Abstract
The restriction fragment length polymorphisms distribution and frequency of dystrophin gene in Chinese were studied by using 14 subclones of the entire 14kb cDNA for the dystrophin as hybridization probes. Allelic fragments were detected in hybridization patterns of PvuII/la, Taq I /2b-3, Taq I/5b-7, arid Xba l /10. Among them, the allelic fragments (26kb and 3.8kb) in PvuII/2b-3 pattern and the allelic fragments (10.0kb and 8.4kb) in Taq I/5b-7 patterns had never been reported previously. Compared with the data from Caucasians and Japanese, it indicated that there was a significant difference (P<0.01) of the allelic fragment frequency in Taq I/2b-3 and Xba I/10 patterns between Chinese and Caucasians. The frequencies of allelic fragments A2 (5.6kb) in Taq I/8 and A2 (10.7kb) in EcoR V/9 were high in Caucasians, yet had not been detected in Chinese, the differences were also highly significant. But in Chinese and Caucasians, the B1B2 allelic frequencies in Taq I/5b-7 are the same. As to the frequency of the allelic fragments A1A2 and B1B2 in Pvu II/la, there was no significant difference between Chinese and Japanese.
Similar content being viewed by others
Introduction
Dystrophin gene is a huge gene with a length of about 2,300kb, and has been mapped on the Xp21.1-21.3 1. The cDNA coding for the protein dystrophin is composed of 13,973bp 2. Dystrophin is a kind of rod-shaped cellular skeletal protein (427kd) distributed on the inner surface the cytoplasmic membrance of skeletal muscle cells, cardiac muscle cells and brain cells3, 4, 5, 6, 7. When deletion and /or duplication take place in the coding sequence of the gene, the normal structure and function of the gene on the membranes will be altered8 and will bring about a severe X-linked recessive disease (Duchenne muscular dystrophy, DMD) with an incidence of approximately 1 in 3,500 newborn males. The affected boys are crippled and usually die in their juvenile life. The mutated dystrophin gene also can bring about another allelic phenotype, that is Becker muscular dystrophy (BMD) with a similar syndrome but a much longer life span.
Since the full-length cDNA coding for dystrophin was cloned in 1987, probing the molecular structure of the dystrophin gene had become one of the most attractive research subjects in the field of human molecular genetics. The purpose of the present paper is to reveal the genetic polymorphism of dystrophin gene at DNA level. Different fragments of dystrophin cDNA were prepared and used as probes to hybridize with genomic DNA isolated from Chinese individuals . The allelic frequency of dystrophin gene in Chinese population was measured and then compared with those in Caucasian and Japanese populations.
Materials and Methods
Samples
Peripheral blood samples were obtained from 100 unrelated Chinese (60 males and 40 females). They were adult students and voluntary donators.
cDNA probes
Six dystrophin cDNA subclones 1-2a, 2b-3, 4-5a, 5b-7, 8 and 9-14 were obtained from the American Type Culture Collection. They were constructed on the bluescribe SK or bluescript KS vectors (M13 derivatives) by Dr. L. M. Kunkel. Some probes were prepared on the basis of the restriction map 9 of dystrophin cDNA in our laboratory. The probes were listed in Tab 1.
DNA analysis
Genomic DNA was prepared according to Kunkel's method10 and low-melting-temperature agarose embedding method. DNA was released from leucocytes in 0.5% low melting temperature agarose blocks and cleaved with restriction enzymes according to the manufacturer's instructions. The digested DNA was run on 0.8% agarose gel (GIBCO-BRL), denatured in 0.5 M NaOH /1.5 M NaCl for 1 h, neutralized in 1 M Tris. HCl /1. 5 M NaCl for 1 h and blotted onto nitrocellulose membrane (Schleicher and Schuell) or Nylon membrane (Boehringer-Mannheim). After baking the membranes at 80-85°C for 2 h, DNA was hybridized to 32p labeled probes. The membranes were washed with 1×SSC / 0.1%SDS, and autoradiography was performed with intensifying screens at-70°C for several days.
Statistical analysis
The data from Japanese and Caucasians were quoted respectively from papers published by Deng et al11; Darras et al12 and Gallati et al13. The comparison of the allelic frequency between either Chinese and Japanese or Chinese and Caucasians was carried out by χ2 test with fourfold table.
Results
The RFLPs detected in Chinese
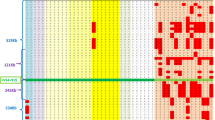
As shown in Tab 2, the 6 pairs of allelic fragments were recognized in the hybridization patterns of Pvu II/la, Taq I/2b-3, Pvu II/2b-3, Taq I/5b-7 and XbaI/10(Fig 1, 2, 3, 4, 6). Among them, the allelic fragments (A1=26.0 kb, A2=3.8 kb) in Pvu II/2b-3 pattern and those (A1=10.0 kb, A2=8.4 kb) in Taq I/5b-7 pattern had never been reported before, and Mendelian inheritance was demonstrated in one two-generation informative family, respectively.
Taq I polymorphism detected by dystrophin cDNA probe 5b-7. Arrow heads and arrows point to two pairs of polymorphism with variant fragments of 10.0 kb(A1) and 8.4 kb(A2), 3.2 kb (B1) and 1.6 kb(B2). Eight constant fragments, 6.4, 4.4, 3.6, 2.9, 2.3, 2.1, 2.0 and 1.8 kb are also detected. Mendelian segregation is observed in one family of A1A2, four families of B1B2.
In addition to the allelic fragments mentioned above, a new constant band with a length of 2.4 kb was identified in Xba I/10 hybridization pattern (Fig 6). The probe in revealing that new fragment was just the same one used in studying the Western populations.
The size of 5th hybridizing band in Taq I/8 pattern was 2.0 kb, and was not2.3 kb as reported previously in Western populations. By studying Taq I/8 and Hind III/8 patterns of 10 DMD patients who have different deletions in the region corresponding to cDMD8, such as deletion 3.7 kb-3.2 kb, 4.5 kb-3.2 kb, 3.7 kb-3.2 kb-4.5 kb, 6.5 kb-2.0 kb (-3.0 kb/cDMD9), and 4.5 kb, we preliminarily confirmed that the order of the 5 constant bands in Taq I/8 pattern was 3.7 kb-3.7 kb-4.5 kb-6.5 kb-2.0 kb and represented the exons No.47, 48, 49, 50+51, 52 of the dystrophin gene.
The hybridization results from Taq I/8 (the pattern photo was not shown here) and EcoR V/9 (Fig 5) indicated .that the allelic fragments reported abroad 11, 12 have not been detected in the 86 X-chromosomes and the 44 X-chromosomes tested respectively.
The hybridization patterns from EcoR V/ 5b-7, EcoR V/11b-12a, EcoR V/12b-13, Pvu II/4-5a, Pvu II/10, Pvu II/14, Taq I/4-5a, TaqI/12-13, Xba I/2b-3, Xba I/5b-7, Xba I/8, Xba I/9, Xba I/12 and Xba I/12a-13 did not show any RFLPs in the 15-30 X-chromosomes tested respectively.
Comparison of the RFLPs between Chinese and Japanese
As shown in Tab 3, the Pvu II/la hybridization patterns (Fig.1) from 43 unrelated Chinese were studied. In 57 X-chromosomes, the low-frequency allelic fragments A1A2 (A1=20.0 kb; A2=5.8 kb) found in Japanese 11 were not detected, while the high-frequency allelic fragments B 1B2 (B1=15 kb, 0.43; B2=8.0 kb, 0.57) were similar to those in Japanese. The difference of the allelic frequencies between Chinese and Japanese was not significant (P>0.05) by χ2 test.
Comparison of the RFLPs between Chinese and Caucasians
As shown in Tab 4, 3 allelic fragments (Taq I/2b-3: A1=3.4 kb, A2=3.2 kb; Taq I/5b-7: B1=3.2 kb, B2=1.6 kb; Xba I/10: A1=7.4 kb, A2=6.7 kb) were detected in unrelated Chinese, while 2 other from EcoR V/9 and Taq I/8 were not found in these Chinese individuals. The results of statistical analysis for the 5 allelic frequencies showed that between Chinese and Caucasian populations, the difference of allelic frequencies from Taq I/2b-3, Taq I/8, EcoR V/9 and Xba I/10 was quite significant (P<0.01). However that from Taq I/5b-7 (B1B2) showed no statistical significance (P>0.05).
Discussion
Two new RFLPs
In dystrophin gene, so far there have been over 30 RFLPs revealed by genomic DNA probes and over 20 RFLPs by cDNA probe. However those RFLPs markers are still far from enough to complete the molecular structure analysis for the huge gene. For instance, there are a number of DMD/BMD families (about 30%-40% of total cases) having no detectable deleted or duplicated mutations. Therefore, the 2 new RFLPs found in Pvu II/2b-3 and Taq I/5b-7 hybridization patterns described in the present paper could enrich the boundary markers within the dystrophin gene. It would definitely be beneficial for carrying out the genetic linkage analysis for DMD or, BMD families.
A new 2.4 kb constant band in Xba I/cDMD10
The hybridization pattern and allelic fragments of the Xba I / 10 were first reported by Gallati et al13. There are 5 constant hybridization bands 8.3 kb, 6.0 kb, 4.2 kb, 2.5 kb, 2.0 kb and one allelic fragment (7.4 kb or 6.7 kb). However, 7 constant Hind III bands were found by Darras et al 12 with the same probe. Because there were no Hind III site and Xba I site in the sequence of probe 10, thus, the difference between the number of Hind III bands and Xba I bands implied that a certain Xba I/10 hybridization band may contain 2 Hind III bands. However, when both of the electrophoretic time and the gel size were lengthened in this study, a new 2.4 kb band (Fig. 6) could be resolved preceding the 2.5 kb band of Xba I/10 constant pattern. This new discovery further supported the viewpoint that cDMD10 involved at least 7 exons.
The size of 5th band in Taq I/cDMD8 pattern
The hybridization pattern of Taq I/8 reported by Daxras et al12 consisted of 4 constant bands and a pair of allelic fragments. The position of the 5th band described as 2.3 kb 12 was parallel to that of the 6th band of lambda bacteriophage DNA/Hind III in all of the 10 autoradiograph films we made. The size of that band we measured was similar to that of 1.9 kb band in Taq I/8 hybridization pattern reported by Zhao, et al14. Furthermore, by using the isosceles triangle radiation comparison method, the molecular weight of the band from the photo of Darras, et al12 and that of ours were compared. The result indicated that the position and size of both bands were identical. Hence, we inferred that the size difference was not a real polymorphism between Chinese and Caucasians, but might be an error in measurement or printing.
The 5 allelic frequencies between Chinese and Caucasian
The allelic frequency of 5 RFLPs from Taq I/2b-3, Taq I/5b-7, Taq I/8, EcoR V/9 and Xba I/10 hybridization reported by Darras et al12 and Gallati et al13 was compared with that of ours in present study. The results showed that there was a significant difference (P <0.01) in the frequency of EcoR V/9, Taq I/2b-3, Taq I/8 and Xba I/10 between the Chinese and Caucasian populations. In Chinese, the allelic frequency of Taq I/2b-3, Taq I/ 8 and EcoR V/9 was low, but that of Xba I/10 was high. The difference between 2 races may be resulted from different neutral mutation rates within introns during evolution. If this kind of molecular variation of dystrophin gene was a common phenomenon in the human genome, we suggested that the testees of “human genomic mapping and sequencing project”15 should include the genomes of the main ethnic populations worldwide.
Potential applications of the RFLPs
Among the RFLPs described in this paper, Taq I/5b-7 (B1B2) and Xba I/10 (A1A2) are 2 flanking markers of the deletion hot spot in the dystrophin gene. The frequency of these 2 kinds of heterozygote (0.46 and 0.48) was very high in Chinese. In addition, the RFLP revealed by Pvu II/la was just at the 5′-end of the gene with a very high heterozygote frequency (0.46), and is a flanking marker of the second deletion hot locus. P vu II/la, Taq I/5b-7 and Xba I/10 could be used as a set of molecular markers of the genetic linkage analysis for 5′ to 3′ end. Taking together these 3 RFLPs would be of great value to the genetic counseling and prenatal diagnosis of DMD or BMD families in Chinese population.
The RFLPs detected by cDMD probes could be more useful than those by genomic DNA in identifying the carriers with DMD or BMD 16, 17, because each hybridization band represented an exon-containing fragment and about 60% of the patients were resulted from deletion or duplication of more than one exon. When the females from DMD or BMD families were analyzed by the RFLPs detected by cDMD probes, they were also detected by dosage effect of each band in hybridization pattern. This method including the analysis of the RFLPs and the detection of deletion or duplication in dystrophin gene could save much material and labour in the research. And it could also improve the accuracy in genetic counseling and prenatal diagnosis.
References
Den Dunnen JT, Bakker E, Van Ommen GJB, Pearson PL . The DMD gene analysed by field inversion gel electrophoresis. Brit Med Bull 1989; 45:644–653.
Koenig M, Monaco AP, Kunkel LM . The complete sequence of dystrophin predicts a rod-shapes cytoskeletal protein. Cell 1988; 53:219–228.
Hoffmann EP, Brown RH, Kunkel LM . Dystrophin: The protein product of the Duchenne muscular dystrophy locus. Cell 1987; 51:919–923.
Hoffmann EP, Knudson CM, Campbell KP, Kunkel LM . Subcellular fractionation of dystrophin to the triads of skeletal muscle. Nature 1987; 330:754–758.
Arahata K, Ishiura S, Ishiguro T, Tsukahara T, Suhara Y, Eguchi C, et al. Immunostaining of skeletal and cardiac muscle surface membrane with antibody against Duchenne muscular dystrophy peptide. Nature 1988; 333:861–863.
Campbell KP, Kahl SD . Association of dystrophin and an integral membrane glycoprotein. Nature 1989; 338:259–262.
Chelly J, Hamard G, Koulakoff, A, Kaplan JC, Kahn A, Berwald NY . Transcription of the dystrophin gene in human muscle and non-muscle tissues. Nature 1988; 333:858–860.
Koenig M, Beggs AH, Moyer M, Scherpf S, Indrichs KH, Bettecken T, et al. The molecular basis for Duchenne versus Becker muscular dystrophy: correlation of severity with type of deletion. Am J Hum Genet 1989; 45:498–506.
Koenig M, Hoffman EP, Bertelson CJ, Monaco AP, Feener C, Kunkel LM . Complete cloning at the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell 1987; 50:509–517.
Ktmkel LM, Smith KD, Bower SH, Borgaonkar DS, Wachtel SS, Miller OJ, et al. Analysis of human Y-chromosome specific reiterated DNA in chromosome variants. Proc Natl Acad Sci U.S.A. 1977; 74:1245–1249.
Deng HX and Niikawa N . Pvu II RFLPs in the DMD gene detected by a subclone (cDMD la) of the cDNA. Nucl Acid Res 1990; 16:9072
Darras BT and Francke U . Normal human genomic restriction fragment patterns and polymor-phisms revealed by hybridization with the entire dystrophin cDNA. Am J Hum Genet 1988; 43:612–619.
Gallati SL, Schneider V, Mullis P and Moser H . RFLPs for Duchenne muscular dystrophy cDNA clone 9 and 10. Am J Hum Genet 1990; 46:1090–1094.
Zhao YS, You WY, Liu WQ, Qi H, Zhuo CC, Chen YF, et al. Preliminary study for detecting deletion of DMD gene by restriction enzyme Taq I (Abstract). Genetic study in China in 1987–1990. 1991, Science and Technology Press of China, 1st edition.
National Research Council. Mapping and sequencing the human genome. National Academy Press 1988.
Zeng YT, Chen MJ, Ren ZR, Qiu XK, Huang SZ . Analysis of RFLPs and DNA deletions in the Chinese Duchenne muscular dystrophy gene. J Med Genet 1991; 28:167–170.
Zeng YT, Zhang ML, Xu YJ, Sheng M, Huang SZ, Huang JH, et al. Prenatal diagnosis f Duchenne muscular dystrophy by DNA RFLP analysis. Natl Med J China 1988; 68:565–567.
Acknowledgements
We thank Dr. Louis M. Kunkel (Harvard Medical School, Boston, USA) and ATCC Corporation of US for the supply of dystrophin gene cDNA probe. The project is supported by Shanghai Joint Laboratory of Life Science, Academia Sinica, and National Post-Doctor Science Foundation of China, National Natural Science Foundation of China.
Author information
Authors and Affiliations
Additional information
*This paper published in Acta Biol Exper Sinica 1992; 25:173-83 in Chinese
Rights and permissions
About this article
Cite this article
Yu, L., Wang, M., Wang, Q. et al. Comparing the frequencies of restriction fragment length polymorphisms for dystrophin gene in Chinese with those from Japanese and Caucasian populations. Cell Res 3, 39–47 (1993). https://doi.org/10.1038/cr.1993.5
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/cr.1993.5