Abstract
Hematopoietic stem and progenitor cells have the capacity to self-renew and differentiate into all blood cell lineages, and thus sustain life-long homeostasis of the hematopoietic system. Although intensive studies have focused on the orchestrated genetic network of hematopoietic stem and progenitor cell specification and expansion, relatively little is known on the regulation of hematopoietic stem and progenitor cell survival during embryogenesis. Here, we generated two types of miR-142a-3p genetic mutants in zebrafish and showed that the loss-of-function mutants displayed severe reduction of hematopoietic stem and progenitor cells. Further analysis showed that the diminished proliferation and excessive apoptosis in miR-142a-3p mutants were attributed to the increased p53 signaling. Mechanistically, we demonstrated that miR-142a-3p directly targets p53 during hematopoietic stem and progenitor cell development, and the hematopoietic stem and progenitor cell survival defect in miR-142a-3p mutants could be rescued by loss of p53. Therefore, our work reveals the significance of the miR-142a-3p-p53 pathway in controlling hematopoietic stem and progenitor cell survival, and thus advances our understanding of the role of p53 in vertebrate hematopoiesis.
Similar content being viewed by others
Introduction
Hematopoiesis in vertebrate is initiated in two distinct and successive processes termed primitive and definitive waves [1]. Hematopoietic stem and progenitor cells (HSPCs), which are generated from definitive hematopoiesis via a specific endothelial-to-hematopoietic transition process [2–5], possess the capacities of self-renewal and differentiation into erythroid, myeloid and lymphoid lineages to sustain blood cells during embryogenesis and adulthood. The emergence of HSPCs is tightly regulated by signaling pathways (Notch, Hedgehog, BMP, Wnt and pro-inflammatory signals) [6–10], transcription factors (Runx1, Cbfb and Scl) [11–13], G-coupled protein receptor 56 [14] and epigenetic regulators [15, 16].
MicroRNAs are reported to be highly expressed in hematopoietic tissues including bone marrow [17] and emerging evidence has shown that they are implicated in hematopoiesis in different organisms [18–20]. In mouse, miR-125 is highly expressed in hematopoietic stem cells (HSCs) to expand HSC numbers in vivo [21]. In vitro, mouse fibroblasts were reprogrammed into hemogenic endothelial cells that differentiated into CD45+ cKit+ cells (HSPCs) between day-20 and day-35 culture; in these HSPCs, the highest enrichment of microRNAs including miR-142 was observed [22]. In Xenopus, miR-142-3p has an essential role in establishing the mesodermal lineage that contributes to both HSC emergence and vasculogenesis [23]. In zebrafish, miR-142-3p modulates neutrophil development by controlling its maturation [24]. MiR-142 is also an essential regulator of lymphocyte ontogenesis and megakaryopoiesis [20, 25]. In our previous study, suppression of miR-142a-3p by antisense morpholinos shows that HSPC development is impaired when miR-142a-3p is knocked down. Further microarray data and functional assays demonstrate that irf7 acts as a direct target of miR-142a-3p and that the miR-142a-3p-irf7 signal pathway regulates the formation and differentiation of definitive HSPCs possibly through the inflammatory signaling [26]. Another group has reported that ectopic expression of miR-142-3p leads to the reduction of primitive erythrocyte progenitor cells and HSCs in zebrafish [27]. Thus, the role of zebrafish miR-142a-3p in definitive hematopoiesis remains elusive, largely because of different methods used by different groups.
P53, a tumor suppressor involved in many disorders including hematological malignancy [28], can activate DNA repair proteins when DNA is under sustained damage, leading to apoptosis and G1/S cell cycle arrest [29–31]. p53 is a critical transcription factor for cell cycle regulation in hematopoietic cells and is involved in the quiescence, self-renewal, senescence and apoptosis of HSCs [32, 33]. HSCs from hypermorphic p53 mice, which constitutively activates P53, fail to expand [34] and HSC self-renewal in p53−/− mice is enhanced [35]. In the zebrafish Diamond-Blackfan model, the downregulation of erythroid marker gata1 is modulated by p53 [36]. Intriguingly, loss of p53 facilitates reprogramming murine fibroblasts to hematopoietic progenitor cells and p53−/− reprogrammed cells efficiently generated erythroid, megakaryocytic, myeloid, and lymphoid lineages [37]. In addition, suppression of p53 can exert hematopoietic rescue effect on genetic alterations including mysm1 deficiency [38], and p53−/− HSCs are more resistant to irradiation [32]. Taken together, p53 may be closely linked to the cell cycle and apoptosis and thus critical for the survival and stemness maintenance of HSPCs. Gfi-1, Necdin and P21 were the downstream targets of P53 in mediating HSC quiescence and Puma was responsible for HSC apoptosis [28, 29]. However, the upstream regulator(s) of p53 governing HSC development is largely unclear.
In this work, we demonstrate that p53 is a direct target of miR-142a-3p during zebrafish definitive hematopoiesis, and the hematopoietic defects in miR-142a-3p mutants can be rescued by loss of p53, which deepens our understanding of the role of p53 in mediating miR-142a-3p function in HSPC development in zebrafish.
Results
Generation of miR-142a-3p mutants by TALENs and CRISPR/Cas9
In order to substantiate our previous findings that loss of miR-142a-3p leads to decreased HSPCs [26], both TALENs and CRISPR/Cas9 were used to generate genetic mutants of miR-142a. First, we designed a pair of TALEN arms that target two sites inside of the miR-142a gene (Figure 1a). The TALEN mRNAs were injected into WT embryos (F0) and after genotyping of F2 embryos, a mutant, named 142T−/−, was identified. 142T−/− embryos were born at the expected Mendel ratio, and they could develop into fertile adults. According to the sequencing result, 13 base pairs were deleted in the locus of the miR-142a gene (Figure 1b and c) and this disrupted the secondary structure of miR-142a as well as the formation of mature miR-142a-3p (Figure 1d). Further analysis revealed that the expression of miR-142a-3p in both the thymus and caudal hematopoietic tissue region was undetectable (Figure 1e). Consistently, real-time quantitative PCR (qPCR) result demonstrated that miR-142a-3p and another member of miR-142a, miR-142a-5p, were severely decreased (Figure 1f), indicating the loss-of-function of miR-142a-3p and miR-142a-5p.
Generation of miR-142a mutants by TALENs. (a) Schematic diagram showed the TALEN target sites at the miR-142a locus. (b and c) Indel mutations induced by TALENs showed a 13-base-pair deletion in the miR-142a locus. Green arrow shows the initiation of the mutation. (d) Predicted RNA secondary structure of wild-type and mutated pri-miR-142a by TALENs using the RNA Structure Software (version 4.6, Rochester, NY, USA). (e) Expression of miR-142a-3p was absent in 142T−/− embryos at 4 days post-fertilization (dpf) by WISH. Red dashed circles mark the expression of miR-142a-3p in the thymus and red arrowheads mark the expression of miR-142a-3p in the CHT. (f) qPCR demonstrated that the expression of miR-142a-3p and miR-142a-5p was nearly undetectable in 142T−/− embryos at 4 dpf. The expression of miR-142a-3p and miR-142a-5p was normalized to U6 (mean±s.d., n=3, **P<0.01).
To generate a large deletion of the miR-142a gene by CRISPR/Cas9, a pair of gRNAs flanking the miR-142a locus was designed and co-injected into zebrafish WT embryos with Cas9 mRNA (Supplementary Figure S1a). After F2 screening, a miR-142a mutant was identified, named 142C−/−, and the sequencing result displayed a 986-base-pair deletion in the locus of miR-142a, which was able to be employed to distinguish the WT siblings and mutants by PCR (Supplementary Figure S1b). Impaired secondary RNA structure of miR-142a and failure to form the mature miR-142a-3p also occurred in 142C−/− embryos (Supplementary Figure S1c and d). Whole-mount in situ hybridization (WISH) result demonstrated that miR-142-3p was absent in both the thymus and caudal hematopoietic tissue region in 142C−/− embryos (Supplementary Figure S1e), suggesting a loss-of-function of miR-142a-3p.
Taken together, two zebrafish genetic mutants of miR-142a were generated in which miR-142a-3p was disrupted and thus facilitated functional investigation of miR-142a in zebrafish hematopoiesis.
Loss of miR-142a-3p leads to decreased HSPCs in zebrafish
Previous studies implicate that loss of miR-142a-3p leads to decreased erythrocytes as well as vascular defects [39, 40]. Thus, we examined whether miR-142a-3p mutants display such developmental defects. With a somitic marker myod to define specific developmental stages, WISH results showed that primitive hematopoiesis including erythroid progenitors (labeled by scl and gata1) and myeloid cells (labeled by lyz and pu.1) was unaffected in 142T−/− embryos at 24 and 36 hours post-fertilization (hpf), respectively (Supplementary Figure S2a and b). Moreover, vascular development was examined and artery–vein specification was relatively normal as expression of arterial markers dll4, dltC and efnb2a and venous markers flt4 and msr at 26 and 36 hpf was unaltered (Supplementary Figure S2c and d). Real-time PCR results also displayed normal expression of dll4 and flt4 in 142T−/− (Supplementary Figure S2e). Taken together, primitive hematopoiesis and vascular development were normal in 142T−/− embryos.
To confirm the HSPC defects in miR-142a-3p morphants as reported in our previous study [26], the HSPC marker runx1 was examined in 142T−/− embryos and WISH result displayed its reduction in the aorta–gonad–mesonephros (AGM) at 26 and 36 hpf (Figure 2a). In addition, qPCR and western blot analyses also showed similar decrease of runx1 in 142T−/− embryos (Figure 2b and c). Consistently, the number of cmyb:EGFP+ cells (HSPCs) in Tg(cmyb:EGFP) embryos was also decreased in 142T−/− embryos (Figure 2d), whereas cmyb-positive cells in the pronephric duct remained unaffected (Figure 2d), demonstrating that the decrease of HSPCs in 142T−/− embryos is specific. To determine the role of miR-142a-3p in the expansion and differentiation of HSPCs at later stages, WISH was performed and the result showed that cmyb was decreased in the caudal hematopoietic tissue (the equivalent of fetal liver in mammals) from 2 to 3.5 dpf in 142T−/− embryos (Figure 2e). Moreover, the differentiated hematopoietic lineages including erythrocytes (labeled by gata1), neutrophils (labeled by lyz) and T cells (labeled by ikaros and rag1) were all decreased in 142T−/− embryos at 4 dpf (Figure 2f), further suggesting that HSPC differentiation was disrupted. Consistent with the results in 142T−/− embryos, 142C−/− embryos displayed similar reduction in HSPC markers runx1 and cmyb (Supplementary Figure S3a) and decreased expression of differentiated hematopoietic markers at 4 dpf (Supplementary Figure S3b). Therefore, loss of miR-142a-3p leads to the decreased HSPCs and their derivatives in both 142T−/− and 142C−/− embryos.
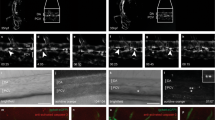
MiR-142a-3p is essential for HSPC emergence. (a) 142T−/− embryos showed decreased expression of HSPC marker runx1 at 26 and 36 hpf by WISH. (b) 142T−/− embryos displayed decreased expression of HSC marker runx1 at 26 and 36 hpf using qPCR. The expression of runx1 was normalized to β-actin (mean±s.d., n=3, **P<0.05). (c) Runx1 was decreased in 142T−/− embryos at 26 and 36 hpf by western blot analysis (left panel) and the quantification (right panel). The protein level of Runx1 was normalized to β-Actin. (d) The number of EGFP-labeled HSPCs (red arrowheads) in Tg (cmyb:EGFP) embryos was decreased in the AGM region in 142T−/− embryos at 36 hpf (mean±s.d., n=5, *P<0.05). White arrowheads indicate cmyb-positive cells in the pronephric duct. cmyb-labeled pronephric duct was counted as an internal control. (e) 142T−/− embryos displayed reduction of HSPC marker cmyb from 2 to 3.5 dpf by WISH. (f) Differentiated hematopoietic lineages were decreased in 142T−/− embryos including erythrocytes (gata1), neutrophils (lyz) and lymphocytes (ikaros and rag1) at 4 dpf by WISH. Red arrowheads mark hematopoietic cells in the caudal hematopoietic tissue (CHT), whereas red circles denote the thymus. (g) Hematoxylin and eosin (HE) staining of the kidney showed that decrease in cell population and the volume of the head kidney greatly in 142−/− adult fish of 12 weeks.
Concerning the absence of miR-142a-3p and miR-142a-5p in 142T−/−, it is critical to determine whether the decrease in HSPCs is also attributed to the absence of miR-142a-5p. In fact, knockdown of miR-142a-5p by antisense morpholino showed normal expression of HSPC marker runx1 (Supplementary Figure S3c), excluding the involvement of miR-142a-5p in HSPC regulation. Therefore, we concluded that the defects of HSPCs in 142T−/− embryos were only attributed to the absence of miR-142a-3p.
To demonstrate whether the defect of HSPCs in 142T−/− can recover during adulthood, the kidney of adult mutant was sectioned and stained with hematoxylin and eosin to test the number of hematopoietic cells. It was observed that both the cellularity and the volume of the pronephros decreased dramatically in 12-week-old 142T−/− adult fish (Figure 2g), confirming the continuous defect of definitive hematopoiesis in adult zebrafish upon miR-142a deletion. To test whether there exists hypermaturation of neutrophils in 142T−/− as reported previous [24], we also sorted myeloid cells using flow cytometry (FACS) and conducted Wright–Giemsa staining of eosinophils, macrophages and neutrophils. However, our analysis showed that the morphology of myeloid cells in between mutants and WT siblings seems no difference (Supplementary Figure S4), suggesting normal maturation of neutrophils in 142T−/− embryos.
Previous work showed that irf7 is a direct target of miR-142a-3p, and miR-142a-3p morphants display increased expression of irf7 [26]. Using WISH and qPCR, we were able to detect that there was an ectopic expression of irf7 in 142T−/− embryos (Figure 3a and b). Moreover, knockdown of irf7 in 142T−/− embryos displayed a partial rescue on HSPCs at both 26 and 36 hpf (Figure 3c), confirming that irf7 functions downstream of miR-142a-3p to mediate HSPC development.
Increased expression of irf7 in 142T−/− embryos and rescue of HSPC defects by irf7 knockdown. (a and b) 142T−/− embryos showed increased expression of irf7 at 4 dpf using WISH and qPCR (mean±s.d., n=3, *P<0.01). (c) Irf7 MO-injected 142T −/− embryos showed the restoration of runx1 expression at 26 and 36 hpf using WISH. The quantification of runx1-positive cells in the AGM was shown in the lower panel (mean±s.d., n=5, *P<0.05).
Taken together, miR-142a-3p is indispensable for the HSPC emergence and differentiation, and its direct target irf7 is upregulated in 142T−/− embryos, confirming the findings in our previous study [26].
Decreased proliferation and increased apoptosis of HSPCs in 142T−/− embryos
To determine how HSPCs were decreased in 142T−/− embryos, we performed 5-Bromo-2-deoxyuridine (BrdU) labeling for cell proliferation and terminal dexynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) staining for apoptosis assays. First, we examined HSPC proliferation by counting the proportion of BrdU immunostaining-positive cells in total HSPCs (labeled by cmyb-positive cells). The number of proliferative HSPCs (BrdU) was decreased in the AGM in 142T−/− embryos compared with WT siblings at 24 and 36 hpf, respectively (Figure 4a). These data together indicated that proliferation of emerging HSPCs in the AGM region of 142T−/− embryos was affected.
142T−/− embryos display decreased proliferation and increased apoptosis of HSPCs. (a) Percentage of the proliferative HSPCs labeled by BrdU (marked by white arrow heads) in the AGM of 142T−/− embryos and wild-type siblings 24 hpf (mean±s.d., n=6, *P<0.001). (b) TUNEL assay showed more apoptotic cells (white box) in the AGM of 142T−/− embryos at 22 and 24 hpf. The number of TUNEL-positive cells in the AGM region was quantified (mean±s.d., n=3, *P<0.05). (c) Transverse section showed increased apoptotic HSPCs in the AGM region of 142T−/− embryos. Yellow dashed circles denote the dorsal aorta and cardinal vein. TUNEL-positive cells were counted in the dorsal aortal region (mean±s.d., n=4, *P<0.05).
Next, we found that 142T−/− embryos showed increased TUNEL-positive cells in the AGM region at 22 and 24 hpf by TUNEL assay (Figure 4b). Frozen section of the AGM region revealed an increased number of TUNEL-positive cells in the dorsal aorta at 24 hpf (Figure 4c). Consistent with that, 142C−/− also displayed excessive TUNEL signals in the AGM region at 24 hpf (Supplementary Figure S5a). Therefore, cells in this region including HSPCs underwent excessive apoptosis in both types of miR-142a mutants.
On the basis of the decrease in proliferation and the increase in apoptosis of HSPCs in 142T−/− embryos, we referred to our previous microarray data that contained upregulated genes in miR-142a-3p morphants [26] and found that apoptotic genes including p53 were greatly upregulated (Supplementary Figure S5b). Because of the dual roles of p53 in both cell cycle arrest and apoptosis and the hint from the microarray, we next focused on the relationship between miR-142a-3p and p53.
142T−/− embryos display ectopic expression of p53
To elucidate whether the expression of p53 in miR-142a mutants is consistent with the microarray data in morphants, we first examined the expression of p53 and its downstream target p21. qPCR showed that the expression levels of p53 and p21 were increased at 26 and 36 hpf (Figure 5a). To find out whether p53 is increased in HSPCs, frozen section of 142T−/− embryos stained with p53 probe was performed and the result displayed that p53 was ectopically expressed especially in the dorsal aortal region (Figure 5b). To further support this notion, we sorted cmyb-positive cells from the trunk region of 142T−/− embryos at 36hpf for qPCR analysis. Result showed that expression of p53 was increased in cmyb-positive cells in 142T−/− (Figure 5c). Taken together, we concluded that p53 was specifically upregulated in HSPCs of 142T−/− embryos.
142T−/− embryos display ectopic expression of p53. (a) The expression of p53 and p21 was increased in 142T−/− embryos at 26 and 36 hpf using qPCR (mean±s.d., n=3, *P<0.05, **P<0.01). (b) P53 was increased in the dorsal aorta region in 142T−/− embryos at 24 and 26 hpf by WISH. (c) The expression of p53 was increased in cmyb-positive cells in the trunk of 142T−/− embryos at 36hpf.
MiR-142a-3p inhibits p53 by direct binding
On the basis of several studies revealing that p53 could act as a direct target of microRNAs [41] and the increase of p53 in 142T−/− embryos, we sought for the possibility that p53 might act as a direct target of miR-142a-3p. Systematic bioinformatic analysis using RNA hybrid website (BiBiServ2) revealed that there is a miR-142a-3p-binding site in the coding sequence (CDS) but not the 3’-UTR of zebrafish p53, suggesting that miR-142a-3p may be a direct regulator of p53.
To confirm the direct targeting of p53 by miR-142a-3p, we cloned the WT p53-coding region as well as mutated sequences (mismatched with miR-142a-3p seed sequence) containing the predicted miR-142a-3p-binding site into luciferase and GFP reporter vector (Figure 6a, Supplementary Figure S6a and b). Luciferase activity and GFP expression were examined, respectively. Overexpression of the miR-142a-3p duplex in HEK293T cells and zebrafish embryos suppressed the luciferase activity of WT but not the mutated p53-coding region in a dose-dependent manner (Figure 6b, Supplementary Figure S6a), indicating that p53 is a direct target of miR-142a-3p. Moreover, injection of GFP reporter fused with the WT p53 but not the mutated p53-coding region together with the miR-142a-3p duplex displayed a marked inhibition of GFP activity (Supplementary Figure S6b). Taken together, these data strongly suggest that p53 acts as a direct target of miR-142a-3p.
MiR-142a-3p binds to the coding region of p53 mRNA. (a) The scheme of the reporter constructs containing the miR-142a-binding site and luciferase reporter. The coding region of p53 was inserted downstream of the luciferase gene. The microRNA-binding sites were predicted by RNA hybrid website. (b) Luciferase reporter assay showed that there was a suppression of the luciferase activity in wild-type but not mutated p53 in a dose-dependent manner in HEK293T cells (mean±s.d., n=3, *P<0.05, ns stands for no significance). (c) Overexpression of the miR-142-3p duplex decreased the protein level of wild-type p53. The construct of Myc-tagged wild-type p53-coding region (CDS) and western blotting result, left panel; Quantification of western blot, right panel. Myc antibody serves as a tag to evaluate the expression of p53. NC stands for negative control of the miR-142-3p duplex. (d) Overexpression of the miR-142-3p duplex had no effect on the protein level of mutated p53. The construct of Myc-tagged mutated p53 CDS and western blot result, left panel; quantification of western blot analysis, right panel.
To further demonstrate the inhibition of p53 by miR-142a-3p directly, we ectopically expressed the full-length p53 cDNA fused with Myc-tag in the HEK293T cells. The WT or mutated p53 CDS was co-transfected with the miR-142a-3p duplex. Consistent with the reporter assays, overexpression of miR-142a-3p could attenuate Myc-P53 expression in WT p53 but not the mutated p53-transfected cells (Figure 6c and d), further supporting the specific binding of miR-142a-3p to p53.
P53 mediates the decrease in HSPCs upon loss of miR-142a-3p
To determine whether p53 upregulation mediated the HSPC defects in 142T−/− embryos, we first injected p53 morpholino (MO) into 142T−/− embryos. WISH and qPCR results showed that p53 MO could partially rescue the decreased expression of runx1 in 142T−/− embryos at 26 and 36 hpf (Figure 7a and b). Furthermore, we generated a double knockout of miR-142a-3p and p53 by outcrossing adult 142T−/− with p53M214K−/−. WISH showed that the expression of runx1 at 36 hpf was markedly restored in 142T−/− p53M214K−/− embryos, compared with 142T−/− p53M214K+/+ embryos (Figure 7c), consistent with the results by p53 MO knockdown. To determine whether there is a synergy effect between irf7 and p53 in miR-142a-3p-mediated HSPC development, we knocked down both irf7 and p53 in 142T−/− embryos. Double knockdown of irf7 and p53 fully rescued the HSPC defect (Figure 8a and b), demonstrating that irf7 and p53 act synergistically downstream of miR-142a-3p in HSPC development. In addition, the expression of p53 and irf7 was examined in miR-142a mutants injected with irf7 MO or p53 MO, respectively. qPCR showed that knockdown of irf7 or p53 had no effect on the expression of each other (Figure 8c and d), revealing that irf7 and p53 are parallel targets of miR-142a-3p.
P53 mediates the decrease of HSPCs in miR-142a-3p mutants. (a) P53 MO-injected 142T−/− embryos showed the restoration of runx1 expression at 26 and 36 hpf by WISH (left panel). The quantification of runx1-positive cells in the AGM was shown in the right panel (mean±s.d., n=3, *P<0.05). (b) qPCR showed that compared with 142T−/− at 36 hpf, the decrease in runx1 expression was rescued in 142T −/− embryos injected with p53 MO (mean±s.d., n=3, *P<0.05). (c) WISH showed the expression of runx1 in 142T+/+ p53+/+, 142T−/− p53+/+ and 142T−/− p53−/− embryos at 36 hpf. There was a rescue effect on the decreased runx1 expression in 142T −/− p53−/− double mutants. The quantification of runx1-positive cells in the AGM was shown in the right panel (mean±s.d., n=60, *P<0.05).
Combined knockdown of irf7 and p53 restores decreased HSPCs in 142T −/− embryos. (a and b) Injection of irf7 MO with p53 MO rescued the decreased expression of HSPCs marker runx1, and the extent of the restoration was greater than the single injection of irf7 MO at 26 and 36 hpf (mean±s.d., n=5, *P<0.05). (c) The increased expression of p53 was not affected in 142T−/− embryos injected with irf7 MO at 26 and 36 hpf by qPCR (each sample was composed at least 40 embryos, mean±s.d., n=3, *P<0.05, **P<0.01, ns stands for no significance). (d) The increased expression of irf7 was not affected in 142T−/− embryos injected with p53 MO at 26 hpf, 2 and 4 dpf by qPCR (mean±s.d., n=3, *P<0.05, **P<0.01, ns stands for no significance).
In order to test that p53 inhibition specifically restored apoptosis in the AGM region, we injected p53 MO into 142T−/− embryos. As expected, p53 MO injection led to alleviated apoptosis in both the trunk and AGM region of 142T−/− embryos (Supplementary Figure S7a), strengthening the cause–consequence relationship between p53-induced apoptosis and HSPC defects in miR-142a-3p mutant.
In addition, we also examined the expression of differentiated hematopoietic markers in p53 MO-injected 142T−/− mutants. As shown in Figure S7b, the defects of differentiated populations including erythrocytes, neutrophils and T cells in 142T−/− embryos were also partially restored by p53 knockdown, supporting that p53 functions downstream of miR-142a-3p in HSPC differentiation.
Discussion
In this work, by generating two types of miR-142a-3p mutants using both TALENs and CRISPR/Cas9 methods, we demonstrate that miR-142a-3p has an essential role in definitive hematopoiesis. Decreased HSPCs and their derivatives, as well as ectopic expression of p53 and irf7, were observed in miR-142a-3p mutants. Loss of p53 could rescue the defects of HSPCs in miR-142a-3p mutants and direct binding assay together support that p53 is a direct target of miR-142a-3p, and the newly identified miR-142a-3p-p53 axis has a critical role in HSPC survival and thus protects emerging HSPCs and their derivatives from apoptosis. In addition, p53 acts synergistically with the previously reported irf7 downstream of miR-142a-3p in controlling HSPC development [26] (Supplementary Figure S8).
Consistent with our previous finding that miR-142a-3p is highly expressed in the runx1+/cmyb+ cells in the trunk region that labels HSPC populations in zebrafish [26], two independent groups also showed evidence that the counterpart of miR-142a-3p is enriched in HSPC population in Xenopus and mice [22, 23]. Studies in Xenopus demonstrated that miR-142a-3p targets TGFβ and functions as a master regulator in HSPC lineage specification and thus sits at the apex of the hierarchy programming definitive hemangioblast [23]. This is consistent with our work and illustrates the crucial function of miR-142a-3p in definitive hematopoiesis across vertebrates.
However, there are also different views on the function of miR-142a-3p in HSPCs based on morpholino or duplex injection [27, 40]. It was reported that ectopic expression of the miR-142-3p duplex (concentration of 20 μmol l−1) suppresses the primitive erythrocyte progenitors (gata1+) and HSC formation (cmyb+) [27]. We also performed miR-142a-3p duplex injection and found that the dose of duplex they used caused nonspecific side effects in embryos, such as heart edema, whereas lower dose (concentration of 5–0 μmol l−1) showed increased expression of HSPC marker runx1 at 26 hpf as well as cmyb and rag1 at 5 dpf. To exclude the delusive dose effect of morpholino or duplex, in this work, we chose to generate miR-142a genetic mutants for functional investigation.
Recently, by targeting two miR-142 genes including miR-142a and miR-142b, the mutants of miR-142 by zinc-finger nucleases (ZFN) method are generated, and these miR-142-3p mutants display neutrophil defects but with relatively normal HSPCs [24]. Consistently, our 142T−/− embryos also displayed reduced number of neutrophils (marked by lyz) by WISH at 4 dpf (Figure 2f). Furthermore, the kidney, which contains more neutrophils compared with other organs [42], was shrunken in 142T−/− adult fish (Figure 2g). Notably, hematoxylin and eosin staining result revealed that the cellular density of neutrophils was decreased during the loss of miR-142. According to the Giemsa staining, the maturation of neutrophil was normal, as different subtypes of neutrophils with various morphologies were all observed (Supplementary Figure S4). This result is discrepant with the previously reported neutrophil phenotypes in miR-142a/b mutants [24]. On the basis of the deficiency of miR-142a and miR-142b genes, there is a possibility that the reported neutrophil hypermaturation in miR-142a/b mutants might be due to the deletion of miR-142b. Moreover, neutrophils at different maturation stages can be observed in WT and 142T−/− embryos; therefore, the ratio of naive versus mature neutrophils varies with development.
As a tumor suppressor, p53 is also a critical transcription factor in hematopoietic cells and it is involved in the quiescence, self-renewal, senescence and apoptosis of HSPCs [32, 33]. It also modulates the cell cycle arrest of HSPCs through p21 [32]. Notably, mounting evidence suggests that HSPCs are highly sensitive to cellular stress, cell cycling and the microenvironment, which are closely related to the function of p53 [43, 44]. In zebrafish, mutation in cleavage and polyadenylation specificity factor 1 (cpsf1) displays definitive HSC defects due to a p53-dependent apoptotic cell death [45]. In mouse, the depletion of HSPCs in mutants of Mysm1 and Pot1b could be restored by loss of the P53 [38, 46]. These findings support that HSPC survival is tightly controlled by various factors and that p53-mediated apoptosis is the key to maintain the survival of HSPCs. Our work here demonstrates that in zebrafish embryogenesis, the activation of P53 in the AGM region leads to excessive apoptosis, which impairs the survival of the earliest specified HSPCs. Consistent with our findings, super-p53 mice, carrying one extra gene dose of p53, results in significant acquisition of repopulating capacity but decreased survival of HSCs in recipients [47]. Direct and indirect evidence sheds light upon the hypothesis that the dosage of p53 should be strictly controlled to maintain the normal HSPC survival. Although several studies have identified the downstream targets of p53 in regulating HSPCs, the direct upstream regulators of p53 in modulating definitive hematopoiesis are not well defined. Here, we show that miR-142a-3p directly binds to the coding region of p53 and blocks its function, and then reveals the crucial role of the miR-142a-3p-p53 axis in HSPC survival.
In our previous study, irf7 knockdown only partially rescued the HSPC defects in miR-142a-3p morphants [26], suggesting that other targets of miR-142a-3p may be involved in this process. In fact, double knockdown of irf7 and p53 could fully rescued the HSPC defects and the rescue effect was better compared with irf7 knockdown, demonstrating that there is a synergistic effect of both irf7 and p53 downstream of miR-142a-3p in HSPC development. Tight control of HSPC survival is crucial for HSPC development at early stages in vertebrates. It has been reported that scl-a-deficient zebrafish embryos display more apoptosis and failure to maintain the survival of nascent HSPCs in the AGM [48]. In mice, knockout of gata2 in hematopoietic cells using Vav-Cre shows increased apoptosis and attenuated survival of HSPCs [49]. Interestingly, a recent report also showed that miR-142−/− CD4+ DCs underwent excessive apoptosis, compared with their littermate counterparts [50]. Although developmental defects in HSPCs were not examined in miR-142−/− mice, we speculate that a similar HSPC phenotype would be expected at the onset of definitive hematopoiesis. Recently, inhibition of p53 and its downstream targets p21 or miR-34a has been demonstrated to enhance the reprogramming efficiency of induced pluripotent stem (iPS) cells generation [51, 52]. Moreover, repression of p53 can efficiently induce CD34+ cord blood cells to iPSCs [53]. These lines of evidence in stem cells give clues that excessive level of p53 might dampen the emergence of stem cells and miR-142a-3p could protect the nascent emerging HSPCs from unwanted apoptosis, and this protection is critical for HSPC development during embryogenesis.
In summary, our studies demonstrate that miR-142a-3p directly targets p53 to regulate HSPC survival in zebrafish embryos. In addition to our previous report in which irf7 is a direct target of miR-142a-3p in controlling HSPC formation and differentiation, here we discover that p53 acts downstream of miR-142a-3p to control the survival of HSPCs.
Materials and Methods
Generation of miR-142a mutants and RNA secondary structure prediction
Zebrafish miR-142a mutants were generated by TALEN and CRISPR/Cas9, as previously described [54, 55]. The injection dose of miR-142a TALEN mRNA was 250 pg per embryo. The injection doses of gRNA and cas9 mRNA were 100 and 500 pg per embryo, respectively. The TALEN-generated miR-142a mutants (142T−/−) had a 13-base-pair deletion and could be verified by sequencing (Supplementary Table S1). The CRISPR/Cas9-generated miR-142a mutants (142C−/−) caused a 986-base-pair deletion, and the structure of mature miR-142a-3p in both mutants was disrupted. The 142C−/− could be genotyped by a combination of different lengths of PCR products (Supplementary Table S2). The microRNA secondary structure was predicted by the RNA Structure software (version 4.6).
Fish strains and embryos
Zebrafish strains including WT Tubingen, 142T−/−, 142C−/−, p53M214K, cmyb:EGFP/142T+/− and 142T+/− p53M214K+/− were raised and maintained in system water at 28.5 °C. Fish line p53M214K [56] was kindly provided by Dr Anming Meng (Tsinghua University). This study was approved by the Ethical Review Committee in the Institute of Zoology, Chinese Academy of Sciences, China.
Morpholinos and microinjection
Antisense morpholinos were ordered from Gene Tools including p53 MO and irf7 MO. The p53 MO: 5′-GCGCCATTGCTTTGCAAGAATTG-3′ was injected at the dose of 4 ng per embryo and the irf7 MO was conducted as previously described [26].
WISH
Antisense RNA probes were synthesized with SP6 or T7 RNA polymerase (Promega, Madison, WI, USA). WISH for miR-142a-3p was conducted at 52 °C and for others were performed at 65 °C with probes including miR-142a-3p, scl, pu.1, msr, flt4, dll4, dltC, efnB2a, runx1, cmyb, gata1, lyz, ikaros, rag1 and p53. Numbers in the bottom right denote the ratio of representative embryos in the total scored embryos. At least three independent experiments were conducted for each analysis.
Quantitative RT-PCR and western blot analyses
Trunk regions of embryos were dissected for qPCR and western blot analyses. Quantitative RT-PCR and western blot analyses were performed as described previously [57]. Antibody used for western blot analysis included anti-Runx1(1:200, AnaSpec, Fermont, CA, USA) and anti-Myc (1:500, Cell Signaling, Danvers, MA, USA). Anti-Myc antibody was used as a tag to test the binding affinity of miR-142a-3p to p53.
Reporter assay
The WT and mutated p53-coding regions were cloned and then inserted into the EcoRI and SpeI sites of the pGL3-luciferase vector, which was kindly provided by Dr Peifeng Li (Institute of Zoology, CAS, Beijing, China). The WT p53-coding region was mutated from ‘CACUGC’ to ‘GUGAUG’. Wild-type or mutated pGL3-p53 CDS co-transfected with negative control or the miR-142-3p duplex (Invitrogen, Shanghai, China) were transfected into HEK293T cells using Lipofectamine2000 (Invitrogen). The luciferase activity was measured using the luciferase activity assay kit (Promega). Luciferase and GFP reporter assay in zebrafish was conducted as described previously [24, 58].
Immunostaining, BrdU incorporation and TUNEL assay and section
Live zebrafish embryos injected with BrdU were fixed in 4% paraformaldehyde (PFA) overnight and then replaced with methanol at −20 °C for 2 hpf. Embryos were permeabilized with proteinase K, refixed in 4% PFA for 20 min and then rinsed in PBST, followed by incubation in 2 n HCl for 1 h. Then, embryos were blocked for 1 h in blocking solution and incubated with primary anti-BrdU antibody (Roche, Mannheim, Germany) and secondary Alexa Fluor 488-conjugated anti-mouse antibody (Invitrogen). The BrdU-labeled embryos were treated with 30% sucrose and then washed three times using PBST and embedded in O.C.T. medium (SAKURA, Torrance, CA, USA). The embryos were sectioned using LEICACM1900 cryostats. Detection of both cmyb RNA and mitosis marker pH3 and BrdU simultaneously was conducted as described previously [59].
Embryos at 24 hpf in methanol were washed with PBST and then permeabilized with proteinase K. After being washed with PBST, embryos were subjected to TUNEL assay according to the instruction of the TUNEL staining kit (Roche) at 4 °C overnight.
Flow cytometry of transgenic fish and whole-kidney marrow
Flow cytometry analysis and cell sorting were based on the forward scatter and side scatter as described previously [42]. All experiments were conducted using the FACSCalibur flow cytometer (BD, San Jose, CA, USA). Data was analyzed using the FlowJo software Version 7.6.1.
Hematoxylin and eosin and Wright–Giemsa
Hematoxylin and eosin staining was performed on the serial sections of larval zebrafish. Wright–Giemsa staining was performed as described previously [42].
Microscopy
Embryos were observed using a Nikon microscope for digital image capture. Confocal images of live cmyb:EGFP embryos, TUNEL and BrdU assays were taken on confocal microscope (Nikon A1 laser microscope, Nikon A1, Melville, NY, USA). All images were assembled using Photoshop (Adobe Systems, Mountain View, CA, USA).
Statistical analysis
Values are presented as mean±s.d. Student’s t-test was used to compare the means of different groups and P-values<0.05 were considered to be significant (*P<0.05, **P<0.01).
References
Wang LD, Wagers AJ . Dynamic niches in the origination and differentiation of haematopoietic stem cells. Nat Rev Mol Cell Biol 2011; 12: 643–655.
Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DY, Traver D . Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 2010; 464: 108–111.
Boisset JC, van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C . In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 2010; 464: 116–120.
Jaffredo T, Gautier R, Eichmann A, Dieterlen-Lievre F . Intraaortic hemopoietic cells are derived from endothelial cells during ontogeny. Development 1998; 125: 4575–4583.
Jaffredo T, Bollerot K, Sugiyama D, Gautier R, Drevon C . Tracing the hemangioblast during embryogenesis: developmental relationships between endothelial and hematopoietic cells. Int J Dev Biol 2005; 49: 269–277.
Clements WK, Kim AD, Ong KG, Moore JC, Lawson ND, Traver D . A somitic Wnt16/Notch pathway specifies haematopoietic stem cells. Nature 2011; 474: 220–262.
Espin-Palazon R, Stachura DL, Campbell CA et al. Proinflammatory signaling regulates hematopoietic stem cell emergence. Cell 2014; 159: 1070–1085.
Kim AD, Melick CH, Clements WK et al. Discrete Notch signaling requirements in the specification of hematopoietic stem cells. EMBO J 2014; 33: 2363–2373.
Kobayashi I, Kobayashi-Sun J, Kim AD et al. Jam1a-Jam2a interactions regulate haematopoietic stem cell fate through Notch signalling. Nature 2014; 512: 319–323.
Wilkinson RN, Pouget C, Gering M et al. Hedgehog and Bmp polarize hematopoietic stem cell emergence in the zebrafish dorsal aorta. Dev Cell 2009; 16: 909–916.
Bresciani E, Carrington B, Wincovitch S et al. CBF beta and RUNX1 are required at 2 different steps during the development of hematopoietic stem cells in zebrafish. Blood 2014; 124: 70–78.
de Pater E, Kaimakis P, Vink CS et al. Gata2 is required for HSC generation and survival. J Exp Med 2013; 210: 2843–2850.
Ren X, Gomez GA, Zhang B, Lin S . Scl isoforms act downstream of etsrp to specify angioblasts and definitive hematopoietic stem cells. Blood 2010; 115: 5338–5346.
Solaimani Kartalaei P, Yamada-Inagawa T, Vink CS et al. Whole-transcriptome analysis of endothelial to hematopoietic stem cell transition reveals a requirement for Gpr56 in HSC generation. J Exp Med 2014; 212: 93–106.
Trowbridge JJ, Snow JW, Kim J, Orkin SH . DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell 2009; 5: 442–449.
Luo M, Jeong M, Sun D et al. Long non-coding RNAs control hematopoietic stem cell function. Cell Stem Cell 2015; 16: 426–438.
Chen CZ, Li L, Lodish HF, Bartel DP . MicroRNAs modulate hematopoietic lineage differentiation. Science 2004; 303: 83–86.
Liu J, Li W, Wang S et al. MiR-142-3p attenuates the migration of CD4(+) T cells through regulating actin cytoskeleton via RAC1 and ROCK2 in arteriosclerosis obliterans. PLoS ONE 2014; 9: e95514.
Lagrange B, Martin RZ, Droin N et al. A role for miR-142-3p in colony-stimulating factor 1-induced monocyte differentiation into macrophages. Biochim Biophys Acta 2013; 1833: 1936–1946.
Kramer NJ, Wang WL, Reyes EY et al. Altered lymphopoiesis and immunodeficiency in miR-142 null mice. Blood 2015; 125: 3720–3730.
Guo S, Lu J, Schlanger R et al. MicroRNA miR-125a controls hematopoietic stem cell number. Proc Natl Acad Sci USA 2010; 107: 14229–14234.
Pereira CF, Chang B, Qiu JJ et al. Induction of a hemogenic program in mouse fibroblasts. Cell Stem Cell 2013; 13: 205–218.
Nimmo R, Ciau-Uitz A, Ruiz-Herguido C et al. MiR-142-3p controls the specification of definitive hemangioblasts during ontogeny. Dev Cell 2013; 26: 237–249.
Fan HB, Liu YJ, Wang L et al. miR-142-3p acts as an essential modulator of neutrophil development in zebrafish. Blood 2014; 124: 1320–1330.
Chapnik E, Rivkin N, Mildner A et al. miR-142 orchestrates a network of actin cytoskeleton regulators during megakaryopoiesis. Elife 2014; 3: e01964.
Lu X, Li X, He Q et al. miR-142-3p regulates the formation and differentiation of hematopoietic stem cells in vertebrates. Cell Res 2013; 23: 1356–1368.
Song BF, Zhang Q, Zhang ZJ et al. Systematic transcriptome analysis of the zebrafish model of diamond-blackfan anemia induced by RPS24 deficiency. BMC Genomics 2014; 15: 759.
Krug U, Ganser A, Koeffler HP . Tumor suppressor genes in normal and malignant hematopoiesis. Oncogene 2002; 21: 3475–3495.
Speidel D . The role of DNA damage responses in p53 biology. Arch Toxicol 2015; 89: 501–517.
Lowe SW, Schmitt EM, Smith SW, Osborne BA, Jacks T . p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature 1993; 362: 847–849.
Kuerbitz SJ, Plunkett BS, Walsh WV, Kastan MB . Wild-type p53 is a cell cycle checkpoint determinant following irradiation. Proc Natl Acad Sci USA 1992; 89: 7491–7495.
Pant V, Quintas-Cardama A, Lozano G . The p53 pathway in hematopoiesis: lessons from mouse models, implications for humans. Blood 2012; 120: 5118–5127.
Nii T, Marumoto T, Tani K . Roles of p53 in various biological aspects of hematopoietic stem cells. J Biomed Biotechnol 2012; 2012: 903435.
Dumble M, Moore L, Chambers SM et al. The impact of altered p53 dosage on hematopoietic stem cell dynamics during aging. Blood 2007; 109: 1736–1742.
Tyner SD, Venkatachalam S, Choi J et al. p53 mutant mice that display early ageing-associated phenotypes. Nature 2002; 415: 45–53.
Bibikova E, Youn MY, Danilova N et al. TNF-mediated inflammation represses GATA1 and activates p38 MAP kinase in RPS19-deficient hematopoietic progenitors. Blood 2014; 124: 3791–3798.
Batta K, Florkowska M, Kouskoff V, Lacaud G . Direct reprogramming of murine fibroblasts to hematopoietic progenitor cells. Cell Rep 2014; 9: 1871–1884.
Belle JI, Langlais D, Petrov JC et al. p53 mediates loss of hematopoietic stem cell function and lymphopenia in Mysm1 deficiency. Blood 2015; 125: 2344–2348.
Lalwani MK, Sharma M, Singh AR et al. Reverse genetics screen in zebrafish identifies a role of miR-142a-3p in vascular development and integrity. PLoS ONE 2012; 7: e52588.
Nishiyama T, Kaneda R, Ono T et al. miR-142-3p is essential for hematopoiesis and affects cardiac cell fate in zebrafish. Biochem Biophys Res Commun 2012; 425: 755–761.
Le MT, Teh C, Shyh-Chang N et al. MicroRNA-125b is a novel negative regulator of p53. Genes Dev 2009; 23: 862–876.
Traver D, Paw BH, Poss KD, Penberthy WT, Lin S, Zon LI . Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat Immunol 2003; 4: 1238–1246.
Mohrin M, Shin J, Liu Y et al. Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science 2015; 347: 1374–1377.
Zhang M, Zhu X, Zhang Y et al. RCAD/Ufl1, a Ufm1 E3 ligase, is essential for hematopoietic stem cell function and murine hematopoiesis. Cell Death Differ (e-pub ahead of print 8 May 2015; doi:10.1038/cdd.2015.51).
Bolli N, Payne EM, Rhodes J et al. cpsf1 is required for definitive HSC survival in zebrafish. Blood 2011; 117: 3996–4007.
Wang Y, Shen MF, Chang S . Essential roles for Pot1b in HSC self-renewal and survival. Blood 2011; 118: 6068–6077.
Herrera-Merchan A, Cerrato C, Luengo G et al. miR-33-mediated downregulation of p53 controls hematopoietic stem cell self-renewal. Cell Cycle 2010; 9: 3277–3285.
Zhen FH, Lan YH, Yan B, Zhang WQ, Wen ZL . Hemogenic endothelium specification and hematopoietic stem cell maintenance employ distinct Scl isoforms. Development 2013; 140: 3977–3985.
de Pater E, Kaimakis P, Vink CS et al. Gata2 is required for HSC generation and survival. J Exp Med 2013; 210: 2843–2850.
Mildner A, Chapnik E, Manor O et al. Mononuclear phagocyte miRNome analysis identifies miR-142 as critical regulator of murine dendritic cell homeostasis. Blood 2013; 121: 1016–1027.
Hong H, Takahashi K, Ichisaka T et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 2009; 460: 1132–1135.
Choi YJ, Lin CP, Ho JJ et al. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol 2011; 13: 1353–1360.
Takenaka C, Nishishita N, Takada N, Jakt LM, Kawamata S . Effective generation of iPS cells from CD34(+) cord blood cells by inhibition of p53. Exp Hematol 2010; 38: 154–162.
Cade L, Reyon D, Hwang WY et al. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res 2012; 40: 8001–8010.
Chang N, Sun C, Gao L et al. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res 2013; 23: 465–472.
Berghmans S, Murphey RD, Wienholds E et al. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc Natl Acad Sci USA 2005; 102: 407–412.
Wei YL, Ma DY, Gao Y, Zhang CX, Wang L, Liu F . Ncor2 is required for hematopoietic stem cell emergence by inhibiting Fos signaling in zebrafish. Blood 2014; 124: 1578–1585.
Alcaraz-Perez F, Mulero V, Cayuela ML . Application of the dual-luciferase reporter assay to the analysis of promoter activity in Zebrafish embryos. BMC Biotechnol 2008; 8: 81.
Jia XE, Ma K, Xu T et al. Mutation of kri1l causes definitive hematopoiesis failure via PERK-dependent excessive autophagy induction. Cell Res 2015; 25: 946–962.
Acknowledgements
We thank the laboratory members for helpful discussions and critical reading of the paper. This work was supported by grants from the National Basic Research Program of China (2010CB945302 and 2011CB943904), the National Natural Science Foundation of China (31271570, 31425016) and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA01010110).
Author information
Authors and Affiliations
Contributions
XL and YW performed the research and contributed to the writing. FL conceived the research, analyzed the data and wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
( Supplementary Information is linked to the online version of the paper on the Cell Discovery website.)
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Lu, X., Wei, Y. & Liu, F. Direct regulation of p53 by miR-142a-3p mediates the survival of hematopoietic stem and progenitor cells in zebrafish. Cell Discov 1, 15027 (2015). https://doi.org/10.1038/celldisc.2015.27
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/celldisc.2015.27