based virtual screening. Biochimie 94, 2457–2460 (2012).
Zhong, H. J. et al. A rhodium(III) complex as an inhibitor of neural precursor cell expressed, developmentally down-regulated 8-activating enzyme with in vivo activity against inflammatory bowel disease. J. Med. Chem. 60, 497–503 (2017).
Wu, K. J. et al. Structure-based identification of a NEDD8-activating enzyme inhibitor via drug repurposing. Eur. J. Med. Chem. 143, 1021–1027 (2018).
Zhong, H. J. et al. Structure-based repurposing of FDA-approved drugs as inhibitors of NEDD8-activating enzyme. Biochimie 102, 211–215 (2014).
Ma, H. et al. Discovery of benzothiazole derivatives as novel non-sulfamide NEDD8 activating enzyme inhibitors by target-based virtual screening. Eur. J. Med. Chem. 133, 174–183 (2017).
Lu, P. et al. Discovery of a novel NEDD8 activating enzyme inhibitor with piperidin-4-amine scaffold by structure-based virtual screening. ACS Chem. Biol. 11, 1901–1907 (2016).
Lu, P. et al. A novel NAE/UAE dual inhibitor LP0040 blocks neddylation and ubiquitination leading to growth inhibition and apoptosis of cancer cells. Eur. J. Med. Chem. 154, 294–304 (2018).
Fu, D. J. et al. Discovery of novel indole derivatives that inhibit NEDDylation and MAPK pathways against gastric cancer MGC803 cells. Bioorg. Chem. 107, 104634 (2021).
Fu, D.-J. et al. Discovery of novel tertiary amide derivatives as NEDDylation pathway activators to inhibit the tumor progression in vitro and in vivo. Eur. J. Med. Chem. 192, 112153 (2020).
Zlotkowski, K. et al. A small-molecule microarray approach for the identification of E2 enzyme inhibitors in ubiquitin-like conjugation pathways. SLAS Discov. 22, 760–766 (2017).
Li, Y. et al. Discovery of a small molecule inhibitor of cullin neddylation that triggers ER stress to induce autophagy. Acta Pharm. Sin. B 11, 3567–3584 (2021).
Xu, T. et al. A small molecule inhibitor of the UBE2F-CRL5 axis induces apoptosis and radiosensitization in lung cancer. Signal Transduct. Target Ther. 7, 354 (2022).
Mamun, M. A. A. et al. Micafungin: a promising inhibitor of UBE2M in cancer cell growth suppression. Eur. J. Med. Chem. 260, 115732 (2023).
Scott, D. C. et al. Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase. Nat. Chem. Biol. 13, 850–857 (2017).
Hammill, J. T. et al. Piperidinyl ureas chemically control defective in cullin neddylation 1 (DCN1)-mediated cullin neddylation. J. Med. Chem. 61, 2680–2693 (2018).
Hammill, J. T. et al. Discovery of an orally bioavailable inhibitor of defective in cullin neddylation 1 (DCN1)-mediated cullin neddylation. J. Med. Chem. 61, 2694–2706 (2018).
Kim, H. S. et al. Discovery of novel pyrazolo-pyridone DCN1 inhibitors controlling cullin neddylation. J. Med. Chem. 62, 8429–8442 (2019).
Kim, H. S. et al. Improvement of oral bioavailability of pyrazolo-pyridone inhibitors of the interaction of DCN1/2 and UBE2M. J. Med. Chem. 64, 5850–5862 (2021).
Zhou, H. et al. A potent small-molecule inhibitor of the DCN1-UBC12 interaction that selectively blocks cullin 3 neddylation. Nat. Commun. 8, 1150 (2017).
Zhou, H. et al. High-affinity peptidomimetic inhibitors of the DCN1-UBC12 protein-protein interaction. J. Med. Chem. 61, 1934–1950 (2018).
Zhou, H. et al. Selective inhibition of cullin 3 neddylation through covalent targeting DCN1 protects mice from acetaminophen-induced liver toxicity. Nat. Commun. 12, 2621 (2021).
Bomprezzi, R. Dimethyl fumarate in the treatment of relapsing-remitting multiple sclerosis: an overview. Ther. Adv. Neurol. Disord. 8, 20–30 (2015).
Zhou, H. et al. Targeting DCN1-UBC12 protein-protein interaction for regulation of neddylation pathway. Adv. Exp. Med. Biol. 1217, 349–362 (2020).
Wang, S. et al. Development of highly potent, selective, and cellular active triazolo[1,5- a]pyrimidine-based inhibitors targeting the DCN1-UBC12 protein-protein interaction. J. Med. Chem. 62, 2772–2797 (2019).
Zhou, W. et al. Potent 5-cyano-6-phenyl-pyrimidin-based derivatives targeting DCN1-UBE2M interaction. J. Med. Chem. 62, 5382–5403 (2019).
He, Z.-X. et al. Discovery of potent and selective 2-(Benzylthio)pyrimidine-based DCN1-UBC12 inhibitors for anticardiac fibrotic effects. J. Med. Chem. 65, 163–190 (2022).
Zhou, W. et al. Development of phenyltriazole thiol-based derivatives as highly potent inhibitors of DCN1-UBC12 interaction. Eur. J. Med. Chem. 217, 113326 (2021).
Huang, X. & Dixit, V. M. Drugging the undruggables: exploring the ubiquitin system for drug development. Cell Res. 26, 484–498 (2016).
Shafique, S., Ali, W., Kanwal, S. & Rashid, S. Structural basis for Cullins and RING component inhibition: targeting E3 ubiquitin pathway conductors for cancer therapeutics. Int. J. Biol. Macromol. 106, 532–543 (2018).
Yu, Q. et al. Gossypol inhibits cullin neddylation by targeting SAG-CUL5 and RBX1-CUL1 complexes. Neoplasia 22, 179–191 (2020).
Barbieri, I. & Kouzarides, T. Role of RNA modifications in cancer. Nat. Rev. Cancer 20, 303–322 (2020).
Lee, J. M., Hammaren, H. M., Savitski, M. M. & Baek, S. H. Control of protein stability by post-translational modifications. Nat. Commun. 14, 201 (2023).
Scherer, P. C. et al. Inositol hexakisphosphate (IP6) generated by IP5K mediates cullin-COP9 signalosome interactions and CRL function. Proc. Natl Acad. Sci. USA 113, 3503–3508 (2016).
Coleman, K. E. et al. SENP8 limits aberrant neddylation of NEDD8 pathway components to promote cullin-RING ubiquitin ligase function. Elife. 6, e24325 (2017).
Wang, B. et al. Liquid-liquid phase separation in human health and diseases. Signal Transduct. Target Ther. 6, 290 (2021).
Shao, X. et al. Deneddylation of PML/RARalpha reconstructs functional PML nuclear bodies via orchestrating phase separation to eradicate APL. Cell Death Differ. 29, 1654–1668 (2022).
Bouchard, J. J. et al. Cancer mutations of the tumor suppressor SPOP disrupt the formation of active, phase-separated compartments. Mol. Cell 72, 19–36.e18 (2018).
Chou, M. C. et al. PAICS ubiquitination recruits UBAP2 to trigger phase separation for purinosome assembly. Mol. Cell 83, 4123–4140.e4112 (2023).
Mattiroli, F. & Penengo, L. Histone ubiquitination: an integrative signaling platform in genome stability. Trends Genet. 37, 566–581 (2021).
Mattiroli, F. et al. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell 150, 1182–1195 (2012).
Liu, J., Jin, J., Liang, T. & Feng, X. H. To Ub or not to Ub: a regulatory question in TGF-beta signaling. Trends Biochem. Sci. 47, 1059–1072 (2022).
Noh, E. H. et al. Covalent NEDD8 conjugation increases RCAN1 protein stability and potentiates its inhibitory action on calcineurin. PloS One 7, e48315 (2012).
Gomarasca, M., Lombardi, G. & Maroni, P. SUMOylation and NEDDylation in primary and metastatic cancers to bone. Front. Cell Dev. Biol. 10, 889002 (2022).
Bian, Y. et al. An enzyme-assisted RP-RPLC approach for in-depth analysis of human liver phosphoproteome. J. Proteom. 96, 253–262 (2014).
Kettenbach, A. N. et al. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci. Signal. 4, rs5 (2011).
Weinert, B. T. et al. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 4, 842–851 (2013).
Rigbolt, K. T. et al. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal 4, rs3 (2011).
Stuber, K. et al. Structural and functional consequences of NEDD8 phosphorylation. Nat. Commun. 12, 5939 (2021).
Kelsall, I. R. et al. HOIL-1 ubiquitin ligase activity targets unbranched glucosaccharides and is required to prevent polyglucosan accumulation. EMBO J. 41, e109700 (2022).
Otten, E. G. et al. Ubiquitylation of lipopolysaccharide by RNF213 during bacterial infection. Nature 594, 111–116 (2021).
Jackson, E. L. et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 15, 3243–3248 (2001).
Marino, S. et al. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 14, 994–1004 (2000).
Groszer, M. et al. Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science 294, 2186–2189 (2001).
Wang, B., Chandrasekera, P. C. & Pippin, J. J. Leptin- and leptin receptor-deficient rodent models: relevance for human type 2 diabetes. Curr. Diabetes Rev. 10, 131–145 (2014).
Dawson, T. M., Golde, T. E. & Lagier-Tourenne, C. Animal models of neurodegenerative diseases. Nat. Neurosci. 21, 1370–1379 (2018).
Wirtz, S. & Neurath, M. F. Mouse models of inflammatory bowel disease. Adv. Drug Deliv. Rev. 59, 1073–1083 (2007).
Choudhary, N., Bhatt, L. K. & Prabhavalkar, K. S. Experimental animal models for rheumatoid arthritis. Immunopharmacol. Immunotoxicol. 40, 193–200 (2018).
Regitz-Zagrosek, V. & Kararigas, G. Mechanistic pathways of sex differences in cardiovascular disease. Physiol. Rev. 97, 1–37 (2017).
Mao, H. & Sun, Y. Neddylation-independent activities of MLN4924. Adv. Exp. Med. Biol. 1217, 363–372 (2020).
Chen, Y. F. et al. Arctigenin impairs UBC12 enzyme activity and cullin neddylation to attenuate cancer cells. Acta Pharm. Sin. 44, 661–669 (2023).
Paul, D. et al. Artificial intelligence in drug discovery and development. Drug Discov. Today 26, 80–93 (2021).
Lee, M. R. et al. Inhibition of APP intracellular domain (AICD) transcriptional activity via covalent conjugation with Nedd8. Biochem. Biophys. Res. Commun. 366, 976–981 (2008).
Liu, M. et al. Ajuba inhibits hepatocellular carcinoma cell growth via targeting of β-catenin and YAP signaling and is regulated by E3 ligase Hakai through neddylation. J. Exp. Clin. Cancer Res. 37, 165 (2018).
Gao, F., Cheng, J., Shi, T. & Yeh, E. T. Neddylation of a breast cancer-associated protein recruits a class III histone deacetylase that represses NFkappaB-dependent transcription. Nat. Cell Biol. 8, 1171–1177 (2006).
Takashima, O. et al. Brap2 regulates temporal control of NF-κB localization mediated by inflammatory response. PloS One 8, e58911 (2013).
Segovia, J. A. et al. Nedd8 regulates inflammasome-dependent caspase-1 activation. Mol. Cell Biol. 35, 582–597 (2015).
Fei, X. et al. Neddylation of Coro1a determines the fate of multivesicular bodies and biogenesis of extracellular vesicles. J. Extracell. Vesicles 10, e12153 (2021).
Lee, G. W. et al. The E3 ligase C-CBL inhibits cancer cell migration by neddylating the proto-oncogene c-Src. Oncogene 37, 5552–5568 (2018).
Bennett, E. J., Rush, J., Gygi, S. P. & Harper, J. W. Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell 143, 951–965 (2010).
Mendoza, H. M. et al. NEDP1, a highly conserved cysteine protease that deNEDDylates Cullins. J. Biol. Chem. 278, 25637–25643 (2003).
Renaudin, X., Guervilly, J. H., Aoufouchi, S. & Rosselli, F. Proteomic analysis reveals a FANCA-modulated neddylation pathway involved in CXCR5 membrane targeting and cell mobility. J. Cell Sci. 127, 3546–3554 (2014).
Aoki, I., Higuchi, M. & Gotoh, Y. NEDDylation controls the target specificity of E2F1 and apoptosis induction. Oncogene 32, 3954–3964 (2013).
Najor, N. A. et al. Epidermal growth factor receptor neddylation is regulated by a desmosomal-COP9 (Constitutive Photomorphogenesis 9) signalosome complex. Elife 6, e22599 (2017).
Li, S. et al. Neddylation promotes protein translocation between the cytoplasm and nucleus. Biochem. Biophys. Res. Commun. 529, 991–997 (2020).
Ryu, J. H. et al. Hypoxia-inducible factor α subunit stabilization by NEDD8 conjugation is reactive oxygen species-dependent. J. Biol. Chem. 286, 6963–6970 (2011).
Ghosh, D. K. & Ranjan, A. HYPK coordinates degradation of polyneddylated proteins by autophagy. Autophagy 18, 1763–1784 (2022).
Zhao, M. et al. Myeloid neddylation targets IRF7 and promotes host innate immunity against RNA viruses. PLoS Pathog. 17, e1009901 (2021).
Park, J. B. et al. Neddylation of insulin receptor substrate acts as a bona fide regulator of insulin signaling and its implications for cancer cell migration. Cancer Gene Ther. (2024).
Li, H. et al. Itch promotes the neddylation of JunB and regulates JunB-dependent transcription. Cell Signal 28, 1186–1195 (2016).
Guo, Y. J. et al. Neddylation-dependent LSD1 destabilization inhibits the stemness and chemoresistance of gastric cancer. Int. J. Biol. Macromol. 254, 126801 (2023).
Yan, F., Guan, J., Peng, Y. & Zheng, X. MyD88 NEDDylation negatively regulates MyD88-dependent NF-κB signaling through antagonizing its ubiquitination. Biochem. Biophys. Res. Commun. 482, 632–637 (2017).
Zhao, J. et al. 20-Hydroxyeicosatetraenoic acid regulates the expression of Nedd4‑2 in kidney and liver through a neddylation modification pathway. Mol. Med. Rep. 16, 9671–9677 (2017).
Tu, J. et al. Neddylation-mediated Nedd4-2 activation regulates ubiquitination modification of renal NBCe1. Exp. Cell Res. 390, 111958 (2020).
Chen, S. M. et al. Targeting inhibitors of apoptosis proteins suppresses medulloblastoma cell proliferation via G2/M phase arrest and attenuated neddylation of p21. Cancer Med. 7, 3988–4003 (2018).
Guan, J., Yu, S. & Zheng, X. NEDDylation antagonizes ubiquitination of proliferating cell nuclear antigen and regulates the recruitment of polymerase η in response to oxidative DNA damage. Protein Cell 9, 365–379 (2018).
Guan, J. & Zheng, X. NEDDylation regulates RAD18 ubiquitination and localization in response to oxidative DNA damage. Biochem. Biophys. Res. Commun. 508, 1240–1244 (2019).
Maghames, C. M. et al. NEDDylation promotes nuclear protein aggregation and protects the Ubiquitin Proteasome System upon proteotoxic stress. Nat. Commun. 9, 4376 (2018).
Shu, J. et al. Nedd8 targets ubiquitin ligase Smurf2 for neddylation and promote its degradation. Biochem. Biophys. Res. Commun. 474, 51–56 (2016).
Singh, R. K., Iyappan, S. & Scheffner, M. Hetero-oligomerization with MdmX rescues the ubiquitin/Nedd8 ligase activity of RING finger mutants of Mdm2. J. Biol. Chem. 282, 10901–10907 (2007).
Watson, I. R. et al. Mdm2-mediated NEDD8 modification of TAp73 regulates its transactivation function. J. Biol. Chem. 281, 34096–34103 (2006).
Liu, K. et al. TRAF6 neddylation drives inflammatory arthritis by increasing NF-κB activation. Lab. Investig. 99, 528–538 (2019).
Jeong, Y. Y., Her, J. & Chung, I. K. NEDD8 ultimate buster-1 regulates the abundance of TRF1 at telomeres by promoting its proteasomal degradation. FEBS Lett. 590, 1776–1790 (2016).
Wang, B. et al. Neddylation is essential for β-catenin degradation in Wnt signaling pathway. Cell Rep. 38, 110538 (2022).
Schulze-Niemand, E. & Naumann, M. The COP9 signalosome: a versatile regulatory hub of Cullin-RING ligases. Trends Biochem. Sci. 48, 82–95 (2023).
Ferro, A. et al. NEDD8: a new ataxin-3 interactor. Biochim. Biophys. Acta 1773, 1619–1627 (2007).
Gong, L., Kamitani, T., Millas, S. & Yeh, E. T. Identification of a novel isopeptidase with dual specificity for ubiquitin- and NEDD8-conjugated proteins. J. Biol. Chem. 275, 14212–14216 (2000).
Hemelaar, J. et al. Specific and covalent targeting of conjugating and deconjugating enzymes of ubiquitin-like proteins. Mol. Cell Biol. 24, 84–95 (2004).
Wada, H. et al. Cleavage of the C-terminus of NEDD8 by UCH-L3. Biochem. Biophys. Res. Commun. 251, 688–692 (1998).
Artavanis-Tsakonas, K. et al. Identification by functional proteomics of a deubiquitinating/deNeddylating enzyme in Plasmodium falciparum. Mol. Microbiol. 61, 1187–1195 (2006).
Acknowledgements
This work was funded in part by the National Key R&D Program of China (grants 2022YFC3401500 to Y.S. and Y.Z., and 2021YFA1101000 to Y.S.), the National Natural Science Foundation of China (grants 92253203 and U22A20317 to Y.S., 92053117 and 81972591 to Y.Z., 82202838 to S.Z., and 82102749 to Q.Y.), and Zhejiang Provincial Natural Science Foundation of China (grant LD22H300003 to Y.S., LZ22H160003 to Y.Z.), and Leading Innovative and Entrepreneur Team Introduction Program of Zhejiang (grant 2022R01002 to Y.S.).
Author information
Authors and Affiliations
Contributions
Y.S. conceived and outlined the manuscript. S.Z. and Q.Y. performed the literature search and wrote the manuscript, S.Z., Q.Y., Y.Z. and Z.L. drafted figures and tables. Y.Z. and Y.S. revised, and Y.S. finalized the manuscript. All authors contributed to the discussions, and have read and approved the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, S., Yu, Q., Li, Z. et al. Protein neddylation and its role in health and diseases. Sig Transduct Target Ther 9, 85 (2024). https://doi.org/10.1038/s41392-024-01800-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41392-024-01800-9