Abstract
The decoding release factor (RF) triggers termination of protein synthesis by functionally mimicking a tRNA to span the decoding centre and the peptidyl transferase centre (PTC) of the ribosome. Structurally, it must fit into a site crafted for a tRNA and surrounded by five other RNAs, namely the adjacent peptidyl tRNA carrying the completed polypeptide, the mRNA and the three rRNAs. This is achieved by extending a structural domain from the body of the protein that results in a critical conformational change allowing it to contact the PTC. A structural model of the bacterial termination complex with the accommodated RF shows that it makes close contact with the first, second and third bases of the stop codon in the mRNA with two separate loops of structure: the anticodon loop and the loop at the tip of helix α5. The anticodon loop also makes contact with the base following the stop codon that is known to strongly influence termination efficiency. It confirms the close contact of domain 3 of the protein with the key RNA structures of the PTC. The mRNA signal for termination includes sequences upstream as well as downstream of the stop codon, and this may reflect structural restrictions for specific combinations of tRNA and RF to be bound onto the ribosome together. An unbiased SELEX approach has been investigated as a tool to identify potential rRNA-binding contacts of the bacterial RF in its different binding conformations within the active centre of the ribosome.
Similar content being viewed by others
Introduction
Decoding of information within mRNA is predominantly a function of a tRNA that interprets each codon via pairing with its three-base anticodon. If the correct pairing is sensed by a process that involves specific recognition of base-pairing geometry by rRNA bases that contact the codon:anticodon pair, then there is a structural transition in the tRNA that brings the amino acid on the accepting arm of the tRNA into the peptidyl transferase centre (PTC) 1, 2. The amino acid can now take part in peptide bond formation involving transfer of the growing polypeptide chain to the incoming amino acid. A high degree of fidelity has been assured. X-ray structures have shown that the active site of the ribosome is devoid of protein 3, 4, 5, implying these events involve RNA almost exclusively with the tRNA, mRNA and rRNA, all playing key roles. It is observations like these that have consolidated the concept that the ribosome is an 'RNA machine' 6. However, each X-ray structure represents a fixed snapshot and it remains a theoretical possibility there are undetected conformational changes that bring a ribosomal protein(s) into the active centre at an individual step of protein synthesis 7.
One signal in the mRNA that does not ultimately result in the incorporation of an amino acid and yet is part of the genetic code is the stop codon responsible for protein synthesis termination 8. Freed from the necessity of bringing in a new amino acid, there is no a priori reason to involve a tRNA in this step; hence, the discovery that the decoding molecules for stop codons were indeed proteins and not RNA, while unexpected, was perhaps not surprising 9. Through to the 1980s, the expectation was that a ribosomal protein would be the enzyme for peptide bond formation, and there were several prime candidates 10. At this time, the fact that an extrinsic decoding protein might join the intrinsic ribosomal proteins to carry out the function of stop signal decoding and assist in the release of the polypeptide seemed unremarkable. A new and more interesting perspective was suggested when X-ray structures revealed that the active centre of the ribosome was devoid of protein since the decoding protein release factor (RF) had to carry out its functions in an RNA environment made up of the mRNA, rRNA and the adjoining peptidyl-tRNA carrying the completed polypeptide. The implication was that the decoding RF somehow mimicked a tRNA in being able to communicate through a distance of ∼70 Å with both the mRNA in the decoding centre (DC) of the small ribosomal subunit and the PTC of the large ribosomal subunit 11, 12. The completed polypeptide is released from the peptidyl-tRNA by hydrolysis, so that it can now, untethered, finish threading its way through the exit tunnel of the ribosome ready for folding and the outside cellular world.
These concepts posed some interesting questions on how the decoding RF might function, for example, whether the factor communicated directly or indirectly with the two active sites on the ribosome for decoding and catalysis. A direct decoding model would imply the decoding RF might have a structural feature similar to the tRNA anticodon to recognize the stop codon directly and, as described below, this appears highly likely. A key question is how the fidelity of recognition is maintained to guard against premature release of a growing polypeptide and this is still unresolved. The most intractable functional question is whether the RF plays a direct role in the hydrolysis reaction by inserting catalytic residues into the PTC or an indirect role by altering the structure of the PTC to allow correct positioning of the water molecule used to mediate the hydrolysis reaction.
The question of how the decoding RF accommodates to a site (the ribosomal A site) that has been sculptured throughout evolution specifically for a tRNA and is lined almost exclusively with rRNA is a fascinating question. Each tRNA enters the A site as a ternary complex with a delivery elongation factor (EF-Tu) and GTP and leaves the A site by cycling through the inter-subunit cleft between the Peptidyl and Exit sites before leaving the ribosome at the opposite side to which it entered. The decoding RF seems not to use the same escape route but rather a complex mechanism to 'back out' of the A site by the route it entered with the help of a second class of RF and guanine nucleotide. Interestingly, not only has this mechanism uniquely different features in eukaryotes and prokaryotes 13 but also the eukaryotic decoding factor (eRF1) 14 and prokaryotic factors 15, 16, 17 are structurally distinct. This implies that the termination mechanism involving these extrinsic factors might have evolved more than once and the similarities observed today are an example of convergent evolution. As a protein-mediated mechanism in an exclusively RNA alien environment, the specific termination event may have evolved when protein synthesis was already well established.
Progress in answering these questions is discussed with new unpublished data added that enhances our understanding of this step of protein synthesis.
Methods
Methods 18, 19 used for obtaining the novel data discussed here and brief strategies for already published experiments are described in Supplementary Information.
Review
This discussion is a synthesis of published data together with new and unpublished experiments to give our best current understanding of the termination mechanism on bacterial ribosomes. Published experiments are referenced and examples of experiments to document these data are shown in some figures, but where the data are novel and unpublished this is noted.
Is the decoding RF a tRNA mimic?
Biochemical data collectively provided compelling evidence that the decoding RF had an important role in both of the key parts of the ribosomal active site, the DC where information in the mRNA was interpreted and the PTC where the catalytic activities of the ribosome were mediated. A tRNA analogue model was proposed 11 that there were at least two structural domains on the RF: one would be involved in decoding the stop signal in the mRNA and the other in the hydrolysis reaction to release the completed polypeptide at the PTC. These domains would be complemented by at least one other domain or structural element to interact with the second class of RFs for recycling the decoding factor. This model implied the decoding RF would be a highly extended molecule like a tRNA that normally occupied the site during sense codon decoding in that it had to span the ∼70 Å between the decoding site and enzyme centre of the two ribosomal subunits (Figure 1A). It was supported by two key observations. Firstly, it was possible to crosslink the RF to a modified base of the stop codon 20, 21. This observation implied the RF was in close contact with the stop codon and, therefore, suggested a direct role for the factor in codon recognition at the DC. Secondly, the hydrolysis function of the PTC could be abolished by cleavage of a single apparently exposed peptide bond within the ribosome-bound factor (determined to be between residues 244 and 245 in bacterial RF2) 11. The single cleavage somewhat enhanced the decoding function of the molecule. This implied not only was the factor in close contact with the enzyme centre and the DC but also that perhaps the two domains were conformationally coupled. The clear implication was that the decoding RF spanned the distance between the decoding and catalytic parts of the active centre of the ribosome just like a tRNA for critical functional roles and, therefore, was a functional mimic of the tRNA.
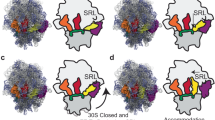
A model indicating the tRNA and RF interactions at the DC and diagrams of structural mimicry between the ternary complex of EF-Tu.tRNA, RF3 and EF-G in a similar orientation. (A) A tRNA analogue model where the RF in the A site spans the ∼70 Å between the decoding site and enzyme centre of the two ribosomal subunits in the same manner as a tRNA interaction with sense codons (left). (B) A comparison of EF-Tu.tRNA, RF3 and EF-G to show possible structural mimicry. The structure of RF3 is based on a threading analysis with the structure of EF-G.
The concept of structural molecular mimicry among protein synthesis factors and their complexes arose as a result of the X-ray structures of several factors with ligands attached. The initial example showed strikingly that the elongation factor (EF-G in bacteria) that translocates the tRNA through the ribosome was a structural mimic of the elongation factor that delivers the aminoacyl tRNA to the ribosome (EF-Tu in bacteria) when that factor was complexed with its tRNA 22, 23, 24, 25. The tRNA bound to EF-Tu was mimicked by domains III, IV and V of the protein structure in EF-G. A similar structural mimicry was subsequently proposed for the two classes of RFs 12, decoding RF (RF1 and RF2 in bacteria) and recycling RF (RF3 in bacteria), although at that time no structures were available. The model proposed that RF3 was like EF-Tu (both are translational G proteins with GTPase activity) and that the decoding RF would mimic a tRNA structure (and thereby by implication domains III, IV and V of EF-G) to give the same overall shape as the EF-G and the EF-Tu-tRNA ternary complex. In reality, this highly attractive idea has proven too simplistic in the case of the RFs. Motifs in RF3 suggest significant homology not only to the G domains of EF-Tu but also to domain IV of EF-G, implying this part of the molecule may reach deep into the DC near the mRNA 26, perhaps disrupting the interactions of the decoding RF with the mRNA and surrounding RNA 27. This is shown with the RF3 structure displayed by homology modelling against the EF-G sequence (Figure 1B). As described below, it is now clear decoding RFs undergo dramatic conformational changes that make structural mimicry less relevant.
Does this mean then the tRNA analogue model 11 was restricted only to functional mimicry and not to structural mimicry? When the first structure of a decoding RF (human eRF1) was published in 2000 14, it was found to be highly extended and 70-80 Å apart at its extremities, consistent with it being a tRNA analogue. One domain, characterized by a GGQ at its tip, was invoked to contact the enzyme centre, and a NIKS motif in a separate domain at the other extremity was invoked to be involved in decoding 14. It was somewhat 'fatter' than a tRNA and had an extra domain now known to interact with the class II factor, eRF3, but still consistent with structural mimicry as well as functional mimicry of the tRNA. What of the bacterial decoding RFs? After many unsuccessful attempts by several groups, a structure of a bacterial factor (RF2) was finally published in 2001 15. Highly surprising was that it did not resemble the human eRF1 structure, nor was it tRNA-like, but rather it was considerably more compact. It certainly did not fit the tRNA analogue proposal. While it was tempting to speculate that this was a rare non-physiological form (a crystallization artefact), the same structure was resolved from crystals with different unit cells, and later the second bacterial factor, RF-1, was shown to have the same overall structure. Puzzlingly, two motifs characterized biochemically and genetically as likely to be involved at the DC 28 and the enzyme centre, respectively 29, were quite close together (27 Å) in the structure and could not span the two parts of the ribosomal active centre. One of the motifs, the tripeptide sequence GGQ 29 (the only sequence in common between the prokaryotic and eukaryotic factors), was expected to be at the PTC, whereas the other motif, PXT in RF1 and SPF in RF2 28 (proposed as the 'anticodon' responsible for discrimination of the second and third bases of the stop codon after an elegant series of genetic studies that achieved codon switching dependent on these bases), was expected to be at the DC. The RF2 structure presented a puzzling paradox as to how the RF functioned on the ribosome. Further experiments have resolved this paradox by revealing that the molecule undergoes a highly significant conformational change 19, 30, 31, 32, 33.
How do the bacterial RFs function at the decoding site?
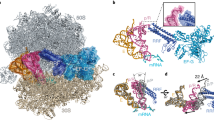
What was the detailed evidence that had placed the RF protein in close contact with the DC of the small ribosomal subunit? When the stop codon was in the decoding site as part of a designed mRNA that contained a unique crosslinking moiety as part of a modified stop codon (a side chain oxygen of the first base U had been substituted with a slightly larger sulphur atom to give 4S U), covalent linkage between bacterial RF2 and the mRNA within a ribosomal termination complex could be activated by UV light at a specific wavelength 20, 21. As the crosslinking was effectively 'zero-length' it occurred between molecules that were in very close contact. When the mRNA was radiolabeled, a new radioactive species could be identified (shown in Figure 2A) and this was reduced to the size of the native protein after RNase T1 digestion that left just a dinucleotide attached to the protein, implying that the contact between the RF and the stop codon was very intimate.
PAGE separation of RF2 crosslinked complexes and an X-ray structure of the bacterial termination complex showing the RF interaction with the stop codon. (A) Analysis of the fragments of mRNA containing thio-UG*AC. Aliquots of the reactions with (+) and without (−) RF2 were subjected to ribonuclease (RNase) T1 digestion (+). The large radioactive band (top left) shows the position of the RF2-mRNA crosslinked species prior to digestion. The band (arrow) shows the position of RF2 with the radioactive dinucleotide (thio-UG*) attached following digestion. *G represents the radioactive nucleotide. (B) A depiction of the X-ray structure (modified from Petry et al. 32) showing the α5 helix/loop of RF2 orientated towards the first base of the stop codon.
A detailed search for the crosslinked site on the bacterial RF2 used in the experiment involved cleavage of the factor by specific proteolysis with chymotrypsin. The peptide fragments were separated by HPLC to detect any that were radiolabeled (that is, had the RNA dinucleotide attached). One radioactive peptide was identified and shown to contain the sequence DIQ. This placed the crosslink within the α5 helix region of RF2 15 towards the N terminus of the molecule (131-133), somewhat distant from the SPF motif (207-209) identified by Ito et al. 28 as discriminatory at the second and third bases of the stop codon, but closer to the site of a number of charge-switch mutations that result in relaxed codon recognition 34, 35. Nevertheless, we did not publish details of the crosslink site on the RF since there was no supporting biochemical evidence or a contextual biological explanation. Recently, however, with the publication of the compelling X-ray structure of the bacterial termination complex by Petry et al. 32, this has been provided. While it was not possible to resolve the electron density of the anticodon loop from that of the stop codon due to the medium resolution of the crystal structures, in this depiction there was an unexpected feature, the α5 helix/loop on the factor containing the DIQ sequence was oriented towards the first base in a manner that could explain why a crosslink directed away from a moiety within this base of the stop codon might have occurred with this sequence on the protein. DIQ is just three amino acids from the GG at the tip of the α5 loop (Figure 2B). Collectively, biochemical data and structural analysis provide strong evidence that the bacterial decoding RFs are intimately involved in stop codon recognition and highlight which part of the RF structure is involved in first base discrimination.
Is the decoding RF more promiscuous than a tRNA in its contacts?
When the stop codon enters the ribosomal A site, the 'last' tRNA carrying the completed polypeptide occupies the P site. Petry et al. 32 were able to obtain stable ribosomal complexes with RF only when a tRNA occupied the P site and mRNA was present in the complex. This implies that the tRNA is contributing to the stability of the RF binding either indirectly by stabilising the conformation of the ribosome or directly by providing a binding face for the RF. We have determined whether the RF makes close contact with the P-site tRNA at the decoding site near the anticodon by determining whether the base adjacent to the anticodon of a P-site tRNAArg (base 32) can crosslink to the RF. Figure 3A shows a cartoon of the orientations of the P-site tRNA with the crosslinking moiety indicated. When crosslinks were activated by light of the appropriate wavelength, the tRNA was able to form crosslinks to the RF as shown in Figure 3B (previously unpublished). This indicates that the RF makes close contact with the P-site tRNA. It may reflect that the RF has structural dimensions when on the ribosome that are somewhat wider than a tRNA and that it 'squeezes' into the A site.
RF crosslinking to the P-site tRNA. (A) An orientation of tRNAArgshowing the anticodon and the orientation of the natural thio-C at position 32. (B) PAGE analysis of complexes with (+) and without (−) RF2. The upper arrow shows the position of the RF crosslinked (XL) to tRNAArg with the position of non-crosslinked RF (lower arrow) indicated.
Ito et al. 28 had identified a tripeptide motif in domain II of the RF proteins that differed between the two bacterial factors and seemed to be the key to discrimination between A and G in the second and third position of the stop codon. A and G are both allowed in the second and third positions of stop signals but only one decoding factor, RF2 (UAA, UGA), can recognize G in position 2 and only the other factor, RF1 (UAA, UAG), can recognize G in position 3. This recognition profile can exclude UGG as a stop codon since neither factor is able to recognize G at both positions. While no definitive evidence for contact between these motifs and the mRNA was provided in these elegant genetic studies, it was strongly implied as switching the unique motifs in each factor was accompanied by a change in their codon specificity. A model for discrimination at the second and third bases was presented 28 (Figure 4A).
Modelling the decoding motifs of bacterial RFs with the mRNA and mapping the SPF motif of RF2 to the DC of the ribosome. (A) The PAT and SPF motifs of E. coli RF1 and RF2, respectively, were modelled as 'anticodons' to explain how the second and third bases of the stop codons could be differentiated by these factors to give their specific recognition patterns (RF1 UAG; RF2 UGA). This diagram was modified from Ito et al. 28. (B) The SPF specific region of RF2 was mapped exclusively to rRNA from one of the two ribosomal subunits. The cleavages in E. coli rRNA derived from hydroxyl radicals generated from near 205SPF207 (Cys 204 or 209 on modified RF2s with unique cysteines have been mapped here on the rRNA structures derived from X-ray structures of 16S rRNA from Thermus thermophilus 36. The black and the solid 'light grey' indicate the cleaved regions of rRNA (to distinguish adjoining cleavage when viewed in two dimensions). The panel was created in Pdb swiss prot viewer.
We engineered RF2 to remove its two cysteines at positions 128 and 274 (replacing them with alanine and serine, respectively) with no significant loss of functional activity and then inserted a cysteine at 204, and at 209, in two different constructs spanning the tripeptide motif (SPF 205-207) identified by Ito et al. 28. The reagent 1-(p-bromoacetamidobenzyl)-EDTA (BABE) was attached to the specific cysteine, newly engineered into the protein so that Fenton chemistry could be used to activate the generation of free hydroxyl radicals at the site. This enabled the mapping of desired sites on the factor (in this case the SPF motif) to ribosomal co-ordinates, after forming a ribosomal complex and activating the radical production. The radicals cleave the rRNA at sites with which they collide and, in principle, the most frequent cleavages represent those parts of the RNA nearest to the site of radical generation. Specific cleavages were found only in the small subunit rRNA, with none identified in the rRNA of the large subunit. When mapped onto the three-dimensional model of the ribosome as determined by X-ray crystallography, they formed a ring delineating the DC of the ribosome 19 (Figure 4B). Collectively, the data from the crosslinking studies and from the subdomain swapping experiments implied that the discriminatory motif was at the DC and must be at least near the mRNA, and the hydroxyl radical mapping provided compelling support for a major role of RF in termination codon recognition.
The X-ray structure of the termination complex 32 modelled the loop containing the SPF motif of RF2 (PXT of RF1) into the decoding site. In the modelled structure, the discriminatory motif was in the near vicinity but not in close contact with the second and third bases of the stop codon as predicted although it is likely the region is restructured when the termination complex forms (see Figure 2B). It was not possible to resolve the merging density between the RF and the mRNA in the ribosome structure to derive the actual contacts in the structure. This loop does not appear to have flexibility in the crystal structures of the RFs 16.
The eukaryotic RF has been shown not to respond to a simple triplet codon, but requires four bases as a minimum for activity in vitro unlike the bacterial factor that has activity with the three base codons as specified in the genetic code 37. Nevertheless, the concept that the stop codon might extend beyond three bases in bacteria as well as in eukaryotes was provided by statistical analyses of the gene regions around stop codons in a wide range of organisms. After correcting for the slight bias in the occurrence of each of the four bases (A, G, C and T) between positions 1 and 3 of sense codons, there is no further bias apparent as one moves through the coding region in genes towards the stop codon. Before the stop codon is reached, however, a clearly identified reproducible bias is revealed and it is still present for a short section of sequence after the stop codon. Then the unbiased pattern returns within the untranslated region. Identified initially in E. coli since it was the first organism where significant numbers of gene sequences became available 38, this presented as the classic signature of a sequence element. Subsequently, it has been found in the genes of almost all organisms examined 39. The pattern revealed that for the genes in many organisms the most striking bias was in the position immediately following the stop codon (+4). This suggested a promiscuity of contacts by the RF with mRNA compared with tRNA and that the RF may make further contacts with bases downstream of the stop codon. This would be part of an extended sequence element for the molecular signature of the termination signal 8. We tested the significance of this experimentally in vivo in bacteria and in biochemical crosslinking studies. Indeed, in E. coli where the +4 base was altered, each of the three stop codons showed a widely differing hierarchy of termination signal efficiencies dependent on the identity of this base (Figure 5A). Strength of 4-base signals, UAAN and UGAN, correlated well with the frequency at which they are found at natural termination sites in E. coli 18. Moreover, if the crosslinking moiety was placed on the +4 base within the designed mRNA, then a crosslink additional to that found from position 1 was obtained, indicating that the decoding RF also made close contact with this base. How does this potential interaction of the decoding RF with the fourth base affect orientation of the protein to the first base? We determined how the fourth base affected the crosslink between the first base of the stop codon and the decoding RF. As an example, as shown in Figure 5B (previously unpublished), a change from +4 G to U following the UAA stop codon enhances the strength of the crosslink between the first base of the stop codon and RF1, which preferentially recognizes these stop codons. Petry et al. 32 observed that electron density attributable to the decoding RF extends to the fourth base of the termination signal, as we predicted from our biochemical studies.
The influence of 4-base stop signals on the efficiency of protein synthesis termination. (A) Results are shown for UGAN signals. The protein products (upper panel lower band: termination product; the upper band: frameshift product) and the termination efficiency for each signal expressed as a percentage are shown (graph). (B) PAGE analysis showing the strength of crosslinks to RF1 for UAAG and UAAU signals. The position of a +1 crosslink (after ribonuclease T1 digestion) is shown. The upper band represents probable crosslinking to ribosomal protein S1.
X-ray studies with mRNA complexed with cognate and non-cognate tRNA on the ribosome gave great insight into how structural changes in nucleotides of the rRNA in particular were important for decoding fidelity, with the rRNA acting as a sensor for correct codon/anticodon pairing that was particularly stringent at the first and second base positions. It was less so at the wobble third position, thus providing an explanation of how a tRNA can recognize more than one codon differing in the third position. The antibiotic paromomycin that causes miscoding was used to trap the flexible rRNA bases in the conformation normally adopted when the codon/anticodon recognition is cognate (and fool the ribosome to incorporate an amino acid in error) 1, 2. After sensing cognate interaction, a more profound conformational change in the tRNA is triggered so that it bends into the PTC for the incoming amino acid to be incorporated into the growing peptide chain. Could this occur with the stop signal? There may be analogies with the recognition of the cognate bases of the stop signal, with the RF fixing parts of the DC in a particular conformation, which then triggers a greater conformational change in the ribosome or RF that facilitates its activities at the PTC. The structural data from the termination complex have not yet reached a high enough resolution to suggest how fidelity of stop signal decoding might be controlled and whether there are indeed structural changes in the rRNA.
In vivo studies suggest there is a very high level of selectivity by RF for genuine stop codons 40, and yet site-directed crosslink studies in vitro with physiologically relevant buffers show the RF can enter a termination complex and make contact with codons to give productive crosslinks when only one of the second and third bases is cognate (the first base U is fixed and contains the crosslinking moiety (Poole and Tate, unpublished)). For example, a crosslink is obtained with UCAG or UAGG and RF2, which are non-cognate in the second and third positions, respectively, but not with UCGG where both the second and third bases are non-cognate. This implies there may be rather loose initial scanning, with the site-directed crosslinks occurring in an initial binding state. Such non-cognate interactions do not lead to termination of the growing polypeptide prematurely in vivo, otherwise a completed protein would never occur. The very low decoding error rates in vivo suggest there must be another step, similar to tRNA accommodation for regular sense codons, that results from this initial scanning of the cognate or non-cognate complex between the RF and stop codon. Resolving this question will be a challenge for the future.
After a comprehensive study of how the sequences upstream and downstream of the stop codons affect the efficiency of termination and the preclusion of readthrough or frameshifting in specific in vivo assays, we defined the sequence element for the E. coli stop signal as a 12-base sequence, of the form NNN NNN STOP NNN 39, 41. The involvement of the downstream nucleotides in the signal may be explained by the interactions these bases make with the RF since not only could we detect crosslinks from the +4, +5 and +6 positions of the mRNA to the RF but not beyond 42, but also bases in these +4 to +6 positions affected the efficiency of the signals when under competition from either non-cognate readthrough in the presence or absence of suppressor tRNAs, or from programmed frameshifting 39, 41. These results were also consistent with the accumulating bioinformatics predictions that indicated a bias beyond the +4 base. The region of the RF forming crosslinks to the +4, +5 and +6 positions is not obvious from the crystal structure of the termination complex 32. However, these crosslinks may occur when the RF is in a different conformational state to the one modelled into the crystal structure, which is a conformational state where scanning of the cognate/non-cognate interaction is still to occur.
The involvement of six upstream bases in the mRNA in the defined signal was not so easily explained as these bases are already occupying the P and E sites and are involved in other interactions. However, as described above, there may be a restriction on the tRNAs that can best be accommodated in the P site (and E site) when RF occupies the A site. This could explain why there is a strong bias in codon pairs involving the last codon and the stop codon 43 with some missing altogether in gene sequences and others occurring at widely differing frequencies, suggesting some tRNAs are restricted from occupying the P site as the last tRNA in a termination complex. The codons that show strong positive selection bias at the last codon position in general are recognized by single species of tRNAs that are hyper modified at position 34 or 32 or 37 or combinations of these positions 39. These modifications could be binding determinants that stabilize the RF-stop codon interaction and increase the rate of decoding of the signal and, thereby, would explain why a crosslink from position 32 of the P-site tRNA is possible (see Figure 3B). This then could provide an explanation for the inclusion of the last codon in the termination signal. Essentially, these upstream sequences may, despite being a linear signal, communicate three-dimensional information that affects the architecture of the ribosomal A site into which the 'alien' RF protein binds.
We tested whether upstream sequences affected the RF orientation to the first base of the stop signal, utilising site-directed crosslinking when different codon/tRNA combinations were in the P site. As shown in Figure 6A (previously unpublished), the crosslink profiles from the first base of the stop codon to the RF in the A site were significantly affected by the specific identity of the tRNA in the P site. The orientation of the factor to the stop signal at this key invariant position was clearly affected by the adjoining tRNA, specified by the codon in the upstream part of the defined sequence element. This can explain why the last codon (the NNN adjacent to the stop codon in the sequence element) was highly influential on the efficiency of stop codon readthrough as determined in a series of studies by Isaksson and colleagues 44, 45. Moreover, if a Shine and Dalgarno element that can base pair with the 16S rRNA is placed upstream of the stop codon, as is found in the frameshift site for the prfB gene encoding RF2, then the orientation of the first base of the stop codon to the decoding RF is again affected as determined from the site-directed crosslinks (Figure 6B). This is of specialized interest for the rare frameshift mechanism that occurs during the translation of the RF2 mRNA. The UGA stop signal at the RF2 frameshift site has been determined to be particularly weak because of its downstream context, CUA, with this being the weakest of the 64 possible combinations for this +4NNN+6. Additionally, however, the stop signal strength may be further compromised by the upstream interaction between the Shine and Dalgarno sequence in the mRNA and the rRNA that clearly has the potential to exert an influence downstream and lower the rate of recognition of the factor for this internal stop signal. This is additional to the major effect of this interaction at the frameshift site that has been shown to destabilize the E-site tRNA leading to the existing frame being maintained only by a single codon/anticodon base pairing and thereby primed for failure 46.
Analysis of RF crosslinking to stop signals when different tRNAs are in the P site and when different upstream sequences are present in the mRNA. (A) The intensity of crosslinks with different P-site tRNAs. The stop signals are UGAG excepting for P-site tRNAAla where it is UGAU. The different strengths of the crosslinks to the +1 and +4 (tRNAAla) thio-Us and to ribosomal protein S1 are indicated. (B) Crosslink intensities with different upstream sequences (left) before and after ribonuclease T1 digestion. Upstream sequences in separate mRNAs comprise a Shine and Dalgarno (SD) element, a nullified SD element (NSD) and an undefined SD element (USD). Arrows denote the positions of RF2 and S1 crosslinks.
Are there interaction sites for the decoding RF at the active centre of the ribosome?
As described above, a paradox existed initially between the crystal structure of the bacterial decoding RF as a compact structure and the function of the protein in spanning the two parts of the ribosomal active centre. Key motifs on the RF off the ribosome were only 20-30 Å apart and yet on the ribosome seemed to be close to the decoding and the catalytic centres that were 70 Å apart. Two cryoelectron microscopy studies resolved this paradox by showing a much more elongated structure than the crystal structure 30, 31. It was clear domain III had undergone a rotation away from the body of the protein like the derrick of a crane, and with this massive conformational change now extended up to the PTC, with the superdomain (domains II and IV) oriented towards the DC. How and when this structural change occurs is still not clear, but it seems likely to occur at some point when the factor is binding or has initially bound to the ribosome. An alternative model has been proposed that both the open and closed forms of the structure occur in solution in equilibrium but only the open form binds to the ribosome. Small-angle X-ray scattering data from E. coli RF1 and a functionally active truncated RF1 derivative have provided evidence for the existence of RF1 in the open cryoelectron microscopy conformation in solution 47. The flexibility of the open form untethered in solution might then explain why this form was not captured in an X-ray structure.
What seems most likely is that there are at least two binding states of the decoding RF on the ribosome and they may have quite different ribosomal footprints. Moreover, even though the two cryoelectron microscopy studies used the same source of RF/ribosome complexes, the density attributed to the decoding RF was not identical, and the orientation of the modelled factors also differed from the X-ray structure of Petry et al. 32. These may simply reflect the limitations of the resolutions of the structures analysed rather than real differences. However, these structures represent snapshots of the decoding factor on the ribosome and, equally likely, there may be a dynamic pattern of ribosomal interactions of the RF with the proteins at the entrance to the active centre of the ribosome, and then with rRNA within the centre, reflective of multiple binding states during the termination process.
The footprint of the bacterial RF on the ribosome
Little is known on the exact details of the RF footprints on the ribosome. How might the subtleties of these footprints be elucidated biochemically? As a start to resolving this problem, we have trialled a modified SELEX procedure that had been used successfully to define more precisely known binding sites of specific ribosomal proteins with rRNA 48, 49, 50 after fragmentation of the rRNA into short sequences. Importantly, the binding sites of these proteins with intact ribosomes were reproduced from the fragmented rRNA, showing that the technique could identify physiologically relevant short binding motifs. The aim in our study, in contrast, was to probe unknown interactions between the RF and rRNAs to see whether this technique might be appropriate to define rRNA contacts made by the decoding RF. In vitro Selection from Randomly Fragmented rRNA (SERF) is described here with the two different bacterial decoding RFs: RF1 and RF2 from E. coli. Each makes functional interactions with the rRNA-rich ribosomal A site of the E. coli ribosome. We have correlated the data with the published literature on known ribosomal regions of interaction.
Interactions between rRNAs of both ribosomal subunits and RFs are likely to be essential for correctly positioning the factors into the ribosomal A site. Indeed, whereas the interaction of RF2 with the E. coli 70S ribosome can be documented readily, for example, on an immunoblot after separation of the ribosome complex from the unbound factor (Figure 7A), in contrast, associations of the factor with either the large subunit or small subunit individually are very weak or almost undetectable (Figure 7A, rows 2 and 3, respectively), even when crosslinking is used to stabilize the interactions before purification 51. Nevertheless, the weak interaction of RF2 with the large ribosomal subunit can be enhanced by an isolated cognate stop codon, in the absence of the small subunit (Figure 7B; Brown and Tate, unpublished). Since the decoding of the stop codon is not a large subunit function, the result in Figure 7B implies that a direct interaction of the codon with the factor does impart some conformational change that strengthens the RF interaction with the 50S subunit. Indeed, [32P-labelled] UGAN has been shown to interact with the RF in the absence of either of the subunits or the intact ribosome, whereas no binding could be detected with a series of non-cognate stop and sense codons tested (McCaughan and Tate, unpublished). A similar enhanced binding of RF to the small ribosomal subunit by cognate codon was also obtained, but since the mRNA binds to this subunit this result was not unexpected.
Binding of RF2 to ribosomes or individual ribosome subunits. (A) Immunodot blots detecting RF2 ribosome/subunit complexes. After binding RF2 to 70S (Row 1), 50S (Row 2) and 30S (Row 3), the complexes were fixed by crosslinking with dimethylsuberimidate before separation on sucrose gradients away from unbound factor. RF2 was detected in each fraction by its immuno reactivity with its specific antibody. (B) Stimulation of RF2 binding to the 50S subunit by the cognate stop codon. The influence of stop codon on the interaction of RF2 with the 50S subunit was detected in a more sensitive ELISA assay following separation from the unbound factor.
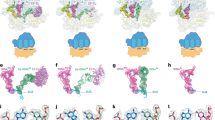
Preliminary results from the new SERF strategy selected two classes of fragments of rRNA for the RF ribosome footprint. Firstly, rRNA fragments were selected that were consistent with previous biochemical or structural studies that implicated a regional involvement in termination; secondly, the strategy selected a small number of rRNA fragments that were unexpected and were from regions not previously supported by the models of the RF on the ribosome. A good example of the first type of rRNA motifs is the protein L11-associated rRNA, where there are already accumulated biochemical data supported by the cryoelectron microscopy and X-ray structures, which not only suggest a close proximity of the bacterial decoding RFs, RF1 and RF2, to this region but also suggest that the orientation of the two factors must be different to the extent of having a profound differential effect on their activities (discussed in more detail below). An example of the second class is a fragment isolated from the region of the L1 stalk. The isolation of fragments from this side of the ribosome, distant from the side that the factors enter, could simply represent RNA fragments that are 'false positives', but the recent evidence of L1 stalk movement towards the active centre during protein synthesis (and the fact that both the bacterial and mtRF1-type factors selected different fragments from this region) means even such unlikely rRNA sequences might be worthy of further investigation.
Significantly, the range of fragments selected by the bacterial RFs did not map in a scatter pattern (Figure 8) over the structures of the rRNAs in a manner that might have indicated significant non-physiological 'noise' in the selections. Additionally, most selected fragments scored positive when tested in a yeast three-hybrid RNA protein interaction system that we used as an independent measure to confirm the interactions. There were only two different fragments isolated from 16S rRNA, and only one of these was isolated multiple times. Five fragments from 23S rRNA were isolated multiple times (and a number of others singly), and a fragment from the 3′ part of 5S rRNA was isolated many times. Most fragments were isolated by both factors, or two RF1s, bacterial RF1 and mRF1 (UAA, UAG specificity). The relatively small number of fragments selected may reflect that the factors actually make relatively few significant contacts with rRNA, or, perhaps as likely, the affinity of each factor for any one site is low and, therefore, the technique provides a selection tool of relatively high stringency.
Mapping the selected fragments of rRNAs bound to RF1 and RF2. (A and B) The backbone structure of Thermus thermophilus 16S ribosomal RNA at 3.31 Å 36. The fragments of rRNA bound to RF1 (A) and RF2 (B) are shown in green and red with h44 in a darker shade of grey for orientation. (C and D) The structures of 23S and 5S rRNA from Deinococcus radiodurans 52. The selected fragments of large subunit rRNA bound to RF1 (C) and RF2 (D) are shown in multiple colours. Nucleotide numbers for each fragment and associated helices in brackets are given. The L1 region has a disordered structure; therefore, all selected nucleotides are not shown in (C) and (D). The figures were created in Pdb swiss prot viewer.
The structure of the backbone of 16S rRNA is shown in Figure 8A and 8B. The results obtained in the SERF selection with the two bacterial factors, RF1 and RF2, can be mapped onto the structure. The regions illustrating the two fragments of the rRNA that have affinity for RF1/RF2 are shown in red and green, respectively. Helix 44, traversing from the decoding site at the top to the bottom of the subunit, is shown in a darker grey as an anchor point in Figure 8A. In this small subunit, both bacterial factors RF1 and RF2 selected multiple times a fragment of helix 21 (red) that is part of the central domain of 16S rRNA. The central domain is dominated by helices 21-23 and helix 21 wraps around the back of the 5′ domain. In addition, a single isolate of a fragment of helix 24 (green) was selected with RF1. Helix 24 contains the conserved 790 loop and its vicinity to RF1 and RF2 was earlier indicated by hydroxyl radical footprinting from specific sites on the factors 19, 53, 54.
From the large subunit rRNA, RF1 and RF2 selected some fragments in common and some uniquely. Figure 8C and 8D show these fragments mapped onto the backbone structure of 23S rRNA from Deinococcus radiodurans at 3.1 Å 52. A common fragment selected by both factors was from domain VI (nucleotides 2640-2670) that comprises the sarcin-ricin loop (helix 95). Previously, we have shown that the RF2 ribosomal interaction affects the chemical reactivity of nucleotides within the loop (Brown and Tate, unpublished). The sequence and overall structure of this loop is critical for its function 55, 56. This loop is located within domain VI of 23S rRNA at the surface of the ribosome below the GTPase-associated centre. The vicinity of RF2 to this loop was also indicated by cryoelectron microscopy 30, 31.
Does this analysis give any insight into initial contacts made by RF during its interaction with the ribosome? The 3′ end of 23S rRNA, exposed at the surface of the ribosome near to the factor entry site to the active centre, was selected by SERF with both bacterial factors. The regions selected by RF1 uniquely included nucleotides from the GTPase-associated centre (helix 42-44). The GTPase-associated centre (including proteins L11 and L7/L12 stalk) is also at the side of the subunit where the RF enters the active centre of the ribosome. Selection of these fragments is consistent with past biochemical studies of factor-dependent termination that implicated the regions in which they reside. The L11 region has long been known as important for both RF1 and RF2 function. Ribosomes lacking L11 (derived from E. coli mutants) 57, 58, 59 were inactive with RF1 in vitro (but hyperactive with RF2) 60 and this phenotype could be simulated by a specific N terminal tyrosine (Y7) modification on L11 that also abolished RF1 function 61. The ribosomal region has been inferred previously to be relatively close to RF1 by hydroxyl radical footprinting 53 as well as to EF-G 54. The cryoelectron microscopy model 30, 31, hydroxyl radical footprinting 19, 51 and genetic analysis 57 all confirmed the importance of this region of 23S rRNA as a site of potential interaction between factor and ribosome during translation termination.
A more centrally located region in domain IV of 23S rRNA was specifically selected by RF2 (Figure 8D). This region is located at the subunit interface and nucleotides in this region are involved in making bridges between the 50S and helix 44 of the 30S subunits 62. This region is relatively close to the GGQ motif of RF2 as shown by hydroxyl radical footprinting 19.
A 5S rRNA fragment (nucleotides 89-120) was the most common fragment repeatedly selected by RF1 and RF2. This region is located in domain IV or loop D of 5S rRNA. From the structure of the large subunit (Figure 8C and 8D), it is clear that these nucleotides make a bridging interaction with domain II and V of 23S rRNA and are critical for ribosomal function 63, 64, 65. Mutation of a conserved nucleotide (U89) in loop D of 5S rRNA disturbs ribosomal function 66, 67.
The global footprint of RF2, from reconstructed cryoelectron microscopy images and X-ray structures of this factor and RF1 on the ribosome in a fixed termination state, assisted greatly in assessing the significance of the fragments selected 30, 31, 32. Those in near proximity to the derived positions are the sarcin-ricin loop, the central region of domain IV of the 23S rRNA, the L11 region and helix 24 in the small subunit (both RF1-derived fragments). Although the 5S rRNA subdomain would appear to be somewhat distant, its position seems quite flexible with crosslinks found from nucleotide U89 to several nucleotides of 23S rRNA quite close to the RF2-derived image 63, 68. Putative interactions of fragments that are quite distant from the imaged RF footprints and located at the extremities of the subunit, such as the exposed 3′ terminus of the 23S rRNA on the factor entry side, the L1 structure (RF1) and the helix 21 region of the small subunit, require further validation.
Two separate models proposing two-stage binding of RF to the ribosome have been previously suggested to explain genetic and biochemical data. The first invokes initial ribosomal binding of RFs prior to codon recognition with the involvement of the N terminal domain (domain I in RF2) 69, and the second is a kinetic model involving two binding states (state 1 facilitating initial ribosome binding, and either a competent state 2 involving formation of a termination complex with cognate codon or a non-competent state 2 with near-cognate or non-cognate codons) 70. Although the existence of folded and unfolded forms of RF2 was not appreciated at the time these models were proposed, unfolding of domain III could be mediated following cognate codon recognition after the codon independent initial binding (state 1). Correct orientation of this domain at the PTC and domain II of RF2 at the decoding site, determined by whether there is a cognate (stop) or non-cognate codon in the A site, would result in the termination competent state (state 2). Hence, some of the exposed outer fragments of rRNA determined to have affinity for RFs in the current study might be important for this initial binding or transition to the termination competent state represented by the cryoelectron microscopy of RF2.
Accommodation of the decoding RF at the PTC
Following cognate tRNA/sense codon recognition in the A site, there is an accommodation of the tRNA as it swings into the PTC. What have the structures suggested regarding how the decoding RF is accommodated following cognate stop codon recognition? The X-ray structures of the ribosome showed that there were no ribosomal proteins within about 18 Å of the site of key rRNA structures thought to be where the catalytic activity of the ribosome resides. This means that it is exclusively an RNA centre, apart from the specific site of peptide bond formation where the growing peptide chain is transferred to the incoming amino acid, during the elongation of the growing polypeptide chain when the tRNA occupies the ribosomal A site. However, in the termination event of protein synthesis, uniquely, a protein decoding RF extends up to the catalytic centre. The crystal structures of the RFs on the ribosome show that the loop at the extremity of domain III (containing the GGQ motif and several other residues flanking this motif) comes into close contact with the PTC 32. It is ordered, faces the last 3′ nucleotide of the P-site tRNA carrying the completed polypeptide and is in the close vicinity of several important 23S rRNA nucleotides of the PTC. Of the nearest nucleotides, A2451 has been implicated as having a catalytic role in peptide bond formation 71, 72, and U2602 has been previously implicated as the most important nucleotide for the hydrolysis event that releases the completed polypeptide 72. These findings are highly provocative to support the contention that the decoding RF might participate quite directly in polypeptide release. However, the resolution of the X-ray structure of the termination complex is not high enough to place the individual side chains of the RF2 loop (residues 244-257) or to observe whether a water molecule is associated with a particular amino acid.
Of interest is that the site in the RF2 protein most sensitive to proteolytic cleavage, identified as between amino acids 244 and 245, is at the start of this loop. Cleavage abolished the peptidyl-tRNA hydrolysis function of the PTC, which led to our proposal of the tRNA analogue hypothesis for the decoding RF in 1994 11. The next residue in the loop at position 246 is also particularly interesting. In most RF2 genes the codon at this position encodes Ala or Ser (the residue at this position in E. coli RF1) 73, 74. Unusually, the E. coli K12 genome has Thr at position 246. When the RF2 protein is expressed in E. coli from this gene, the recombinant protein binds to the ribosome with the expected specific activity for codon recognition but with a low or sometimes no specific activity for hydrolysis. In contrast, the RF1 gene can be expressed to give a protein with the expected specific activities for the two main functions, codon recognition and peptidyl-tRNA hydrolysis. This loss of activity in RF2 seems to be solely dependent on amino acid 246 since if the Thr is substituted by the typical amino acids in these positions (Ser or Ala) specific activity is restored to normal levels. A lack of a methyl modification on Q252 of the GGQ loop in the highly expressed protein may, however, be the tipping point, perhaps because the RF2 concentration exceeds the capacity for the modifying enzyme. Now, the slightly longer side chain of Thr over Ser or Ala apparently can no longer be accommodated at the 'hydrolysis site' leading to dramatic loss of activity. Of interest is that U2602 was cleaved at high frequency when hydroxyl radicals were released from amino acid 246 in the Thr246Cys protein variant 19. These data also point to the RF having an intimate relationship with the PTC.
To further test this we systematically changed amino acids 238-273 from the existing residues to cysteine rather than alanine as cysteine can potentially be used as a 'launching site' for hydroxyl radicals to facilitate mapping the positions of these residues relative to ribosome structures. Interestingly, the codon-dependent binding activity of these variants to the ribosome was mostly enhanced by changes to domain III amino acids (Figure 9A). A small number of the variants had modest reduction in their binding activity but none of these included the 11 residues within the 'GGQ' loop. The hydrolysis function of these variant RF2s was measured both with the cognate codon, UGA, and with the non-cognate stop codon, UAG, as a control. The variants involving loop residues all had severely reduced activity with the cognate codon (and specificity was maintained – in no case was there activity with the non-cognate codon) (Figure 9B). Interestingly, an exception for the severe effect of changes in loop residues was the variant T246C. Changing the Thr to Cys produced the least affected variant, perhaps reflecting that the Thr in this position was already significantly compromising the activity of the hydrolysis function of recombinant RF2. In our hands, the variant Q252C of the GGQ loop could not be isolated without incurring other mutations in the adjoining region (70 clones screened) and no data are shown for this particular variant. These mutagenic studies highlighted that all of the amino acids in the loop region of RF2 are important for sustaining the hydrolysis function. Amino acids close to the beginning and the end of the loop in the helix extending it to the catalytic centre are also quite sensitive to changes (residues 240 and 241 before the loop; and residues 259, 260, and 262 after the loop) as significant loss of hydrolysis activity was observed when these residues were substituted, but beyond that the substitutions can be made generally without significant loss of the hydrolysis function.
Ribosome dependent functions of variants of RF2 with indicated nutations in domain III. (A) Cognate stop codon-dependent ribosome binding of RF2 variants. Ribosome binding assays were performed with cognate codon UGA (closed bars) and non-cognate codon UAG as a control (open bars) using equal ratios of ribosome to RF variant. Experiments were repeated in triplicate with duplicate samples in each case and binding activities were expressed as the average RF·70S ribosome· [32P] stop codon complex formed (pmol) plus the SEM. *p < 0.05 and **p < 0.01 (Student two-tailed t-test) change in activity with respect to the unmodified RF2. (B) Peptidyl-tRNA hydrolysis activities of the RF2 variants. Peptide release assays were performed with the cognate codon UGA (closed bars) and the non-cognate codon UAG (open bars) using 5 pmol of the variant RF2s. Experiments were repeated in triplicate with duplicate samples in each case and hydrolysis activities were expressed as the average release of f[3H]Met (fmol) plus the SEM. *p < 0.05 and **p < 0.01 change in activity with respect to the unmodified RF2. The position of the domain III loop is shown by the red bar (amino acids 244-255).
The recent observation of the intimate contact between residue 246 and nucleotide 2602 is particularly important (as described above U2602 has been previously implicated as the most important nucleotide for the hydrolysis event that releases the completed polypeptide 72). This nucleotide has a water molecule positioned close by in structures of the large ribosomal subunit complexed with novel transition state analogues, aimed at unravelling the mechanism of peptide bond formation 75. The water molecule interacts with the oxyanion of the transition state tetrahedral intermediate. This could also be 'the water molecule' that is the acceptor for the completed polypeptide at hydrolysis during peptide chain termination, perhaps hydrogen bonded to amino acid 246 of RF2.
Conclusion
Three major questions remain in our understanding of how the decoding RF can functionally mimic a tRNA in the RNA-rich active centre of the ribosome. Considerable progress has been made with the comprehensive biochemical and structural evidence supporting a functional mimicry. The idea of structural mimicry as well is now blurred by the knowledge that the decoding protein undergoes major conformational changes to fulfil its funtion. One question to be resolved is how the factor, mRNA and rRNA achieve a high fidelity recognition of the stop signal and what microstructural changes occur when a cognate stop signal is detected. The biochemical and structural data, in particular, have recently provided an excellent platform to advance this knowledge. The second major question is how and at what point in the termination mechanism the critical structural change in the factor occurs as well as what are other structural changes that accommodate the factor at the PTC. Novel approaches and existing strategies perhaps drawn from other systems will likely give insight into these processes. Finally, the question of how the PTC carries out its catalysis functions (including hydrolysis of the completed polypeptide away from the last tRNA) is still to be resolved although there are now some tantalizing hints. Does the RF co-ordinate a water molecule at the site for the reaction; or does it carry a water molecule in with it or provide an electrostatically acceptable channel for water to enter and participate in the hydrolysis of the polypeptide from the tRNA, with the water molecule as the acceptor rather than the alpha amino group of an incoming amino acid?
Note added in proof: A just published crystal structure of RF3 76 does not support a role for RF3 extending down to the decoding site despite the observed sequence homology with domain IV of EF-G. Additionally, it has now been established residue A2541 of the 23SrRNA is unlikely to have a catalytic function in peptide bond formation as previously thought, rather its likely role is in structural ordering of the peptidyl transferase centre and participating in a network of hydrogen bonds at the site 77.
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
References
Ogle JM, Carter AP, Ramakrishnan V . Insights into the decoding mechanism from recent ribosome structures. Trends Biochem Sci 2003; 28:259–266.
Ogle JM, Ramakrishnan V . Structural insights into translational fidelity. Annu Rev Biochem 2005; 74:129–177.
Cech TR . The ribosome is a ribozyme. Science 2000; 289:878–879.
Ban N, Nissen P, Hansen J, Moore PB, Steitz TA . The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 2000; 289:905–920.
Nissen P, Hansen J, Ban N, Moore PB, Steitz TA . The structural basis of ribosome activity in peptide bond synthesis. Science 2000; 289:920–930.
Wilson KS, Noller HF . Molecular movement inside the translational engine. Cell 1998; 92:337–349.
Maguire BA, Beniaminov AD, Ramu H, Mankin AS, Zimmermann RA . A protein component at the heart of an RNA machine: the importance of protein L27 for the function of the bacterial ribosome. Mol Cell 2005; 20:427–435.
Tate WP, Mannering SA . Three, four or more: the translational stop signal at length. Mol Microbiol 1996; 21:213–219.
Capecchi MR . Polypeptide chain termination in vitro: isolation of a release factor. Proc Natl Acad Sci USA 1967; 58:1144–1151.
Hampl H, Schulze H, Nierhaus KH . Ribosomal components from Escherichia coli 50S subunits involved in the reconstitution of peptidyltransferase activity. J Biol Chem 1981; 256:2284–2288.
Moffat JG, Tate WP . A single proteolytic cleavage in release factor 2 stabilises ribosome binding and abolishes peptidyl-tRNA hydrolysis activity. J Biol Chem 1994; 269:18899–18903.
Nakamura Y, Ito K, Isaksson LA . Emerging understanding of translation termination. Cell 1996; 87:147–150.
Kisselev L, Ehrenberg M, Frolova L . Termination of translation: interplay of mRNA, rRNAs and release factors? EMBO J 2003; 22:175–182.
Song H, Mugnier P, Das AK, et al. The crystal structure of human eukaryotic release factor eRF1-mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 2000; 100:311–321.
Vestergaard B, Van LB, Andersen GR, Nyborg J, Buckingham RH, Kjeldgaard M . Bacterial polypeptide release factor RF2 is structurally distinct from eukaryotic eRF1. Mol Cell 2001; 8:1375–1382.
Shin DH, Brandsen J, Jancarik J, Yokota H, Kim R, Kim S-H . Structural analyses of peptide release factor 1 from Thermotoga maritima reveal domain flexibility required for its interaction with the ribosome. J Mol Biol 2004; 341:227–239.
Graille M, Heurgué-Hamard V, Champ S, et al. Molecular basis for bacterial class I release factor methylation by PrmC. Mol Cell 2005; 20:917–927.
Poole ES, Brown CM, Tate WP . The identity of the base following the stop codon determines the efficiency of in vivo translation termination in Escherichia coli. EMBO J 1995; 14:151–158.
Scarlett D-JG, McCaughan KK, Wilson DN, Tate WP . Mapping functionally important motifs SPF and GGQ of the decoding release factor RF2 to the Escherichia coli ribosome by hydroxyl radical footprinting. Implications for macromolecular mimicry and structural changes in RF2. J Biol Chem 2003; 278:15095–15104.
Tate W, Greuer B, Brimacombe R . Codon recognition in polypeptide chain termination: site directed crosslinking of termination codon to Escherichia coli release factor 2. Nucleic Acids Res 1990; 25:6537–6544.
Brown CM, Tate WP . Direct recognition of mRNA stop signals by Escherichia coli polypeptide chain release factor two. J Biol Chem 1994; 269:33164–33170.
Czworkowski J, Wang J, Steitz TA, Moore PB . The crystal structure of elongation factor G complexed with GDP, at 2.7 Å resolution. EMBO J 1994; 13:3661–3668.
AEvarsson A, Brazhnikov E, Garber M, et al. Three-dimensional structure of the ribosomal translocase: elongation factor G from Thermus thermophilus. EMBO J 1994; 13:3669–3677.
Nissen P, Kjeldgaard M, Thirup S, et al. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 1995; 270:1464–1472.
Nissen P, Kjeldgaard M, Nyborg J . Macromolecular mimicry. EMBO J 2000; 19:489–495.
Klaholz BP, Myasnikov AG, van Heel M . Visualization of release factor 3 on the ribosome during termination of protein synthesis. Nature 2004; 427:862–865.
Wilson DN, Dalphin ME, Pel HJ, Major LL, Mansell JB, Tate WP . Factor-mediated termination of protein synthesis: a welcome return to the mainstream of translation. In: Garrett RA, Douthwaite SR, Liljas A, Matheson AT, Moore PB, Noller HF, eds. The ribosome: structure, function, antibiotics, and cellular interactions. Washington, DC: ASM Press, 2000; 495–508.
Ito K, Uno M, Nakamura Y . A tripeptide 'anticodon' deciphers stop codons in messenger RNA. Nature 2000; 403:680–684.
Frolova LY, Tsivkovskii RY, Sivolobova GF, et al. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA 1999; 5:1014–1020.
Rawat UBS, Zavialov AV, Sengupta J, et al. A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature 2003; 421:87–90.
Klaholz BP, Pape T, Zavialov AV, et al. Structure of the Escherichia coli ribosomal termination complex with release factor 2. Nature 2003; 421:90–94.
Petry S, Brodersen DE, Murphy IV FV, et al. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell 2005; 123:1255–1266.
Rawat U, Gao H, Zavialov A, Gursky R, Ehrenberg M, Frank J . Interactions of the release factor RF1 with the ribosome as revealed by cryo-EM. J Mol Biol 2006; 357:1144–1153.
Ito K, Uno M, Nakamura Y . Single amino acid substitution in prokaryote polypeptide release factor 2 permits it to terminate translation at all three stop codons. Proc Natl Acad Sci USA 1998; 95:8165–8169.
Uno M, Ito K, Nakamura Y . Polypeptide release at sense and noncognate stop codons by localized charge-exchange alterations in translational release factors. Proc Natl Acad Sci USA 2002; 99:1819–1824.
Ogle JM, Brodersen DE, Clemons WM, Tarry MJ, Carter AP, Ramakrishnan V . Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 2000; 292:897–902.
Konecki DS, Aune KC, Tate W, Caskey CT . Characterization of reticulocyte release factor. J Biol Chem 1977; 252:4514–4520.
Brown CM, Stockwell PA, Trotman CN, Tate WP . The signal for the termination of protein synthesis in prokaryotes. Nucleic Acids Res 1990; 18:2079–2086.
Cridge AG, Major LL, Mahagaonkar AA, Poole ES, Isaksson LA, Tate WP . Comparison of characteristics and function of translation termination signals between and within prokaryotic and eukaryotic organisms. Nucleic Acids Res 2006; 34:1959–1973.
Freistroffer DV, Kwiatkowski M, Buckingham RH, Ehrenberg M . The accuracy of codon recognition by polypeptide release factors. Proc Natl Acad Sci USA 2000; 97:2046–2051.
Major LL . Is the prokaryotic termination signal a simple triplet codon or an extended sequence element? PhD thesis, University of Otago, 2001.
Poole ES, Major LL, Mannering SA, Tate WP . Translational termination in Escherichia coli: three bases following the stop codon crosslink to release factor 2 and affect the decoding efficiency of UGA-containing signals. Nucleic Acids Res 1998; 26:954–960.
Boycheva S, Chkodrov G, Ivanov I . Codon pairs in the genome of Escherichia coli. Bioinformatics 2003; 19:987–998.
Mottagui-Tabar S, Isaksson LA . Only the last amino acids in the nascent peptide influence translation termination in Escherichia coli genes. FEBS Lett 1997; 414:165–170.
Mottagui-Tabar S, Bjornsson A, Isaksson LA . The second to last amino acid in the nascent chain as a codon context determinant. EMBO J 1994; 13:249–257.
Marquez V, Wilson DN, Tate WP, Triana-Alonso F, Nierhaus KH . Maintaining the ribosomal reading frame: the influence of the E site during translational regulation of release factor 2. Cell 2004; 118:45–55.
Vestergaard B, Sanyal S, Roessle M, et al. The SAXS solution structure of RF1 differs from its crystal structure and is similar to its ribosome bound cryo-EM structure. Mol Cell 2005; 20:929–938.
Stelzl U, Nierhaus KH . SERF: In vitro selection of random RNA fragments to identify protein binding sites within large RNAs. Methods (Duluth) 2001; 25:351–357.
Stelzl U, Spahn CMT, Nierhaus KH . Selecting rRNA binding sites for the ribosomal proteins L4 and L6 from randomly fragmented rRNA: Application of a method called SERF. Proc Natl Acad Sci USA 2000; 97:4597–4602.
Stelzl U, Nierhaus KH . A short fragment of 23S rRNA containing the binding sites for two ribosomal proteins, L24 and L4 is a key element for rRNA folding during early assembly. RNA 2001; 7:598–609.
Tate WP, Brown CM, Kastner B . Codon recognition by the polypeptide release factor. In: Hill WE, Dahlberg AE, Garrett RA, Moore PB, Schlessinger D, Warner JR, eds. The ribosome: structure, function, and evolution. Washington, DC: ASM Press, 1990; 393–401.
Harms J, Schluenzen F, Zarivach R, et al. High resolution structure of the large ribosomal subunit from a mesophilic Eubacterium. Cell 2001; 107:679–688.
Wilson KS, Ito K, Noller HF, Nakamura Y . Functional sites of interaction between release factor RF1 and the ribosome. Natl Struct Biol 2000; 7:866–870.
Wilson KS, Noller HF . Mapping the position of translational elongation factor EF-G in the ribosome by directed hydroxyl radical probing. Cell 1998; 92:131–139.
Chan YL, Sitikov AS, Wool IG . The phenotype of mutations of the base-pair C2658.G2663 that closes the tetraloop in the sarcin/ricin domain of Escherichia coli 23S ribosomal RNA. J Mol Biol 2000; 298:795–805.
Szewczak AA, Moore PB . The sarcin/ricin loop, a modular RNA. J Mol Biol 1995; 247:81–98.
Xu WB, Pagel FT, Murgola EJ . Mutations in the GTPase center of Escherichia coli 23S rRNA indicate release factor 2-interactive sites. J Bacteriol 2002; 184:1200–1203.
Arkov AL, Mankin A, Murgola EJ . An rRNA fragment and its antisense can alter decoding of genetic information. J Bacteriol 1998; 180:2744–2748.
Stoffler G, Cundliffe E, Stoffler-Meilicke M, Dabbs ER . Mutants of Escherichia coli lacking ribosomal protein-L11. J Biol Chem 1980; 255:10517–10522.
Tate WP, Dognin J, Noah M, Stoffler-Meilicke M, Stoffler G . The NH2-terminal domain of Escherichia coli ribosomal protein L11: its three dimensional location and its role in the binding of release factors 1 and 2. J Biol Chem 1984; 259:7317–7324.
Tate WP, McCaughan KK, Ward CD, et al. The ribosomal binding domain of the Escherichia coli release factors: modification of tyrosine in the N terminal domain of ribosomal protein L11 affects release factors 1 and 2 differentially. J Biol Chem 1986; 261:2289–2293.
Yusupova MM, Yusupova GZ, Baucom A, et al. Crystal structure of the ribosome at 5.5 angstrom resolution. Science 2001; 292:883–896.
Osswald M, Brimacombe R . The environment of 5S rRNA in the ribosome: cross-links to 23S rRNA from sites within helices II and III of the 5S molecule. Nucleic Acids Res 1999; 27:2283–2290.
Herr W, Noller HF . Protection of specific sites in 23S and 5S RNA from chemical modification by association of 30S and 50S ribosomes. J Mol Biol 1979; 130:421–432.
Ko J, Lee Y, Park I, Cho B . Identification of a structural motif of 23S rRNA interacting with 5S rRNA. FEBS Lett 2001; 508:300–304.
Ammons D, Rampersad J . An E. coli 5S rRNA deletion mutant useful for the study of 5S rRNA structure/function relationships. Curr Microbiol 2001; 43:89–92.
Ammons D, Rampersad J, Fox GE . 5S rRNA gene deletions cause an unexpectedly high fitness loss in Escherichia coli. Nucleic Acids Res 1999; 27:637–642.
Kohrer C, Mayer C, Neumair O, Grobner P, Piendl W . Interaction of ribosomal L1 proteins from mesophilic and thermophilic Archaea and bacteria with specific L1-binding sites on 23S rRNA and mRNA. Eur J Biochem 1998; 256:97–105.
Yoshimura K, Ito K, Nakamura Y . Amber (UAG) suppressors affected in UGA/UAA-specific polypeptide release factor 2 of bacteria: genetic prediction of initial binding to ribosome preceding stop codon recognition. Genes Cells 1999; 4:253–266.
Poole ES, Tate WP . Release factors and their role as decoding proteins: specificity and fidelity for termination of protein synthesis. Biochim Biophys Acta 2000; 1493:1–11.
Polacek N, Gomez MJ, Ito K, Xiong L, Nakamura Y, Mankin A . The critical role of the universally conserved A2602 of 23S ribosomal RNA in the release of the nascent peptide during translation termination. Mol Cell 2003; 11:103–112.
Youngman EM, Brunelle JL, Kochaniak AB, Green R . The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell 2004; 117:589–599.
Dinçbas-Renqvist V, Engström A, Mora L, Heurgué-Hamard V, Buckingham R, Ehrenberg M . A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J 2000; 19:6900–6907.
Wilson DN, Guévremont D, Tate WP . The ribosomal binding and peptidyl-tRNA hydrolysis functions of Escherichia coli release factor 2 are linked through residue 246. RNA 2000; 6:1704–1713.
Schmeing TM, Huang KS, Kitchen DE, Strobel SA, Steitz TA . Structural insights into the roles of water and the 2¢ hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol Cell 2005; 20:437–448.
Gao H, Zhou Z, Rawat U, et al. RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors. Cell 2007; 129:929–941.
Beringer M, Rodnina MV . The ribosomal peptidyl transferase. Mol Cell 2007; 26:311–321.
Acknowledgements
We acknowledge the earlier studies of Drs Chris Brown (University of Otago, Dunedin, New Zealand) and Berthold Kastner (Max Planck Institute for Biophysical Chemistry, Goettingen, Germany) on the RF interactions with the ribosome and subunits illustrated in Figure 7. This SERF study and the site-direct mutagenesis analyses with RF2 domain III were supported by postgraduate scholarships and bridging grants from the University of Otago to Askarian-Amiri ME and Scarlett DJ, respectively, and, from the Marsden Fund of New Zealand, a postgraduate scholarship to Young DJ, and a research grant to Tate WP and Poole ES.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Rights and permissions
About this article
Cite this article
Poole, E., Young, D., Askarian-Amiri, M. et al. Accommodating the bacterial decoding release factor as an alien protein among the RNAs at the active site of the ribosome. Cell Res 17, 591–607 (2007). https://doi.org/10.1038/cr.2007.56
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2007.56