Abstract
Osteoporotic fractures are an increasing cause of mortality and morbidity in ageing populations. A major risk determinant for these fractures is bone mineral density (BMD). Variation on BMD is thought, on the basis of twin and family studies, to be subject to a large amount of genetic variation and it has been hypothesised that this may be due to the influence of multiple genes. However, in families showing segregation of low or high BMD, single major genes have been shown to play a crucial role. We performed a genome-wide screen using 380 microsatellite markers in a single extended family (n=34) in which early-onset low spinal areal BMD segregates in an autosomal dominant-like fashion. A two-point linkage analysis was performed, revealing a maximum LOD score of 3.07 on 1p36.3 (D1S468), confirming results of previous linkage studies of BMD, while no other suggestive linkage peaks (LOD>2.2) were detected elsewhere in the genome. Microsatellite markers were subsequently genotyped for a ±6.9 Mb region surrounding D1S468. This revealed critical recombination events restricting the candidate region to 1.2 Mb and 19 genes. Sequencing analysis of the coding region of candidate genes WDR8 and EGFL3 revealed no mutations or disease-associated polymorphisms. Our results provide some evidence supporting the hypothesis that there are genetic determinants for spinal BMD on 1p36.3. Although no specific disease causing mutation has yet been found, the delineation of a relatively small candidate region in a single extended family opens perspectives to identify a major gene for spinal BMD.
Similar content being viewed by others
Introduction
Osteoporosis is mainly defined by a generalised reduction in bone mineral density (BMD) leading to an increased risk of fracture. Next to environmental factors, such as lifestyle and diet, a variety of genetic factors, each with relatively modest effects, contribute to the regulation of bone mass.1 Therefore, BMD is assumed to be multifactorially determined. In family and twin studies, heritability of BMD has been estimated to range from 50 to 90%, with strongest heritability values reported for the spine.2, 3, 4, 5, 6 Several gene variants and quantitative trait loci (QTLs) related to BMD have been identified by association studies and genome-wide scanning approaches.7 However, replication of these findings has been very inconsistent. It has been proposed that this is due to the complexity of the genetic inheritance of BMD, ethnic heterogeneity, the diversity in study designs and the use of inadequate sample sizes to provide sufficient power.8 Considering these confounding factors, false linkage results may be generated across linkage and association studies.
In general, gene-mapping has been far more successful for rare Mendelian inherited bone diseases both for high and low bone mass disorders. Indeed, in rare instances, low BMD is inherited in a Mendelian manner. Examples include osteogenesis imperfecta caused by mutations in one of the two genes encoding collagen type 1 and osteoporosis-pseudoglioma syndrome caused by inactivating mutations in the lipoprotein receptor-related protein 5 gene.9, 10 The mapping of causal genes in these rare monogenic diseases has contributed to the identification of pathways underlying bone metabolism and osteoporosis as a common disease. In addition, several studies have shown evidence for a major gene mode of inheritance of BMD. These major genes are single genes for which there are variants associated with a large effect on individual risk and as such they may be sufficiently strong to generate a detectable Mendelian-like pattern of inheritance of risk. This is particularly so in pedigrees selected via probands with extremely low or high BMD values.11, 12, 13, 14 Unfortunately, single extended families in which low BMD shows a Mendelian-like inheritance pattern are rare and consequently not many linkage studies have been performed in such families.
In this study, we performed a genome-wide linkage analysis in a single extended (n=34) family in which segregation of low spinal BMD values according to an autosomal dominant-like mode of inheritance favours a major gene locus effect (Figure 1). The pedigree studied here was ascertained through a proband with low spinal BMD and the presence of recurrent fractures, with several relatives also presenting low BMD values. By its large size, this family provides an opportunity to map a major gene for osteoporosis susceptibility, while avoiding the problem of locus heterogeneity incumbent to studies that combine data from numerous small families. In addition, with such a family, one can look for recombinant haplotypes that define a specific genetic interval for a BMD locus.
Pedigree structure showing haplotypes across the chromosome 1p36.3 region. White symbols represent ‘non-affected’ individuals (Z(aBMD)≥−1), black symbols represent ‘affected’ individuals (Z(aBMD)≤−2 and/or treated (T) with bisphosphonates because of an initial BMD T-score ≤−2.5 at the spine and/or hip), and grey symbols represent individuals which are coded as unknown due to intermediate spinal BMD values (−2<Z(aBMD)<−1). ‘Excl’ indicates individuals who are excluded from the linkage analysis (coded unknown) according to the reported exclusion criteria. Symbols containing a question mark indicate that there is no information on the phenotype and no DNA available for this individual. Patient identification number, Z(aBMDL1L4) and age at the time of BMD measurement are indicated beneath each symbol. The arrow marks the proband in this family. Underlined marker names indicate that these markers belong to the Linkage Mapping Kit used for the initial genome-wide linkage screen. A box indicates the chromosome carrying the linked haplotype. Genotypes within parentheses are inferred.
Materials and methods
Subjects
This family came to medical attention because of the presence of recurrent fractures and low BMD in individual II:16 (Figure 1; Supplementary Table 1). Family history revealed the occurrence of fractures in several family members. Consequently, an extensive clinical analysis was conducted on all available individuals in this family (n=34). BMD was measured using dual X-ray absorptiometry (see below). To identify secondary forms of osteoporosis, data on medical history, use of medication, alcohol consumption, smoking behaviour and history of fractures was obtained using self-administered questionnaires and fasting blood samples were collected.15 The following exclusion criteria were applied: a current or past history of hyperthyroidism, metabolic disorders known to affect bone, delayed puberty, alcoholism (current or past alcohol consumption of ≥5 U a day more than once a week), heavy smoking (≥40 pack-years), malabsorption, hemochromatosis, renal or gonadal dysfunction, malignancy and subjects chronically treated with glucocorticoids, synthetic thyroxine, or (anti)androgens. According to these exclusion criteria individuals II:6, II:12 (hyperthyroidism) and II:13 (delayed puberty) were excluded (phenotype ‘unknown’ in linkage analysis). The presence of a type I collagen mutation, suggesting Osteogenesis Imperfecta, or the presence of a heterozygous LRP5 mutation, causing low BMD, was excluded by mutational and linkage analysis.16 Because of an initial BMD T-score ≤−2.5 at the spine and/or hip, which is defined as osteoporosis according to criteria of the World Health Organization, individuals II:5, II:7, II:9, II:16 and II:18 have received a specific osteoporosis treatment with bisphosphonates, which will generally lead to a significant increase of BMD values.17 However, no data about quantitative BMD measurements before these treatments were available for these individuals. The study was approved by the Ethical Committee of the Ghent University Hospital and written informed consent was obtained from all study subjects.
BMD
Areal BMD (aBMD) (g/cm2) was calculated from the bone mineral content (g) divided by its projected area (cm2). aBMD at the lumbar spine (L1–L4) and the hip (femoral neck, trochanter and total hip) was measured using dual X-ray absorptiometry (DXA) with a Hologic QDR 4500C device (Software version 8.26a; Hologic Inc., Bedford, MA, USA). Lumbar spine aBMD was taken as the mean of L1–L4 measurements, unless deformity was present, in which case the mean value of undeformed lumbar vertebrae was used. Differences from the mean aBMD for an age- and gender-matched reference population expressed as units of SD below or above the population mean (Z-scores) for aBMD were calculated. Age- and gender-matched controls provided by the National Health and Nutrition Examination Survey (NHANES) III study group for the hip and by the manufacturer for the lumbar spine were used. T-scores express the units of SD below or above the expected bone density value of a healthy young adult of the same sex.
Volumetric BMD (vBMD) (g/cm3) at L1–L4 was calculated from the bone mineral content divided by the vertebral body volume (VV). VV of L1–L4 was calculated using the projected area of L1–L4 obtained by anteroposterior DXA and assuming the vertebral body resembles a cube (VV=(anteroposterior area)3/2).18, 19
Genotyping
EDTA blood was collected and DNA was extracted by use of a PUREGENE DNA blood purification kit (Gentra Systems). A genome-wide linkage analysis was performed using 380 microsatellite markers of the ABI Prism Linkage Mapping Set MD-10 Version 2.5 (Applied Biosystems). An improved protocol from the ‘Centre National de Génotypage’ (Evry, France) was used for pooling an average of four markers per PCR to carry out all reactions. Additional microsatellite markers were taken from the Marshfield map or were designed based on the simple-tandem-repeat finder in the University of California Santa Cruz genome browser. Microsatellite markers were analysed on a Applied Biosystems Prism 3100 Genetic Analyser running Genemapper v2.0 software (Applied Biosystems).
Linkage analysis
Since the pattern of inheritance of the low lumbar spine BMD in this family is most suggestive for autosomal dominant-like inheritance (Figure 1), a parametric linkage analysis making use of an affected/unaffected-trait model was performed. To perform linkage analysis by affection status, individuals were categorised into two groups. The first group comprises of ‘affected subjects’ which include six individuals (III:5, III:7, III:8, III:9, III:13, III:16) with severely low spinal aBMD (Z(aBMD)≤−2) and five individuals (II:5, II:7, II:9, II:16, II:18) who were treated with bisphosphonates because of an initial BMD T-score ≤−2,5 at the spine and/or hip. The second group comprises the ‘non-affected’ subjects which include nine individuals (II:15, III:1, III:2, III:3, III:4, III:10, III:11, III:12, III:14) with normal spinal aBMD (Z(aBMD)≥−1) and four unrelated spouses (II:4, II:10, II:14, II:17). Because individuals I:2, II:1, II:2, II:3, II:6, II:11, II:12, II:13, III:6 and III:15 could not be assigned to one of these groups they were coded as ‘phenotype unknown’ in the linkage analysis. Individuals under the age of 15 were not incorporated in the genome-wide search.
Prior to linkage analysis, genotyped markers were checked for Mendelian inconsistencies using the PedCheck program. Two-point parametric LOD scores were calculated using the Fastlink program in the Easylinkage software package.20 A disease allele frequency of 0.001 was assumed since familial inheritance of severely low BMD is rare in the population and since the trait shows genetic heterogeneity. Equal frequencies for each allele of a marker were assumed, since population-based frequencies were not available for all markers. An autosomal dominant model was used with a reduced penetrance in a range from 0.60 to 0.90 and a phenocopy rate of 2.5%. A non-parametric linkage analysis was conducted using Genehunter v2.1r5 (Easylinkage) under the assumption of equal distribution of marker allele frequencies and using an Sall scoring function to capture the information about allele sharing between all affected individuals in the pedigree. Haplotypes were created using the Cyrillic 2.1 software.
Sequencing candidate genes
Protein coding regions and splice sites of candidate genes WD repeat-containing protein 8 (WDR8) and epidermal growth factor-like 3 (EGFL3) were amplified from DNA of affected family members II:5 and II:9 using primers designed from intronic sequence adjacent to intron–exon boundaries or exonic DNA when internal primers were required for larger exons. Sequencing reactions employed the same primers used for PCR. Primer sequences and PCR conditions are available on request. Sequence reactions were analysed on an Applied Biosystems Prism 3100 Genetic Analyser. These sequences were compared to the wild-type sequence as submitted to GenBank Accession numbers NM_017818 (WDR8) and NM_001409 (EGFL3). Sequence variants, identified in individuals II:5 and II:9, were analysed in all family members by direct sequencing.
Statistical analysis
Correlations were estimated by Pearson correlation coefficients. Differences in correlations were judged statistically significant when P<0.05. Statistical analyses were performed using SPSS software version 12.
Results
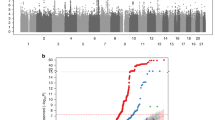
To determine BMD status in this family, areal BMD Z-scores for lumbar spine, femoral neck and total hip were obtained by means of DXA (Supplementary Table 1). Although BMD Z-scores at these different skeletal sites are highly correlated (P<0.01), the segregation of severe low values is restricted to lumbar spine BMD which was subsequently used as the phenotype to perform linkage analysis. In the genome-wide linkage study, the highest two-point parametric LOD-score is generated on chromosome 1 at marker D1S468 (LOD=3.07) for a dominant mode of inheritance with 70 or 80% penetrance (Table 1). Except for D1S243, flanking D1S468, no other marker of the Linkage Mapping Set Kit exceeds the threshold LOD score of 2.2, supposing suggestive linkage according to the criteria of Lander and Kruglyak.21 Because a genetically heterogeneous phenotype such as low BMD makes it difficult to choose the exact parameter set, also a non-parametric linkage analysis (NPL) was conducted. Hereby, the same region was identified yielding a minimum genome-wide NPL P-value (P=0.0013) for marker D1S468 (Table 1).
Recombinations between the disease and marker D1S214 in individuals III:4 and III:8 are locating the disease gene telomeric to this marker, whereas recombination events in individuals III:11 and III:12 are placing the disease gene centromeric to the additionally tested marker D1S243 (Figure 1). Haplotype analysis in this family indicates that the disease gene is most likely located between marker D1S214 and D1S243, an interval estimated to be approximately 4.75 Mb. In the four individuals harbouring a critical recombination event, an additional marker D1S2845 and four unpublished microsatellite markers were tested in an attempt to narrow down the candidate region (Figure 2). Recombinations at μsat1 and μsat4 reduced the candidate interval to 1.2 Mb. All ‘affected’ individuals in this pedigree are carrying the linked haplotype surrounding the candidate region. In all ‘non-affected’ individuals, except for II:15, the linked haplotype is absent.
The candidate region contains 19 genes according to the September 2006 NCBI Genome Browser database Build 36.2 (Supplementary Table 2). Among these, two candidate genes were selected for further mutation screening because of known functionality in acquisition of bone or expression pattern in bone tissue. The WD repeat-containing protein 8 was shown to be expressed in bone and cartilage and also in bone-forming cells including osteoblasts and chondrocytes, especially during endochondral ossification.22 Another positional candidate gene encodes EGFL3. The predicted EGFL3 protein contains 30 EGF-like domains, three of which are of the calcium-binding type and 23 of which contain two additional cysteine residues within the consensus EGF-like motif.23 The N-terminal portion of EGFL3 shows significant sequence similarity to fibrillin, in which mutations are known to cause Marfan syndrome, a disease with important skeletal involvement. Mutation analysis of the coding regions of WDR8 and EGFL3 in all family members revealed no gene variants, which are uniquely residing on the disease-linked haplotype in this family (data not shown).
The measured aBMD values, used as the phenotype for the genome-wide screen, are dependent of both bone size and true volumetric bone density. Therefore, as aBMD is a measurement that is partially influenced by bone size, it was possible that our linkage data resulted from differences in vertebral size between individuals rather than from variation in true volumetric bone density. This was further investigated by calculating volumetric BMDs (vBMD). Individuals with lower spinal Z(aBMD) also had lower vBMD (Supplementary Table 1). The coefficient of determination in regression (r2) suggests that about 81% of the variation in Z(aBMD) in this kindred can be explained by variation in vBMD.
Discussion
A genome-wide scan in this large family presenting with autosomal dominant-like inherited low spinal bone mass reveals a single, relatively small candidate interval on chromosome 1p36.3. In contrast to most genome-wide linkage studies for low BMD, for which a large number of small families were used, this approach based on one large family avoids the problem of locus heterogeneity. In addition, this strategy enables us to delineate a candidate region by looking for recombination events. The candidate interval of 1.2 Mb on 1p36.3, for which suggestive linkage for low BMD at the lumbar spine was found, suggests that a single major gene in this region strongly influences variation in BMD. After estimation of spine vBMD values, the coefficient of determination in regression (r2) between Z(aBMD) and vBMD was calculated to be 0.81 in this kindred. This indicates that variation in Z(aBMD) is mainly determined by variation in vBMD leading to the conclusion that the assumed major gene largely affects vBMD rather than bone size.
All ‘affected’ individuals in this pedigree are carriers of the linked haplotype on 1p36.3, whereas all ‘non-affected’ individuals, except for II:15, do not carry this haplotype (Figure 1). Individual II:15, carrying the linked haplotype, should therefore be considered as a non-penetrant individual. The individuals in this family to which no affection status was assigned show either carriage (II:1, II:2, II:3) or non-carriage (I:2, II:11, III:6, III:15) for the linked haplotype. The intermediate spinal BMD values (−2<Z(aBMD)<−1) observed in these individuals and the non-penetrance observed in II:15 may be due to the influence of environmental or other genetic modifying factors.
Because only suggestive linkage was found (LOD=3.07) according to the criteria of Lander and Kruglyak (1995), it cannot be excluded that our findings were due to chance. The possible occurrence of false-positive results in linkage analyses for osteoporosis susceptibility is illustrated by the inconsistent results among genome-wide linkage studies, disclosing many loci that may contain genes that regulate BMD. Only few regions have been consistently linked to BMD in several different studies, that is, 1p36, 1q21-24, 4q31-34 and 12q23-24.7 Thus, the chromosomal region at 1p36, identified in this study, is one of the few regions which previously emerged in several other linkage studies for BMD (Table 2).24, 25, 26, 27, 28, 29, 30, 31 This strengthens our hypothesis that we have identified true linkage. Recently, a meta-analysis by Lee et al,32 performed to assess evidence for linkage of BMD across whole genome scans also revealed, among other loci, strong evidence of linkage for 1p36, although this locus was not identified in another meta-analysis performed by Ioannidis et al.33 These differences are likely due to the use of different study designs. Lee et al32 used the maximum of published linkage scores for any skeletal site while Ioannidis et al33 analysed whole genome data for lumbar spine BMD, femoral neck BMD and sex separately to perform their meta-analysis. In addition, both studies did not include exactly the same genome-wide scans. Both approaches, although generating partially different linkage peaks, could be complementary in detecting loci that regulate BMD across whole genome scan studies. Quantitative trait loci mapping in animal models provided additional evidence for the involvement of a candidate locus for BMD on 1p36, leading to the assumption that major regulatory genes are shared among different species. Bouxsein et al34 identified a chromosomal region on mouse chromosome 4, synthenic to human 1p36, which is associated with several vertebral trabecular bone traits. Furthermore, Beamer et al35 identified linkage to the same chromosomal mouse region for vertebral vBMD as measured by pQCT which is in accordance with our data that a genetic defect on 1p36 affects vBMD rather than bone size. All these data support the hypothesis that a major locus controlling BMD is located on chromosome 1p36. This locus seems to have pleiotropic effects on bone, as indicated by the various skeletal sites reported to be linked with 1p36. This is further supported by the findings of Karasik et al29 (Table 2), in which principal component analysis of several BMD and quantitative ultrasound (QUS) phenotypes detected 1p36 as a shared genetic locus determining BMD variation at various skeletal sites. In the family presented here, the genetic determinant at 1p36 also affects both hip and spinal BMD as these measurements are highly correlated, although the lumbar spine is most severely affected. Although we have detected the same chromosomal region as in several previous linkage studies in both human and mice, this does not necessarily mean that we are picking up the same genetic variant. The more severe bone phenotype in this family, compared to most of the previous linkage studies, could point to the presence of a different disease allele. The two sequenced candidate genes, WDR8 and EGFL3, appeared to be the most promising candidates, based on published expression and functional data, although no disease causing sequence variations are found at this moment. This does not exclude that mutations are present in non-coding regions of these genes or in other genes in the delineated candidate region. Therefore, further studies are ongoing to identify the putative gene for the low BMD trait in this family.
In conclusion, the localisation of a major locus for BMD on 1p36.3 was replicated in this study, although only suggestive linkage was found. In this single extended family this genetic factor seems to have pleiotropic effects on bone and most explicitly on lumbar spine vBMD. Our findings open perspectives for further identification of a spinal BMD determining gene in a relatively small candidate region. However, at present, sequencing analysis of the coding region of candidate genes WDR8 and EGFL3 revealed no mutations or disease-associated polymorphisms.
References
Deng HW, Chen WM, Conway T et al: Determination of bone mineral density of the hip and spine in human pedigrees by genetic and life-style factors. Genet Epidemiol 2000; 19: 160–177.
Christian JC, Yu PL, Slemenda CW, Johnston Jr CC : Heritability of bone mass: a longitudinal study in aging male twins. Am J Hum Genet 1989; 44: 429–433.
Flicker L, Hopper JL, Rodgers L, Kaymakci B, Green RM, Wark JD : Bone density determinants in elderly women: a twin study. J Bone Miner Res 1995; 10: 1607–1613.
Gueguen R, Jouanny P, Guillemin F, Kuntz C, Pourel J, Siest G : Segregation analysis and variance components analysis of bone mineral density in healthy families. J Bone Miner Res 1995; 10: 2017–2022.
Pocock NA, Eisman JA, Hopper JL, Yeates MG, Sambrook PN, Eberl S : Genetic determinants of bone mass in adults. A twin study. J Clin Invest 1987; 80: 706–710.
Slemenda CW, Christian JC, Williams CJ, Norton JA, Johnston Jr CC : Genetic determinants of bone mass in adult women: a re-evaluation of the twin model and the potential importance of gene interaction on heritability estimates. J Bone Miner Res 1991; 6: 561–567.
Liu YJ, Shen H, Xiao P et al: Molecular genetic studies of gene identification for osteoporosis: a 2004 update. J Bone Miner Res 2006; 21: 1511–1535.
Shen H, Liu Y, Liu P, Recker RR, Deng HW : Nonreplication in genetic studies of complex diseases – lessons learned from studies of osteoporosis and tentative remedies. J Bone Miner Res 2005; 20: 365–376.
Byers PH : Brittle bones – fragile molecules: disorders of collagen gene structure and expression. Trends Genet 1990; 6: 293–300.
Gong Y, Slee RB, Fukai N et al: LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell 2001; 107: 513–523.
Cardon LR, Garner C, Bennett ST et al: Evidence for a major gene for bone mineral density in idiopathic osteoporotic families. J Bone Miner Res 2000; 15: 1132–1137.
Deng HW, Livshits G, Yakovenko K et al: Evidence for a major gene for bone mineral density/content in human pedigrees identified via probands with extreme bone mineral density. Ann Hum Genet 2002; 66: 61–74.
Livshits G, Deng HW, Nguyen TV, Yakovenko K, Recker RR, Eisman JA : Genetics of bone mineral density: evidence for a major pleiotropic effect from an intercontinental study. J Bone Miner Res 2004; 19: 914–923.
Nguyen TV, Livshits G, Center JR, Yakovenko K, Eisman JA : Genetic determination of bone mineral density: evidence for a major gene. J Clin Endocrinol Metab 2003; 88: 3614–3620.
Van Pottelbergh I, Goemaere S, Zmierczak H, De Bacquer D, Kaufman JM : Deficient acquisition of bone during maturation underlies idiopathic osteoporosis in men: evidence from a three-generation family study. J Bone Miner Res 2003; 18: 303–311.
Crabbe P, Balemans W, Willaert A et al: Missense mutations in LRP5 are not a common cause of idiopathic osteoporosis in adult men. J Bone Miner Res 2005; 20: 1951–1959.
Boivin G, Meunier PJ : Effects of bisphosphonates on matrix mineralization. J Musculoskelet Neuronal Interact 2002; 2: 538–543.
Carter DR, Bouxsein ML, Marcus R : New approaches for interpreting projected bone densitometry data. J Bone Miner Res 1992; 7: 137–145.
Tabensky AD, Williams J, DeLuca V, Briganti E, Seeman E : Bone mass, areal, and volumetric bone density are equally accurate, sensitive, and specific surrogates of the breaking strength of the vertebral body: an in vitro study. J Bone Miner Res 1996; 11: 1981–1988.
Lindner TH, Hoffmann K : easyLINKAGE: a PERL script for easy and automated two-/multi-point linkage analyses. Bioinformatics 2005; 21: 405–407.
Lander E, Kruglyak L : Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 1995; 11: 241–247.
Koshizuka Y, Ikegawa S, Sano M, Nakamura K, Nakamura Y : Isolation, characterization, and mapping of the mouse and human WDR8 genes, members of a novel WD-repeat gene family. Genomics 2001; 72: 252–259.
Nakayama M, Nakajima D, Nagase T, Nomura N, Seki N, Ohara O : Identification of high-molecular-weight proteins with multiple EGF-like motifs by motif-trap screening. Genomics 1998; 51: 27–34.
Devoto M, Shimoya K, Caminis J et al: First-stage autosomal genome screen in extended pedigrees suggests genes predisposing to low bone mineral density on chromosomes 1p, 2p and 4q. Eur J Hum Genet 1998; 6: 151–157.
Devoto M, Specchia C, Li HH et al: Variance component linkage analysis indicates a QTL for femoral neck bone mineral density on chromosome 1p36. Hum Mol Genet 2001; 10: 2447–2452.
Karasik D, Myers RH, Hannan MT et al: Mapping of quantitative ultrasound of the calcaneus bone to chromosome 1 by genome-wide linkage analysis. Osteoporos Int 2002; 13: 796–802.
Wynne F, Drummond FJ, Daly M et al: Suggestive linkage of 2p22-25 and 11q12-13 with low bone mineral density at the lumbar spine in the Irish population. Calcif Tissue Int 2003; 72: 651–658.
Wilson SG, Reed PW, Bansal A et al: Comparison of genome screens for two independent cohorts provides replication of suggestive linkage of bone mineral density to 3p21 and 1p36. Am J Hum Genet 2003; 72: 144–155.
Karasik D, Cupples LA, Hannan MT, Kiel DP : Genome screen for a combined bone phenotype using principal component analysis: The Framingham Study. Bone 2004; 34: 547–556.
Xiao P, Shen H, Guo YF et al: Genomic regions identified for BMD in a large sample including epistatic interactions and gender-specific effects. J Bone Miner Res 2006; 21: 1536–1544.
Streeten EA, McBride DJ, Pollin TI et al: Quantitative trait loci for BMD identified by autosome-wide linkage scan to chromosomes 7q and 21q in men from the Amish family Osteoporosis study. J Bone Miner Res 2006; 21: 1433–1442.
Lee YH, Rho YH, Choi SJ, Ji JD, Song GG : Meta-analysis of genome-wide linkage studies for bone mineral density. J Hum Genet 2006; 51: 480–486.
Ioannidis JP, Ng MY, Sham PC et al: Meta-analysis of genome-wide scans provides evidence for sex- and site-specific regulation of bone mass. J Bone Miner Res 2007; 22: 173–183.
Bouxsein ML, Uchiyama T, Rosen CJ et al: Mapping quantitative trait loci for vertebral trabecular bone volume fraction and microarchitecture in mice. J Bone Miner Res 2004; 19: 587–599.
Beamer WG, Shultz KL, Donahue LR et al: Quantitative trait loci for femoral and lumbar vertebral bone mineral density in C57BL/6J and C3H/HeJ inbred strains of mice. J Bone Miner Res 2001; 16: 1195–1206.
Acknowledgements
We are grateful to the family for participating in this study. We thank Drs Moerman, Vanneste and Fryns for referral of the family and K Toye and B Lapauw for expert technical help in collecting the samples.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary information
Rights and permissions
About this article
Cite this article
Willaert, A., Van Pottelbergh, I., Zmierczak, H. et al. A genome-wide linkage scan for low spinal bone mineral density in a single extended family confirms linkage to 1p36.3. Eur J Hum Genet 16, 970–976 (2008). https://doi.org/10.1038/ejhg.2008.31
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2008.31
Keywords
This article is cited by
-
Bone mineral density is linked to 1p36 and 7p15-13 in a southern Chinese population
Journal of Bone and Mineral Metabolism (2011)