Abstract
Tumor suppressor p53 has a key role in maintaining genomic stability and preventing tumorigenesis through its regulation of cellular stress responses, including apoptosis, cell cycle arrest and senescence. To ensure its proper levels and functions in cells, p53 is tightly regulated mainly through post-translational modifications, such as ubiquitination. Here, we identified E3 ubiquitin ligase TRIM32 as a novel p53 target gene and negative regulator to regulate p53-mediated stress responses. In response to stress, such as DNA damage, p53 binds to the p53 responsive element in the promoter of the TRIM32 gene and transcriptionally induces the expression of TRIM32 in cells. In turn, TRIM32 interacts with p53 and promotes p53 degradation through ubiquitination. Thus, TRIM32 negatively regulates p53-mediated apoptosis, cell cycle arrest and senescence in response to stress. TRIM32 is frequently overexpressed in different types of human tumors. TRIM32 overexpression promotes cell oncogenic transformation and tumorigenesis in mice in a largely p53-dependent manner. Taken together, our results demonstrated that as a novel p53 target and a novel negative regulator for p53, TRIM32 has an important role in regulation of p53 and p53-mediated cellular stress responses. Furthermore, our results also revealed that impairing p53 function is a novel mechanism for TRIM32 in tumorigenesis.
Similar content being viewed by others
Main
p53 has a critical role in tumor prevention.1, 2, 3 p53 is frequently inactivated through DNA mutations and other mechanisms in human cancers. In response to stress, p53 binds to the p53 responsive elements (p53 REs) in its target genes and transcriptionally regulates gene expression, which in turn initiates various cellular responses, such as apoptosis, cell cycle arrest and senescence.1, 3, 4 The p53 protein levels are tightly regulated in cells to maintain its proper function, which is primarily achieved through post-translational modifications, especially ubiquitination and resultant proteasomal degradation.5, 6 The E3 ubiquitin (Ub) ligase MDM2 is a p53 target and a critical negative regulator for p53 that degrades p53 through ubiquitination, and thus forms a negative feedback loop with p53 to regulate the p53 protein levels and functions.7, 8 Recently, E3 Ub ligases Pirh2 and Cop1 were identified as p53 targets that can degrade p53 through ubiquitination.9, 10
TRIM32 is a protein of the tripartite motif (TRIM) family.11, 12 The proteins of TRIM family have been reported to be involved in different biological processes, including cell growth, differentiation, development, muscular physiology, innate immune response and cancer.11, 12 The biological function of TRIM32 is not well understood. TRIM32 regulates neuronal and skeletal muscle cell differentiation.13, 14 TRIM32 overexpression was frequently observed in skin carcinoma, and head and neck squamous cell carcinoma, thereby suggesting a potential role of TRIM32 in tumorigenesis.15, 16 TRIM32 was reported to possess E3 Ub ligase activity, attributable to its RING finger. TRIM32 ubiquitinates dysbindin that may contribute to TRIM32’s role in skeletal muscle and neuronal cell differentiation.17 TRIM32 ubiquitinates NF-κB inhibitor Piasy and tumor suppressor Abi2, which may contribute to TRIM32’s role in tumorigenesis.16, 18
In this study, we identified TRIM32 as a novel p53 target and negative regulator for p53. TRIM32 forms a negative feedback loop with p53 to negatively regulate p53 functions in response to stress. TRIM32 is frequently overexpressed in tumors. TRIM32 overexpression promotes cell oncogenic transformation and tumorigenesis in a largely p53-dependent manner. These results demonstrated a novel function of TRIM32 in regulating p53 and p53-mediated stress responses, and revealed that impairing p53 function is an important mechanism for TRIM32 in tumorigenesis.
Results
p53 induces TRIM32 in response to stress
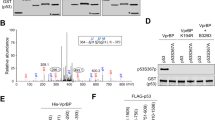
The typical p53 RE is composed of two half-sites (Figure 1a). Some p53 REs have more than two half-sites, referred to as cluster sites, that can increase p53 binding affinity.19, 20 The p53 MH algorithm is a computer program designed to screen for potential p53 target genes by identifying the potential p53 REs in genes.20, 21 The algorithm computes a score between 0 and 100. The sequence with a score >85 is considered as a potential p53 RE.21 Using this algorithm, a list of p53 target genes have been recently identified.21, 22, 23, 24 This algorithm predicted two potential p53 REs in the promoter and intron 1 of TRIM32, respectively (Figure 1a).
p53 transcriptionally induces TRIM32 in response to stress. (a) The potential p53 REs in the promoter and intron 1 of the TRIM32 gene predicted by p53 MH algorithm. Upper panel: the consensus sequence of the typical p53 RE. N, any nucleotide; Pu, purine; Py, pyrimidine; TSS, transcription start site. The scores for the potential p53 REs were labeled. (b and c) p53 expression induced TRIM32 expression at protein (b) and mRNA levels (c) in Tet-on p53-inducible LN-2024 cells treated with doxycycline. The TRIM32 mRNA levels were measured by Taqman real-time PCR assays and normalized with actin. (d–g) Etoposide and 5-fluorouracil (5-FU) induced TRIM32 at both protein (d and f) and mRNA levels (e and g) in a p53-dependent manner in HCT116 and RKO cells. (h) p53 bound to the putative p53 RE in the TRIM32 promoter in HCT116 p53+/+ cells treated with etoposide as detected by ChIP assays. (i) Ectopic p53 expression activated the luciferase activity of the luciferase reporter vector containing the p53 RE in the TRIM32 promoter in HCT116 p53−/− cells. Data are presented as mean±S.D. (n=4). **P<0.001
We first investigated whether p53 regulates TRIM32 expression in LN-2024 cells and its parental LN-Z308 cells. LN-2024 cells are p53-null human glioblastoma LN-Z308 cells stably transfected with a Tet-on p53 expression vector that express wild-type (WT) p53 in the presence of doxycycline.25 This pair of cell lines has been widely used for identifying new p53 target genes.23, 25, 26, 27 Doxycycline induced the expression of p53 and its target MDM2 in LN-2024 cells but not in LN-Z308 cells (Figure 1b). Notably, doxycycline induced TRIM32 at both protein and RNA levels in LN-2024 cells but not in LN-Z308 cells (Figures 1b and c).
To investigate whether TRIM32 is induced by p53 activation in response to physiological stress, HCT116 p53+/+, HCT116 p53−/−, RKO p53+/+ and RKO p53−/− cells were treated with chemotherapeutic agents etoposide or 5-fluorouracil (5-FU). These two pairs of isogenic human colorectal cell lines with or without expression of WT p53 have been widely used for p53 target gene identification and p53 function study.22, 23, 28, 29 Etoposide and 5-FU clearly induced TRIM32 at protein and mRNA levels in p53+/+ HCT116 and RKO but not in p53−/− HCT116 or RKO cells, indicating that p53 activation induces TRIM32 (Figures 1d–g). Interestingly, compared with the induction of MDM2 by p53 that occurred at early time points after treatment (e.g., at 3 h), the induction of TRIM32 by p53 was observed at later time points (e.g., at 15 and 24 h) (Supplementary Figure 1). p53 did not regulate the basal levels of TRIM32 as shown in untreated p53+/+ and p53−/− HCT116 and RKO cells (Figures 1d–g). We further investigated whether TRIM32 can be induced by p63 and p73, two p53 family member proteins, in these cell lines in response to stress. It has been reported that TA-p63α and TA-p73α are the main p63 and p73 isoforms expressed in HCT116 and RKO cells,30, 31, 32 which was confirmed in Supplementary Figure 2a. Etoposide and 5-FU induced TA-p63α and TA-p73α but not TRIM32 in p53−/− HCT116 and RKO cells (Figures 1d and f). Consistently, knockdown of TA-p63α and TA-p73α by siRNA did not clearly affect the induction of TRIM32 by etoposide and 5-FU in p53+/+ HCT116 and RKO cells (Supplementary Figures 2b–e).
In addition to the above-mentioned cell lines, p53-dependent induction of TRIM32 in response to stress (e.g., etoposide) was also observed in normal human neuronal CRL-10742 and colorectal CRL-1831 cells (Supplementary Figures 3 and 4), as well as p53 WT human lung cancer H460 and breast cancer MCF7 cells (Supplementary Figures 5a and b). These results demonstrated that TRIM32 is transcriptionally induced by p53 in response to stress in various types of cells.
The TRIM32 gene contains a functional p53 RE
To investigate whether p53 induces TRIM32 in response to stress through binding to these two putative p53 REs in TRIM32 (Figure 1a), HCT116 p53+/+ and p53−/− cells were treated with etoposide for chromatin immunoprecipitation (ChIP) assays. The p53 antibody specifically pulled down the DNA fragment containing the putative p53 RE in the promoter but not intron 1 of TRIM32 in etoposide-treated p53+/+ cells but not in p53−/− cells or untreated p53+/+ cells (Figure 1h). This result indicates that activated p53 binds to the p53 RE in the TRIM32 promoter in response to stress to regulate TRIM32 expression.
To investigate whether these putative p53 REs confer the p53-dependent transcriptional activity, HCT116 p53−/− cells were transfected with the luciferase reporter vectors containing the above two putative p53 REs in TRIM32 together with control vectors or vectors expressing WT or R273H mutant p53. Compared with cells transfected with control vectors, WT p53 greatly enhanced luciferase activities (by ∼50-fold) of the reporter vectors containing the putative p53 RE in the TRIM32 promoter but not in the intron 1 (Figure 1i). R273H mutant p53 only showed a marginal effect (by ∼threefold) on the luciferase activities, which could be owing to the residual transcriptional activity of the mutant p53.33 These results together demonstrated that TRIM32 is a p53 target gene.
TRIM32 negatively regulates p53 protein levels
TRIM32 was reported to possess the E3 Ub ligase activity.16, 17, 18 These findings raised an interesting question that whether TRIM32 is an E3 Ub ligase for p53. To test this hypothesis, p53-null human lung H1299 cells were co-transfected with vectors expressing WT p53 and TRIM32. TRIM32 overexpression downregulated the exogenous p53 in H1299 cells (Figure 2a). Similarly, ectopic TRIM32 downregulated the endogenous p53 in both etoposide-untreated and -treated p53+/+ HCT116 and RKO cells (Figure 2b).
TRIM32 negatively regulates p53 protein levels under both non-stressed and stressed conditions. (a) Ectopic expression of TRIM32 negatively regulated the exogenous p53 protein in p53-null H1299. Cells were transfected with pCMV-TRIM32-Flag vectors (1, 3 and 6 μg) together with pRC-WTp53 vectors (0.3 μg). (b) Ectopic expression of TRIM32 reduced the levels of endogenous p53 protein in HCT116 p53+/+ and RKO p53+/+ cells with or without etoposide treatment (20 μM for 15 h). (c) Knockdown of endogenous TRIM32 by siRNA (TRIM32-siR) increased the levels of endogenous p53 protein in HCT116 p53+/+ and RKO p53+/+ cells treated with or without etoposide (20 μM for 15 h). TRIM32 knockdown was confirmed by western blot (left panels) and Taqman real-time PCR assays (right panel), respectively. The TRIM32 mRNA levels were normalized with actin. (d) Ectopic TRIM32 expression or TRIM32 knockdown did not affect the p53 mRNA levels in cells. The p53 mRNA levels were measured by real-time PCR and normalized with actin. (e) TRIM32 negatively regulated p53 in an MDM2-independent manner in p53−/− MDM2−/− 2KO cells. Left panels: cells were transfected with pCMV-TRIM32-Flag vectors (1, 3 and 6 μg) together with pRC-WTp53 vectors (0.3 μg). Right panels: the endogenous TRIM32 was knocked down by siRNA in 2KO cells transfected with pRC-WTp53 vectors. (f) TRIM32 negatively regulated the endogenous p53 in an MDM2-independent manner in HCT116 p53+/+ cells. Cells were transfected with TRIM32 siRNA and MDM2 siRNA, respectively, or two siRNAs together. Two different siRNA against TRIM32 and MDM2 were used and similar results were observed. In c and d, data are presented as mean±S.D. (n = 3). **P<0.001
To investigate whether endogenous TRIM32 regulates p53 levels in cells, TRIM32 was knocked down by siRNA oligos in p53+/+ HCT116 and RKO cells (Figure 2c). TRIM32 knockdown induced p53 in both etoposide-untreated and -treated cells (Figure 2c). Furthermore, TRIM32 overexpression or knockdown did not affect p53 mRNA levels (Figure 2d). Consistently, TRIM32 knockdown induced p53 in H460 and MCF7 cells (Supplementary Figures 5c and d).
To investigate whether the downregulation of p53 by TRIM32 is MDM2 dependent, p53−/− MDM2−/− MEFs (mouse embryonic fibroblasts) (2KO) were used. While ectopic TRIM32 clearly downregulated the ectopic p53, knockdown of endogenous TRIM32 upregulated the ectopic p53 in 2KO cells (Figure 2e). Consistently, simultaneous knockdown of endogenous TRIM32 and MDM2 by siRNA in HCT116 p53+/+ cells displayed an addictive effect on the induction of endogenous p53 (Figure 2f). These results indicate that TRIM32 can downregulate p53 independently of MDM2.
TRIM32 interacts with p53
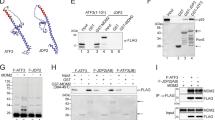
To investigate whether TRIM32 is an E3 Ub ligase for p53, we first examined the interaction between TRIM32 and p53. H1299 cells co-transfected with TRIM32-Flag and p53 vectors were used for immunoprecipitation (IP) assays. TRIM32-Flag was co-precipitated by the p53 antibody, and vice versa, indicating that the ectopic TRIM32 interacts with p53 in cells (Figure 3a). The interaction between endogenous TRIM32 and p53 was also observed in HCT116 p53+/+ cells (Figure 3b).
TRIM32 interacts with p53 and promotes p53 degradation through ubiquitination. (a) The interaction between exogenous TRIM32 and p53 in p53-null H1299 cells co-transfected with the pCMV-TRIM32-Flag and pRC-WT p53 vectors. IP, immunoprecipitation; IB, immunoblotting. (b) The interaction between endogenous TRIM32 and p53 in HCT116 p53+/+ cells. (c) Schematic representation of pCMV-Flag vectors expressing WT or serial deletion mutants of TRIM32. (d) TRIM32 interacted with p53 through the NHL domain. H1299 cells were transfected with the indicated pCMV-Flag vectors, followed by IP to map the interaction domain of TRIM32 with p53. (e and f). Ectopic expression of WT TRIM32 but not the TRIM32 ΔRING mutant promoted the ubiquitination of p53 in H1299 cells. In e, cells were transfected with 2 or 6 μg pCMV-TRIM32-Flag vectors. Cell lysates were immunoprecipitated with the p53 antibody and then immunoblotted with the HA antibody. In f, His-Ub was pulled down by Ni-NTA beads and immunoblotted with the p53 antibody. p53-(His-Ub)n: ubiquitinated p53. (g) Ectopic expression of WT TRIM32 but not the TRIM32 ΔRING mutant reduced the levels of exogenous p53 protein in p53-null H1299 and p53−/− MDM2−/− 2KO cells co-transfected with p53 and TRIM32 expression vectors. (h) MG132 inhibited the p53 protein degradation induced by WT TRIM32 in H1299 and 2KO cells. (i) WT TRIM32 but not TRIM32 ΔRING mutant promoted the degradation of endogenous p53, which can be blocked by MG132, in HCT116 p53+/+ cells. (j) Ectopic TRIM32 expression decreased the p53 protein half-life. HCT116 p53+/+ cells transfected with pCMV-TRIM32-Flag were treated with cycloheximide (CHX; 50 μg/ml) for indicated time periods before assays. Data are presented as mean±S.D. (n=3)
TRIM32 contains several important domains, including an N-terminal RING finger, a B-box type 1 and a B-box type 2, a coiled-coil region and a C-terminal NHL domain (Figure 3c). The NHL domain was reported to be required for the interactions of TRIM32 with other proteins.16, 34 To define the binding domain of TRIM32 with p53, a series of Flag-tagged deletion mutants of TRIM32 were constructed, including ΔRING, ΔRINGΔB-box, NHL and ΔNHL (Figure 3c). They were transfected into H1299 cells together with p53 vectors, and their binding to p53 were analyzed by IP assays. The WT, ΔRING and ΔRINGΔB-box mutants of TRIM32 all interacted with p53, whereas deletion of NHL domain (ΔNHL) largely abolished the TRIM32-p53 binding (Figure 3d). Furthermore, the NHL domain itself interacted with p53 (Figure 3d). These results demonstrated that TRIM32 interacts with p53 in cells, and furthermore, the NHL domain of TRIM32 is required for the binding.
TRIM32 promotes ubiquitination and degradation of p53
We further investigated whether TRIM32 promotes p53 degradation through uniquitination. The RING finger domain of E3 Ub ligases is critical for their E3 Ub ligase activity.5, 35 Here, vectors expressing the WT or ΔRING TRIM32 were transfected into H1299 cells together with p53 vectors and Ub vectors. The ectopic WT TRIM32 but not TRIM32 ΔRING mutant clearly promoted p53 ubiquitination (Figures 3e and f).
To investigate whether p53 ubiquitination by TRIM32 promotes p53 degradation, H1299 cells were transfected with p53 vectors together with vectors expressing the WT or ΔRING TRIM32. The WT but not ΔRING TRIM32 downregulated the ectopic p53 in H1299 cells (Figure 3g). Consistent results were observed in 2KO MEFs, indicating that TRIM32’s effect on p53 ubiquitination and degradation is MDM2 independent (Figure 3g). Notably, MDM2 exhibited a stronger effect on p53 ubiquitination and degradation than TRIM32 as shown in H1299 cells co-transfected with vectors expressing p53 and TRIM32 or MDM2 (Supplementary Figure 6). These results are consistent with the result in Figure 2f, showing that MDM2 knockdown resulted in higher p53 accumulation than TRIM32 knockdown in HCT116 p53+/+ cells. Furthermore, the downregulation of p53 by TRIM32 was largely abolished when H1299 and 2KO cells were treated with proteasome inhibitor MG132 (Figure 3h). Consistently, WT TRIM32 but not ΔRING TRIM32 reduced the endogenous p53 in HCT116 p53+/+ cells, which was largely abolished by MG132 (Figure 3i). These results indicated that TRIM32’s function in promoting p53 ubiquitination and degradation mainly accounts for its activity in downregulating p53.
In non-stressed cells, p53 protein half-life is short, as p53 protein is quickly degraded mainly through ubiquitination. To determine whether TRIM32 decreases the p53 protein half-life, HCT116 p53+/+ cells transfected with WT TRIM32 vectors were treated with protein synthesis inhibitor cyclohexamide (CHX) for different time periods. Ectopic TRIM32 clearly reduced the half-life of p53 compared with cells transfected with control vectors (16 min versus 28 min; Figure 3j).
Recent studies demonstrated that MDM2 can also ubiquitinate and degrade mutant p53.36, 37 Similarly, TRIM32 can degrade mutant p53 (Supplementary Figure 7). It appears that TRIM32 degraded WT p53 more efficiently than mutant p53, which suggests that TRIM32 may have a more important role in regulation of WT p53 than mutant p53 in cells.
TRIM32 negatively regulates the p53 transcriptional activity
To determine whether the downregulation of p53 by TRIM32 reduces p53 transcriptional activity, p21 luciferase reporter (p21-Luc) containing the p53 RE in p21 promoter was used. Ectopic expression of p53 markedly activated the luciferase activity of the p21-Luc in HCT116 p53−/− cells. Notably, co-expression of WT but not ΔRING TRIM32 greatly reduced p53’s function in activating the luciferase activity of the p21-Luc (Figure 4a).
TRIM32 negatively regulates p53 transcriptional activity in response to stress. (a) TRIM32 downregulated the p53 transcriptional activity toward p21-Luc in HCT116 p53−/− cells. Cells were transfected with p21 luciferase vectors (p21-Luc) together with indicated vectors for 24 h before assays. Data are presented as mean±S.D. (n = 4). **P<0.001. (b and c) TRIM32 negatively regulated the p53 transcriptional activity toward its target genes in HCT116 cells measured by real-time PCR (b) and western blot assays (c). HCT116 p53+/+ and p53−/− cells transfected with control or pCMV-TRIM32-Flag vectors were treated with or without etoposide (20 μM for 15 h). (d and e) Knockdown of endogenous TRIM32 enhanced the p53 transcriptional activities toward its target genes in HCT116 cells measured by real-time PCR (d) and western blot assays (e). HCT116 p53+/+ and p53−/− cells transfected with two TRIM32-siR or control-siR were treated with or without etoposide (20 μM for 15 h) before assays. In b and d, data are presented as mean±S.D. (n=3)
The impact of TRIM32 upon p53 transcriptional activity was further investigated by measuring the mRNA levels of a group of well-known p53 targets in cells, including MDM2, p21, Puma, Fas and Bax. Cells transfected with control or TRIM32 vectors were treated with etoposide, and the mRNA levels of these genes were measured by real-time PCR assays. TRIM32 overexpression clearly reduced the induction of these genes by etoposide in p53+/+ but not in p53−/− HCT116 cells (Figure 4b). These changes were confirmed by western blot assays (Figure 4c). Consistently, knockdown of endogenous TRIM32 induced the expression of the above-mentioned genes in p53+/+ but not in p53−/− HCT116 cells treated with etoposide (Figures 4d and e).
TRIM32 negatively regulates p53-mediated apoptosis, cell cycle arrest and senescence
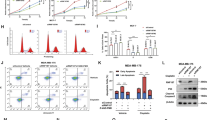
The p53-mediated apoptosis, cell cycle arrest and senescence are important functions of p53 in tumor suppression.1, 2 To investigate whether TRIM32 affects p53 functions in mediating apoptosis, HCT116 and RKO cells were transfected with TRIM32 vectors or siRNA against TRIM32 and then treated with 5-FU. It has been reported that 5-FU induces apoptosis in a highly p53-dependent manner in colorectal cells,38 which was confirmed in Figures 5a and b. TRIM32 overexpression reduced 5-FU-induced apoptosis in p53+/+ but not in p53−/− cells (Figure 5a), whereas TRIM32 knockdown promoted 5-FU-induced apoptosis in p53+/+ but not in p53−/− cells (Figure 5b).
TRIM32 negatively regulates p53-mediated apoptosis, cell cycle arrest and senescence. (a and b) TRIM32 negatively regulated p53 function in apoptosis. HCT116 p53+/+ and p53−/− cells (left), as well as RKO p53+/+ and p53−/− cells (right) were transfected with pCMV-TRIM32-Flag vectors (a) or TRIM32-siR (b). Cells were then treated with 5-FU (300 μM), and apoptosis was measured. The presented is apoptosis measured at 24 and 48 h after treatment in HCT116 and RKO cells, respectively. (c) TRIM32 knockdown by siRNA induced p53-mediated cell cycle arrest in cells. HCT116 p53+/+ and p53−/− cells (left), as well as RKO p53+/+ and p53−/− cells (right), were transfected with control-siR or two different TRIM32-siR for 48 h before cell cycle distribution was analyzed. (d and e) TRIM32 negatively regulated p53 function in senescence. In d, HCT116 p53+/+ and p53−/− cells transfected with pCMV-TRIM32-Flag vectors were treated with doxorubicin (100 nM) for 3 days before senescent cells were detected by SA-β-gal staining. In e, HCT116 p53+/+ and HCT116 p53−/− cells were transfected with TRIM32-siR for 3 days before senescent cells were detected. Left panels: represented images of SA-β-gal staining of senescent cells. Right panels: the percentage of SA-β-gal-positive cells. Data are presented as mean±S.D. (n=4). #P<0.05; *P<0.01
It has been well established that p53 activation induces mainly G1 cell cycle arrest.1, 39 TRIM32 knockdown induced G1 arrest in a p53-dependent manner; it induced G1 arrest in p53+/+ HCT116 and RKO cells but not in p53−/− cells (Figure 5c).
Doxorubicin induces senescence in a p53-dependent manner in different types of cells.40 To investigate whether TRIM32 affects p53-mediated senescence, HCT116 p53+/+ and p53−/− cells transfected with TRIM32 vectors were treated with doxorubicin, and senescent cells were detected by senescence-associated β-galactosidase (SA-β-gal) staining. Doxorubicin induced senescence in a largely p53-dependent manner in HCT116 cells (Figure 5d). While ectopic TRIM32 reduced doxorubicin-induced senescence in HCT116 p53+/+ but not in p53−/− cells, TRIM32 knockdown induced senescence in HCT116 p53+/+ but not in p53−/− cells (Figures 5d and e). Interestingly, knockdown of endogenous TA-p63 or TA-p73 in HCT116 p53+/+ cells did not clearly affect TRIM32’s effects on apoptosis, cell cycle arrest or senescence (Supplementary Figure 8). Notably, TRIM32 overexpression downregulated TA-p63α and TA-p73α protein, whereas TRIM32 knockdown upregulated TA-p63α and TA-p73α in HCT116 p53+/+ cells (Supplementary Figures 9a and b). This result is consistent with a recent report showing that TRIM32 degrades p73 in neuronal cells.41 Furthermore, TRIM32 displayed a stronger effect on p53 than on TA-p63α or TA-p73α as shown in both HCT116 p53+/+ and H1299 cells (Supplementary Figures 9a and b). These results suggested that the downregulation of p53 by TRIM32 mainly contributes to TRIM32’s function in downregulating apoptosis, cell cycle arrest and senescence in response to stress.
TRIM32 is overexpressed in different types of tumors
TRIM32 has been reported to be frequently overexpressed in head and neck cancer, and skin cancer.15, 16 To investigate whether TRIM32 is also overexpressed in other types of tumors, the mRNA levels of TRIM32 were measured in colorectal cancer (n=24), lung cancer (n=24) and hepatocellular carcinoma (n=21), as well as their matched adjacent non-tumor tissues by real-time PCR assays. Compared with their adjacent non-tumor tissues, TRIM32 overexpression (the cutoff is more than twofold higher expression) was observed in a high percentage of tumors, including 8/24 colorectal cancer, 10/24 lung cancer and 10/21 hepatocellular carcinoma (Figures 6a–c). These results demonstrate that TRIM32 is frequently overexpressed in different types of tumors. Considering the important role of TRIM32 in negative regulation of p53, TRIM32 overexpression in tumors may contribute to tumorigenesis.
The overexpression of TRIM32 in human colorectal, lung and liver cancers. (a–c) TRIM32 mRNA levels were measured by Taqman real-time PCR assays and normalized with actin in colorectal cancer (a; n=24), lung cancer (b; n=24), hepatocellular carcinoma (c; n=21), and their matched adjacent non-tumor tissues. The relative TRIM32 levels in samples were calculated by comparing with the TRIM32 levels in #1 non-tumor tissue for each group, which were designated as 1. Tumor samples with TRIM32 overexpression (>twofold higher expression in tumor samples versus matched non-tumor tissues) were labeled with *. Tumor stages were labeled. Data are presented as mean±S.D. (n=3)
TRIM32 promotes cell transformation through downregulation of p53
p53 deficiency promotes oncogenic transformation of primary MEFs.42 To investigate the effect of TRIM32 overexpression on MEF transformation, E1A/Ras-expressing p53+/+ and p53−/− MEFs were transduced with pLPCX-TRIM32-Flag retroviral vectors. TRIM32 overexpression downregulated p53 in E1A/Ras-expressing p53+/+ MEFs (Figure 7a). p53−/− MEFs formed much more colonies in soft agar than p53+/+ MEFs. TRIM32 overexpression greatly promoted the colony formation in p53+/+ MEFs. This promoting effect was much less pronounced in p53−/− MEFs (Figure 7b). Knockdown of TRIM32 by shRNA vectors activated p53 in p53+/+ MEFs (Figure 7c), and greatly reduced colony formation in p53+/+ but not in p53−/− MEFs (Figure 7d). Furthermore, TRIM32’s effect on cell transformation was largely p63- and p73 independent as shown in E1A/Ras-expressing p63−/− or p73−/− MEFs (Supplementary Figure 10). These results demonstrated that TRIM32 promotes cell transformation through downregulation of p53.
TRIM32 promotes cell oncogenic transformation and tumorigenesis in a p53-dependent manner. (a) Overexpression of TRIM32 by pLPCX-TRIM32-Flag vectors decreased p53 protein levels in E1A/Ras-expressing p53+/+ MEFs. (b) Overexpression of TRIM32 promoted the cell oncogenic transformation in a p53-dependent manner in E1A/Ras-expressing MEFs. Left panels: represented images of transformed colonies in soft agar. Right panel: number of transformed colonies per well. (c) Knockdown of endogenous TRIM32 by stable transduction of shRNA vectors increased the p53 protein levels in E1A/Ras-expressing p53+/+ MEFs. The knockdown of TRIM32 was confirmed by Taqman real-time PCR assays (right panel). (d) Knockdown of endogenous TRIM32 by shRNA vectors reduced cell oncogenic transformation in a p53-dependent manner in E1A/Ras-expressing MEFs. In b and d, data are presented as mean±S.D. (n=6). *P<0.01; **P<0.001. (e and f) TRIM32 overexpression promoted the growth of xenograft tumors formed by HCT116 (e) and RKO (f) cells in a p53-dependent manner. Left panel in e: represented images of xenograft tumors at day 25 for HCT116 tumors (e). Right panel in e and f: the growth curves of xenograft tumors. Data are presented as mean±S.D. (n= 12). *P<0.01; **P<0.001. (g) TRIM32 overexpression reduced p53 protein levels in xenograft tumors formed by p53+/+ HCT116 and RKO cells. Four pairs of tumors for each group were examined and very similar results were observed. A pair of tumors for each group was presented. (h) A model illustrating the regulation of p53 by E3 Ub ligases. p53 transcriptionally regulates a group of E3 Ub ligases, such as TRIM32, MDM2, Cop1 and Pirh2, which in turn negatively regulates p53 through ubiquitination. p53 is also negatively regulated by other E3 Ub ligases, such as ARF-BP1, CHIP, synoviolin and Cul1/4
TRIM32 promotes the growth of xenograft tumors through downregulation of p53
We further investigated the effect of TRIM32 overexpression on tumorigenesis. Consistent with previous reports,43, 44, 45 p53 loss promoted the growth of xenograft tumors formed by HCT116 and RKO cells (Figures 7e and f). Notably, TRIM32 overexpression by stable transduction of pLPCX-TRIM32-Flag vectors in cells clearly promoted the growth of p53+/+ HCT116 and RKO tumors. This promoting effect was much less pronounced in p53−/− HCT116 and RKO tumors (Figures 7e and f). Western blot assays confirmed that TRIM32 overexpression downregulated p53 in p53+/+ HCT116 and RKO tumors (Figure 7g). These results strongly suggested that TRIM32 overexpression promotes tumorigenesis through the downregulation of p53 (Figure 7h).
Discussion
In this study, we identified TRIM32 as a novel p53 target and a novel negative regulator for p53. In response to stress, p53 binds to the TRIM32 promoter and induces TRIM32. In turn, TRIM32 promotes p53 degradation through ubiquitination. Thus, TRIM32 negatively regulates p53 levels and functions in cell cycle arrest, senescence and apoptosis in response to stress.
MDM2 has a critical role in negative regulation of p53, which was demonstrated by the embryonic lethality caused by MDM2 knockout in mice that can be rescued by p53 knockout.46, 47 Our results indicated that TRIM32 downregulates p53 in an MDM2-independent manner. In addition to MDM2 and TRIM32, some other Ub ligases have been reported to regulate p53 stability, such as COP1, Pirh2, ARF-BP1, CHIP, synoviolin and Cul1/4.5, 35, 48 Similar to TRIM32, Cop1 and Pirh2 were reported to regulate p53 in an MDM2-independent manner.9, 10 Our results in this study showed that MDM2 is a stronger negative regulator for p53 than TRIM32. Furthermore, TRIM32 and MDM2 showed different kinetics of induction in response to p53 activation. These results suggest that TRIM32 may coordinate with MDM2 in cells in response to stress to regulate p53 levels and functions. It is still unclear why so many E3 Ub ligases in addition to MDM2 are involved in the regulation of p53, and how these E3 Ub ligases, including TRIM32, cross talk and cooperate with MDM2 to regulate p53 in cells. It is possible that these negative regulators modulate p53 in a highly cell type-, stress signal- and developmental stage-specific manners, which needs to be addressed in future studies.
The biological function of TRIM32 is poorly understood. Recent studies suggest that TRIM32 could be involved in tumorigenesis.15, 16 p53 is a haplo-insufficient gene. Slight changes in p53 levels and function could significantly impact tumorigenesis.49 The overexpression of p53 negative regulators in tumors, such as MDM2, MDM4, Pirh2 and cyclin G1, have been reported to contribute to the loss of p53 function in tumors.1, 6, 50 Our finding that TRIM32 negatively regulates p53 strongly supports a potential role of TRIM32 in tumorigenesis. Consistently, we found that TRIM32 mRNA is frequently overexpressed in different types of tumors, although its mechanism is unclear. p53 is frequently mutated in these cancers, and furthermore, mutant p53 proteins largely lose the transcriptional activity of WT p53 toward TRIM32. Other mechanisms independent of p53 should contribute to the TRIM32 overexpression in tumors, which should be addressed in future. The results in this study further showed that TRIM32 overexpression promoted the MEF transformation and tumor growth through the downregulation of p53. These results together strongly suggest that TRIM32 overexpression in tumors leads to the impairment of p53 function that promotes tumorigenesis.
In addition to its role in tumorigenesis, TRIM32 is involved in the regulation of neuronal and muscle cell development, and differentiation.13, 14 TRIM32 ubiquitinates dysbindin that may contribute to TRIM32’s role in skeletal muscle and neuronal cell differentiation.17 Most recently, p73 was reported to upregulate the basal levels of TRIM32, and furthermore, TRIM32 ubiquitinates p73 in neuronal cells, which might contribute to TRIM32’s function in neuronal development and differentiation.41 The regulation of TRIM32 by p53 and p73 appears to be different and highly context dependent. For instance, the induction of TRIM32 by p73 was observed in neuronal cells, but not in the cancer cell lines that showed the upregulation of TRIM32 by p53 activation in response to stress in this study. p53 regulates the inducible levels of TRIM32 in response to stress but not the basal levels of TRIM32 under the non-stressed condition, whereas p73 only regulates the basal levels of TRIM32 in neuronal cells. TRIM32 could be involved in the different cellular processes in different tissues. For instance, TRIM32 regulates cellular stress responses and tumorigenesis through the regulation of p53, whereas TRIM32 might regulate neuronal development and differentiation through the regulation of p73. Future studies should shed further light on how TRIM32 regulates different cellular processes through its regulation of different targets.
Materials and Methods
Cell lines, vectors and siRNA
Human colorectal HCT116 p53+/+, HCT116 p53−/−, RKO p53+/+ and RKO p53−/− cancer cell lines were generous gifts from Dr. Bert Vogelstein. Human glioblastoma p53 Tet-on LN-2024 cells and p53-null LN-Z308 cells were generous gifts from Dr. Erwin Van Meir. p53−/− MDM2−/− 2KO cells were a generous gift from Dr. Gigi Lozano. H1299, H460, MCF7, CRL-1831 and CRL-10742 cells were obtained from ATCC. Cells with stable p53 knockdown were established as we previously described.51 p63−/− and p73−/− MEFs were established from the p63 global knockout mice and p73 knockout mice, respectively (generous gifts from Dr. F McKeon).52, 53 The human TRIM32 cDNA and its deletion mutants were amplified by PCR from the pcDNA-TRIM32-HA plasmid (a generous gift from Dr. Ki-Sun Kwon), and subcloned into the pCMV-Flag vectors or pLPCX-Flag retroviral vectors. Sequences for siRNA oligos against TRIM32, p53, p63 and p73 (Integrated DNA Technologies, Coralville, IA, USA), respectively, were as follows: For human TRIM32: 5′-GGUGGAAAGCUUUGGUGUU-3′ and 5′-GAAGUUGAGAAGUCCAAUA-3′; For p53: 5′-GACUCCAGUGGUAAUCUACUU-3′ and 5′-GUCCAGAUGAAGCUCCCAGAA-3′; For p63: 5′-ACAAGAUUGAGAUUAGCAUGGACTG-3′ and 5′-ACAGUACAAAGAACGGUGAUGGUAC-3′; For p73: 5′-GCCAGCAAGCGUGCCUUCAAGCAGA-3′ and 5′-CGGCAUGGAGACGAGGACACGUACU-3′; For mouse TRIM32: 5′-GGCGGGAAGCTCTGGTGTT-3′ and 5′-GAGGTTGAGAAGTCCAACA-3′. The shRNA vectors against mouse TRIM32 were constructed by inserting the above sequences for mouse TRIM32 siRNA into the GIPZ lentiviral shRNA vector.
ChIP assays
ChIP assays were performed using the ChIP Assay Kit (EMD Millipore, Billerica, MA, USA) as previously described.23 HCT116 p53+/+ and p53−/− cells were treated with etoposide (20 μM) for 15 h before ChIP assays. The DO-1 anti-p53 antibody and mouse IgG (Santa Cruz Biotechnology, Dallas, TX, USA) were used for the assays. The following primer sets were designed to encompass the putative p53 REs in the promoter and intron 1 regions of human TRIM32 gene. For the RE in the promoter: forward primer: 5′-CGGGGTACCTGGGCGTCGGTTTCTTCATCTGC-3′; reverse primer: 5′-CCGCTCGAGACCCGCCTGCCTCCCTCTGC-3′. For the RE in intron 1: forward primer: 5′-CGGGGTACCGTGCAACGGACTATAGGGTGAGGTA-3′; reverse primer: 5′-CCGCTCGAGATTCCCATGACATATTTTCTTTTAGGGAT-3′.
Luciferase reporter assays
For luciferase reporter vector construction, the DNA fragments containing the putative p53 REs in the promoter and intron 1 of human TRIM32 gene were PCR amplified by using the PCR primers for ChIP assays. The PCR fragments were cloned into the promoter region of pGL2 luciferase reporter vector (Promega, Madison, WI, USA) at KpnI and XhoI sites. The pGL2 p21 luciferase reporter vector (p21-Luc), which contains the p53 RE in the p21 promoter, was provided by Promega. Luciferase activity assays were performed as previously described.22 In brief, the reporter vectors were transfected into HCT116 p53−/− cells along with vectors expressing WT p53 (pRC-WT p53), R273H mutant p53 (pRC-mut p53) or control vectors using Lipofectamine 2000 (Invitrogen, Life Technologies, Carlsbad, CA, USA). pRL-SV40 vector expressing renilla luciferase was co-transfected as an internal control to normalize transfection efficiency. Luciferase activities were measured 24 h after transfection. The relative luciferase activity was calculated as luciferase activity of reporter vectors in cells co-transfected with WT p53 compared with that in cells co-transfected with empty vector.
Western blot assays
Standard western blot assays were used to analyze protein expression. Following antibodies were used for assays: anti-TRIM32 (mouse polyclonal, Abnova, Taipei, Taiwan), anti-p21 (Ab-6, Oncogene Research Products, La Jolla, CA, USA), anti-Flag M2 (F3165, Sigma, St. Louis, MO, USA), anti-actin (A5441, Sigma), anti-p53 (DO-1 and FL-393, Santa Cruz Biotechnology), anti-p63 (4A4, Santa Cruz Biotechnology); anti-p73 (ER-15, NeoMarkers, Labvision, Fremant, CA, USA), anti-HA antibody (3F10, Roche, Nutley, NJ, USA), anti-Bax (sc-493, Santa Cruz Biotechnology) and anti-PUMA (no. 4976, Cell Signaling, Danvers, MA, USA). The band intensity was quantified by digitalization of the X-ray film and analyzed with the ImageJ software (NIH, Bethesda, MD, USA).
Analysis of gene expression in cells and tissues
The expression of genes in cells and tumor samples were analyzed by quantitative Taqman real-time PCR assays with TaqMan PCR mixture (Applied Biosystems, Life Technologies) as previously described.23 The expression of genes was normalized to actin. The cDNA arrays containing cDNA from colorectal tumors (n=24), lung tumors (n=24) and their matched adjacent non-tumor tissues were provided by OriGene (Rockville, MD, USA). The formalin-fixed and paraffin-embedded primary hepatocellular carcinoma samples (n=21) and their matched adjacent non-tumor liver tissues were collected at the University of Texas MD Anderson Cancer Center with approved IRB.
IP assays
For co-IP of exogenous TRIM32 and p53 proteins, untreated H1299 cells were co-transfected with pCMV-TRIM32-Flag and pRC-WT p53 vectors. Cells were harvested at 24 h after transfection and lysed in NP-40 buffer. Anti-p53 (DO-1) and anti-Flag (Sigma) agarose beads were used to pull down WT p53 and TRIM32, respectively. The immunonoprecipitates were subjected to western blot assays with the anti-Flag or anti-p53 antibodies (FL-393). For co-IP of endogenous proteins, cell lysates of HCT116 p53+/+ and HCT116 p53−/− cells were incubated with an anti-TRIM32 or anti-p53 (DO-1) antibodies followed by incubation with Protein A/G agarose beads. The immunoprecipitates were subjected to western blot assays with the anti-TRIM32 or anti-p53 antibodies (FL-393).
In vivo ubiquitination assays
In vivo ubiquitination assays were performed as previously described.54 In brief, H1299 cells were transfected with expression vectors expressing WT p53 (pRC-WT p53, 0.3 μg), WT TRIM32 (pCMV-TRIM32-Flag, 6 μg) or TRIM32 ΔRING (pCMV-TRIM32 ΔRING, 6 μg) and HA-tagged Ub or His-tagged Ub (pCDNA-HA-Ub or pCDNA-His-Ub, 1 μg) either alone or in combination by using Lipofectamine 2000 (Invitrogen). Twenty-four hours after transfection, cells were treated with MG132 (20 μM) for 6 h to block proteasome-mediated degradation and cell lysate was prepared. To analyze the ubiquitination of p53, p53 in cell lysates was immunoprecipitated by a monoclonal anti-p53 antibody (DO-1) followed by immunoblotting (IB) with an anti-HA monoclonal antibody, or His-Ub in cell lysate was pulled down by incubation with Ni-NTA beads followed by IB with the anti-p53 (DO-1) antibody.
p53 protein half-life analysis
HCT116 p53+/+ cells were transfected with the pCMV-TRIM32-Flag vector expressing WT TRIM32 or control pCMV-Flag vectors. At 24 h after transfection, cells were treated with CHX (50 μg/ml) for different time periods before cells were collected for western blot analysis of p53 protein. Protein levels were quantified and plotted as percent protein left against time of CHX treatment to calculate p53 protein half-life as previously described.37
Cell cycle, senescence and apoptosis analysis
Cell cycle analysis was performed using a FITC BrdU Flow Kit (BD Biosciences, San Jose, CA, USA) and analyzed using a flow cytometer. Senescent cells were detected by SA-β-gal assays using a Senescence β-Galactosidase Staining Kit (Cell Signaling). For apoptosis analysis, cells were treated with 5-FU (300 μM) for 24, 36 or 48 h. Cells were stained with Annexin V (25 mg/ml) and propidium iodide (50 mg/ml) and analyzed in a flow cytometer.
Cell transformation
WT, p53−/−, p63−/− and p73−/− MEF cells transduced with pBabe-RasV12 and pBabe-E1A retroviral vectors (Addgene, Cambridge, MA, USA) were transduced with pLPCX-TRIM32-Flag retroviral vectors or control pLPCX vectors. Equal amounts of infected cells (4x104/well) were plated in soft agar in six-well plates. Transformed foci were scored 2 weeks later.
In vivo tumorigenecity assays
Xenograft tumorigenicity assays were performed as previously described with the approval of the Institutional Animal Care and Use Committee.51 In brief, 4 × 106 HCT116 cells and 6 × 106 RKO cells were injected subcutaneously into 7-week-old BALB/c nu/nu male athymic nude mice (Taconic, Germantown, NY, USA). Mice were examined and tumor volumes were measured three times/week for 3–4 weeks after injection. Tumor volume = 1/2 (length × width2).
Statistical analysis
The differences in xenograft tumor growth among groups were analyzed for statistical significance by ANOVA, followed by Student’s t-tests using a GraphPad Prism software (San Diego, CA, USA). All other P-values were obtained using two-tailed Student t-tests.
Abbreviations
- p53 RE:
-
p53 responsive element
- TRIM:
-
tripartite motif
- WT:
-
wild type
- Ub:
-
Ubiquitin
- 5-FU:
-
5-fluorouracil
- ChIP:
-
chromatin immunoprecipitation
- IP:
-
immunoprecipitation
- IB:
-
immunoblotting
- CHX:
-
cyclohexamide
- MEF:
-
mouse embryonic fibroblast
- SA-β-gal:
-
senescence-associated β-galactosidase
References
Vousden KH, Prives C . Blinded by the light: the growing complexity of p53. Cell 2009; 137: 413–431.
Levine AJ, Oren M . The first 30 years of p53: growing ever more complex. Nat Rev Cancer 2009; 9: 749–758.
Vogelstein B, Lane D, Levine AJ . Surfing the p53 network. Nature 2000; 408: 307–310.
Feng Z, Levine AJ . The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends Cell Biol 2010; 20: 427–434.
Brooks CL, Gu W . p53 ubiquitination: Mdm2 and beyond. Mol Cell 2006; 21: 307–315.
Harris SL, Levine AJ . The p53 pathway: positive and negative feedback loops. Oncogene 2005; 24: 2899–2908.
Momand J, Zambetti GP, Olson DC, George D, Levine AJ . The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 1992; 69: 1237–1245.
Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B . Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 1992; 358: 80–83.
Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 2003; 112: 779–791.
Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 2004; 429: 86–92.
Meroni G . Genomics and evolution of the TRIM gene family. Adv Exp Med Biol 2012; 770: 1–9.
Cambiaghi V, Giuliani V, Lombardi S, Marinelli C, Toffalorio F, Pelicci PG . TRIM proteins in cancer. Adv Exp Med Biol 2012; 770: 77–91.
Nicklas S, Otto A, Wu X, Miller P, Stelzer S, Wen Y et al. TRIM32 regulates skeletal muscle stem cell differentiation and is necessary for normal adult muscle regeneration. PloS One 2012; 7: e30445.
Schwamborn JC, Berezikov E, Knoblich JA . The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 2009; 136: 913–925.
Horn EJ, Albor A, Liu Y, El-Hizawi S, Vanderbeek GE, Babcock M et al. RING protein Trim32 associated with skin carcinogenesis has anti-apoptotic and E3-ubiquitin ligase properties. Carcinogenesis 2004; 25: 157–167.
Kano S, Miyajima N, Fukuda S, Hatakeyama S . Tripartite motif protein 32 facilitates cell growth and migration via degradation of Abl-interactor 2. Cancer Res 2008; 68: 5572–5580.
Locke M, Tinsley CL, Benson MA, Blake DJ . TRIM32 is an E3 ubiquitin ligase for dysbindin. Hum Mol Genet 2009; 18: 2344–2358.
Albor A, El-Hizawi S, Horn EJ, Laederich M, Frosk P, Wrogemann K et al. The interaction of Piasy with Trim32, an E3-ubiquitin ligase mutated in limb-girdle muscular dystrophy type 2H, promotes Piasy degradation and regulates UVB-induced keratinocyte apoptosis through NFkappaB. J Biol Chem 2006; 281: 25850–25866.
el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B . Definition of a consensus binding site for p53. Nat Genet 1992; 1: 45–49.
Riley T, Sontag E, Chen P, Levine A . Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 2008; 9: 402–412.
Hoh J, Jin S, Parrado T, Edington J, Levine AJ, Ott J . The p53MH algorithm and its application in detecting p53-responsive genes. Proc Natl Acad Sci USA 2002; 99: 8467–8472.
Hu W, Feng Z, Teresky AK, Levine AJ . p53 regulates maternal reproduction through LIF. Nature 2007; 450: 721–724.
Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z . Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci USA 2010; 107: 7455–7460.
Zhang C, Lin M, Wu R, Wang X, Yang B, Levine AJ et al. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc Natl Acad Sci USA 2011; 108: 16259–16264.
Albertoni M, Shaw PH, Nozaki M, Godard S, Tenan M, Hamou MF et al. Anoxia induces macrophage inhibitory cytokine-1 (MIC-1) in glioblastoma cells independently of p53 and HIF-1. Oncogene 2002; 21: 4212–4219.
Khwaja FW, Svoboda P, Reed M, Pohl J, Pyrzynska B, Van Meir EG . Proteomic identification of the wt-p53-regulated tumor cell secretome. Oncogene 2006; 25: 7650–7661.
Kim E, Gunther W, Yoshizato K, Meissner H, Zapf S, Nusing RM et al. Tumor suppressor p53 inhibits transcriptional activation of invasion gene thromboxane synthase mediated by the proto-oncogenic factor ets-1. Oncogene 2003; 22: 7716–7727.
Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 1998; 282: 1497–1501.
Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O et al. p53 regulates mitochondrial respiration. Science 2006; 312: 1650–1653.
Jung YS, Qian Y, Yan W, Chen X . Pirh2 E3 ubiquitin ligase modulates keratinocyte differentiation through p63. J Invest Dermatol 2013; 133: 1178–1187.
Urist M, Tanaka T, Poyurovsky MV, Prives C . p73 induction after DNA damage is regulated by checkpoint kinases Chk1 and Chk2. Genes Dev 2004; 18: 3041–3054.
Vilgelm AE, Washington MK, Wei J, Chen H, Prassolov VS, Zaika AI . Interactions of the p53 protein family in cellular stress response in gastrointestinal tumors. Mol Cancer Ther 2010; 9: 693–705.
Weisz L, Oren M, Rotter V . Transcription regulation by mutant p53. Oncogene 2007; 26: 2202–2211.
Zhang J, Hu MM, Wang YY, Shu HB . TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J Biol Chem 2012; 287: 28646–28655.
Hock A, Vousden KH . Regulation of the p53 pathway by ubiquitin and related proteins. Int J Biochem Cell Biol 2010; 42: 1618–1621.
Lukashchuk N, Vousden KH . Ubiquitination and degradation of mutant p53. Mol Cell Biol 2007; 27: 8284–8295.
Zheng T, Wang J, Zhao Y, Zhang C, Lin M, Wang X et al. Spliced MDM2 isoforms promote mutant p53 accumulation and gain-of-function in tumorigenesis. Nat Commun 2013; 4: 2996.
Hwang PM, Bunz F, Yu J, Rago C, Chan TA, Murphy MP et al. Ferredoxin reductase affects p53-dependent, 5-fluorouracil-induced apoptosis in colorectal cancer cells. Nat Med 2001; 7: 1111–1117.
Amundson SA, Myers TG, Fornace AJ Jr . Roles for p53 in growth arrest and apoptosis: putting on the brakes after genotoxic stress. Oncogene 1998; 17: 3287–3299.
Elmore LW, Rehder CW, Di X, McChesney PA, Jackson-Cook CK, Gewirtz DA et al. Adriamycin-induced senescence in breast tumor cells involves functional p53 and telomere dysfunction. J Biol Chem 2002; 277: 35509–35515.
Gonzalez-Cano L, Hillje AL, Fuertes-Alvarez S, Marques MM, Blanch A, Ian RW et al. Regulatory feedback loop between TP73 and TRIM32. Cell Death Dis 2013; 4: e704.
Lowe SW, Jacks T, Housman DE, Ruley HE . Abrogation of oncogene-associated apoptosis allows transformation of p53-deficient cells. Proc Natl Acad Sci USA 1994; 91: 2026–2030.
Buzzai M, Jones RG, Amaravadi RK, Lum JJ, DeBerardinis RJ, Zhao F et al. Systemic treatment with the antidiabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer Res 2007; 67: 6745–6752.
Yoon CH, Lee ES, Lim DS, Bae YS . PKR, a p53 target gene, plays a crucial role in the tumor-suppressor function of p53. Proc Natl Acad Sci USA 2009; 106: 7852–7857.
Hu W, Chan CS, Wu R, Zhang C, Sun Y, Song JS et al. Negative regulation of tumor suppressor p53 by microRNA miR-504. Mol Cell 2010; 38: 689–699.
Montes de Oca Luna R, Wagner DS, Lozano G . Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 1995; 378: 203–206.
Jones SN, Roe AE, Donehower LA, Bradley A . Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 1995; 378: 206–208.
Wade M, Li YC, Wahl GM . MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer 2013; 13: 83–96.
Levine AJ, Hu W, Feng Z . The P53 pathway: what questions remain to be explored? Cell Death Differ 2006; 13: 1027–1036.
Chen J . The Roles of MDM2 and MDMX phosphorylation in stress signaling to p53. Genes Cancer 2012; 3: 274–282.
Zhang C, Liu J, Liang Y, Wu R, Zhao Y, Hong X et al. Tumour-associated mutant p53 drives the Warburg effect. Nature Commun 2013; 4: 2935.
Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 1999; 398: 714–718.
Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 2000; 404: 99–103.
Sun XX, Challagundla KB, Dai MS . Positive regulation of p53 stability and activity by the deubiquitinating enzyme Otubain 1. EMBO J 2012; 31 (3): 576–592.
Acknowledgements
We thank Dr. Arnold Levine for very helpful discussion and comments. We thank Dr. Bert Vogelstein for providing p53 WT and deficient HCT116 and RKO cells, Dr. Erwin G Van Meir for providing LN-2024 and LN-Z308 cells. This work was supported by grants from the NIH (R01CA143204) and CINJ Foundation (to ZF), and by the grants from NIH (R01CA160558), DOD (W81XWH-10-1-0435) and Ellison Medical Foundation (to WH). JL was supported by the NJCCR post-doctoral fellowship.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by S Kumar
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Liu, J., Zhang, C., Wang, X. et al. E3 ubiquitin ligase TRIM32 negatively regulates tumor suppressor p53 to promote tumorigenesis. Cell Death Differ 21, 1792–1804 (2014). https://doi.org/10.1038/cdd.2014.121
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2014.121
This article is cited by
-
Deciphering the role of neddylation in tumor microenvironment modulation: common outcome of multiple signaling pathways
Biomarker Research (2024)
-
Gene losses may contribute to subterranean adaptations in naked mole-rat and blind mole-rat
BMC Biology (2022)
-
TRIM family contribute to tumorigenesis, cancer development, and drug resistance
Experimental Hematology & Oncology (2022)
-
Pan-cancer illumination of TRIM gene family reveals immunology regulation and potential therapeutic implications
Human Genomics (2022)
-
TRIM32 promotes radioresistance by disrupting TC45-STAT3 interaction in triple-negative breast cancer
Oncogene (2022)