Abstract
Drosophila Nedd2-like caspase (DRONC), an initiator caspase in Drosophila melanogaster and ortholog of human caspase-9, is cleaved during its activation in vitro and in vivo. We show that, in contrast to conclusions from previous studies, cleavage is neither necessary nor sufficient for DRONC activation. Instead, our data suggest that DRONC is activated by dimerization, a mechanism used by its counterparts in humans. Subsequent cleavage at Glu352 stabilizes the active dimer. Since cleavage is at a Glu residue, it has been proposed that DRONC is a dual Asp- and Glu-specific caspase. We used positional-scanning peptide libraries to define the P1–P4 peptide sequence preferences of DRONC, and show that it is indeed equally active on optimized tetrapeptides containing either Asp or Glu in P1. Furthermore, mutagenesis reveals that Asp and Glu residues are equally tolerated at the primary autoprocessing site of DRONC itself. However, when its specificity is tested on a natural substrate, the Drosophila executioner caspase DRICE, a clear preference for Asp emerges. The formerly proposed Glu preference is thus incorrect. DRONC does not differentiate between Asp and Glu in poor substrates, but prefers Asp when tested on a good substrate.
Similar content being viewed by others
Main
Apoptosis is a highly regulated mechanism used by multicellular organisms to eliminate unneeded or damaged cells. Important in the apoptotic demise of these cells are a family of proteases, the caspases, that initiate and execute the apoptotic program.1 In human cell lines, where caspase mechanisms have been most extensively studied, initiator caspases integrate the several cell death signals that begin the apoptotic program, and pass on the signal by directly activating executioner caspases.2, 3 The activated executioners in turn cleave a number of proteins to drive forward the apoptotic phenotype.
To date, seven caspases have been identified in Drosophila melanogaster. DRONC, Dredd and Strica are predicted to be initiator caspases and Drosophila interleukin-1B-converting enzyme (DRICE), death caspase-1 (DCP-1), DAMM and DECAY are predicted to be executioner caspases.4 Like the mammalian initiator caspase-9, DRONC possesses a long prodomain that includes a caspase recruitment domain (CARD). In human caspase-9, the CARD allows recruitment of the caspase zymogen to its activator apoptosis protease-activating factor-1 (Apaf-1), which serves as the caspase-9 activator, or apoptosome.5 Similarly, the CARD of DRONC is thought to direct this initiator caspase to Drosophila Apaf-1-related killer (DARK), the Drosophila counterpart of Apaf-1, forming the Drosophila apoptosome.6, 7
Despite the evident similarities between the human and Drosophila apoptosomes both in composition and structure,8, 9 there are important distinctions between them. Specifically, formation of the Drosophila apoptosome is independent of cytochrome c, at least in some cells.10, 11, 12 Moreover, unlike human caspase-9, which is activated by dimerization within the apoptosome,13 DRONC is reported to require proteolytic autoprocessing for activation,14 although this has recently been disputed.15 Finally, in normal living S2 cells, DRONC is processed between the large and small subunits at Glu352, implying that DRONC can cleave C-terminally to a Glu residue.14, 16 The requirement for autoprocessing and specificity for Glu residues thus places DRONC in a separate category mechanistically from its human ortholog caspase-9, and thereby violates the principle of conservation of mechanism.
To test the hypothesis that DRONC requires autoprocessing for its activation, we obtained uncleavable mutants, analyzed their mechanism of activation and used a specific protease that cleaves within the inter-chain linker segment that defines the autoprocessing site. We also evaluated the cleavage specificity of DRONC by using positional-scanning peptide libraries, and importantly, also targeted mutational analysis of its natural substrate, the Drospohila executioner caspase DRICE.17
Results
Activation mechanism of DRONC
We first sought to determine which of the two mechanisms of activation, dimerization or cleavage, is required for DRONC activation. Full-length DRONC containing a C-terminal His6 tag was expressed in Escherichia coli and purified essentially as described18 (Figure 1). Material was recovered as the two-chain form, and Edman degradation and mass spectrometry analysis demonstrated the expected cleavage at Glu352 to separate the large and small subunits of the catalytic domain, as well as cleavage at Asp135 to remove the CARD. As previously demonstrated, E. coli expression of the Glu352Ala mutant resulted in material that was primarily full length when expressed in E. coli,14 although there was partial CARD removal (Figure 1b).
Constructs and proteins used in the study. (a) DRONC is composed of two domains, the CARD and the catalytic domain. The catalytic domain itself is composed of a large and a small subunit joined by the inter-domain linker segment that is a substrate for autolytic cleavage. (b) Expression of full-length DRONC in E. coli leads to processing that removes the CARD and generates large and small subunits of the catalytic domain. MALDI-TOF and N-terminal sequence analysis revealed that the double bands corresponding to the wt DRONC large subunit are a mixture of cleavage at Glu343 and Glu352. Mutation at Glu352 produces a single-chain enzyme that removes its CARD and undergoes processing at Glu344 when incubated in 1.0 M NaCit. The triple Glu/Ala mutant has a small amount of CARD removal, but is prevented from further autoprocessing in the presence of 1.0 M NaCit. FL, full length; ΔC, CARD removed; LS, large subunit; SS, small subunit
Human initiator caspases can be activated by high concentrations of sodium citrate (NaCit), probably via dimerization.19 We tested NaCit for its ability to activate DRONC and found that the extent of activation depended on both the concentration of DRONC and that of NaCit (data not shown). Maximum activation was at 1.4 M NaCit, and DRONC Glu352Ala activity was enhanced by up to 1000-fold. However, for some experiments we were not able to use 1.4 M NaCit due to protein concentration issues, and so in this study we used 0.7, 1.0 or 1.4 M NaCit as appropriate for the experimental conditions. Each concentration substantially activated DRONC.
We incubated the Glu352Ala mutant for 30 min at 37°C with 1.0 M NaCit, which resulted in complete processing within the inter-chain linker and quantitative removal of the CARD (Figure 1b). Sequence analysis and mass spectroscopy revealed that the site of cleavage had been shifted to Glu344. To prevent any cleavage in the linker, we mutated the Glu residues at positions 343, 344 and 352 to Ala, and expressed and purified the material from E. coli. When the triple mutant was incubated with 1.0 M NaCit, as before no inter-chain cleavage could be observed (Figure 1b).
Now that we had intact full-length material that could not be processed, we tested whether active DRONC is a monomer or dimer, and whether it can be activated by proteolysis. When analyzed by size exclusion on Superose 200, two-chain DRONC eluted with an apparent size of 75.6 kDa, which, given that the monomeric size of this material that has lost its CARD is about 38 kDa, is consistent with the size of a dimer (Figure 2a). We tested a number of commercially available caspase substrates and found that acetyl-Ile–Glu–Thr–Asp–7-amino-4-trifluoromethylcoumarin (Ac-IETD-AFC) was the most effective for measuring DRONC. Interestingly, the bulk of activity against Ac-IETD-AFC was associated with fractions corresponding to the leading edge of the peak when measured in caspase buffer (see Materials and Methods), but the activity mirrored the main part of the protein peak when measured in buffer containing 1.0 M NaCit. The Glu352Ala mutant eluted as a bimodal peak, with the first part of the peak at 70.8 kDa corresponding to material that was full length, and a shoulder at 40 kDa corresponds to material that had lost its CARD (Figure 2b). The elution profile of the 40-kDa CARD-deleted material is consistent with that of a monomer (predicted size 45 kDa), and the 70.8-kDa elution position of the full-length material is between a monomer (predicted size 48 kDa) and a dimer (predicted size 96 kDa). It most likely represents a monomer, and the larger than expected size in size-exclusion chromatography is probably the result of an asymmetric Stoke's radius due to the non-ideal behavior of a protein composed of two globular domains (caspase domain plus CARD). Consequently, in contrast to the two-chain form, the majority of single-chain protein, whether on not it contained a CARD, appeared to be monomeric. Again, the bulk of activity associated with fractions corresponding to the leading edge of the peak when measured in caspase buffer, but the activity mirrored the main part of the protein peak when measured in NaCit.
Size-exclusion chromatography of wt DRONC and DRONC Glu352Ala mutant. Purified wt DRONC (11 μM) (a) or DRONC Glu352Ala mutant (b) were analyzed by size exclusion on Superose 200 (upper traces). Fractions were diluted twofold into caspase buffer (black bars) or 1.0 M NaCit buffer (white bars) and assayed on Ac-IETD-AFC (middle panels). Samples from each fraction were also analyzed by SDS-PAGE (lower panels). Activity is expressed as RFU/min, relative fluorescence units per minute
From this we conclude that, when expressed and purified from E. coli, the two-chain form of DRONC (cleaved at Glu352 in the inter-chain linker) has lost its CARD and is primarily dimeric. In contrast, the single-chain form (mutated from Glu to Ala at residue 352) is primarily monomeric, irrespective of whether it retains the CARD. Thus, cleavage in the inter-domain region stabilizes dimer formation and this dimerization is apparently independent of the CARD.
Cleavage does not activate DRONC
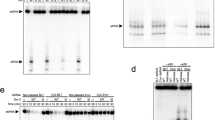
Because two-chain DRONC demonstrated higher activity than the single-chain enzyme, we investigated whether simply cleaving the linker region of a single-chain form was the mechanism of activation. For this we utilized the Glu352Ala mutant to obtain single-chain material, and analyzed several endopeptidases that could in principle cleave in the linker region to generate the two-chain form. Of those tested, GluC protease from Streptomyces griseus, commonly used in protein fragmentation strategies, cleaved primarily in the selected region, specifically at Glu344, the same position cleaved during DRONC-mediated auto-cleavage of the Glu352Ala mutant (Figure 3a). GluC also removed the CARD, and therefore its activity essentially mimicked NaCit-mediated auto-proteolysis of the Glu352Ala mutant. However, we observed no increase in activity above background following cleavage, and in comparison 1.4 M NaCit activated this material about 50-fold (Figure 3b). We can therefore conclude that cleavage in the inter-chain linker segment is, in itself, insufficient to activate DRONC.
Endoproteinase Glu-C cleavage of DRONC Glu352Ala. DRONC (4 μM) Glu352Ala was cleaved with the indicated range of endoproteinase Glu-C concentrations and activity assessed on Ac-IETD-AFC. Samples were analyzed by SDS-PAGE (a) and the activity of DRONC Glu352Ala from which the background of Glu-C activity has been subtracted was determined (b). The NaCit control represents maximum activity in the presence of 1.4 M NaCit. (c) Upon completion of Glu-C cleavage, one-tenth of each sample was assayed again in the presence of 1.4 M NaCit to determine whether endoproteinase Glu-C inactivates DRONC. The result reveals that endoproteinase Glu-C above 0.24 μg inactivate DRONC, but below this there is little inactivation but almost complete processing of DRONC. The fraction containing 0.03 μg of Glu-C was analyzed by MALDI/TOF, which identified fragments corresponding to the CARD, and large and small subunits of DRONC Glu352Ala, cleaved at Glu344
Activity of single-chain versus two-chain DRONC
The activity of two-chain DRONC increased by about fourfold, and that of the Glu352Ala single-chain form by about 32-fold, when measured using Ac-IETD-AFC in buffer containing 1.0 M NaCit (Figure 2). However, Ac-IETD-AFC was a relatively poor substrate for all three forms of DRONC, and we decided to identify a better substrate that would allow more accurate comparison of the activities of the differentially processed forms of the enzyme. Because DRONC has been reported to possess activity against Asp and Glu in P1, we first determined the preferred P1 residue by positional-scanning (Figure 4a). DRONC and human caspase-9 were assayed for activity on 18 individual substrate pools containing P1 residues fixed as one of the 20 natural amino acids (with the absence of Cys and Met), and each pool contains a mixture of these 18 amino acids in the P2, P3 and P4 positions. This allowed us to determine the optimal P1 position for each protease, and we found that DRONC had appreciable activity in pools containing a P1 Glu (as did caspase-9), but substantially more activity on pools containing a P1 Asp, indicating a P1 preference for Asp.
Substrate specificity of wt DRONC. (a) wt DRONC was added to a final concentration 26 μM in 0.7 M NaCit buffer pH 7.4 incubated at 37°C for 1 h and assayed for 2 h on a fluorimeter using excitation 320 emission 460 filter pairs. Caspase-9 was added to a final concentration of 7 μM in 0.7 M NaCit buffer pH 7.4 incubated at 37°C for 30 min and assayed for 15 min. Recombinant wt DRONC and delta CARD caspase-9 were assayed for activity on a P1 substrate scanning library, where P1 is fixed and P2, P3 and P4 are a mixture of 18 amino acids. (b) wt DRONC was incubated at a final concentration of 19 μM in the presence of 1.4 M NaCit and assayed with a positional-scanning substrate library with P1 fixed at aspartic acid. The y-axis represents the hydrolysis rate presented as a percentage of the maximal rate observed. The x-axis provides the positionally defined L-amino acid (single-letter code, d is D-alanine)
We next optimized the P4–P2 preference by using a second positional-scanning library where these positions are varied, with P1 fixed at Asp, as previously described.19 DRONC showed surprisingly little selectivity in P2 and P4, although there was some preference for hydrophobes in P4, but with a stricter preference in P2 for hydrophobes (Figure 4b). On the basis of these two libraries, the sequence Leu–Ala–Leu–Asp (P4–P1) was judged to represent the optimal substrate. This result is very close to a similar approach used by Hawkins et al.16 who determined the optimal P2–P4 sequence as Thr–Ala–Thr, and inspection of Figure 4b reveals that this sequence is also close to optimal.
Armed with this information, we synthesized two substrates, Ac-LALD-AFC and Ac-LALE-AFC, with which we characterized the forms of DRONC. Even though we used the optimal tetrapeptide sequence, we could not achieve saturating conditions for any form of DRONC with Ac-LALD-AFC, preventing us from determining individual kcat and KM values. Consequently, we used the linear portion of a substrate/velocity plot to determine the ratio of kcat/KM, which defines the specificity of each form for the substrate (see Materials and Methods; Equation (1)). Although we could determine kcat/KM for wild-type (wt) DRONC in caspase buffer, we were unable to determine any reliable values for the Glu352Ala and E343/344/352A mutants in the absence of NaCit (Table 1). However, in the presence of 1.4 M NaCit, we obtained catalytic parameters on both substrates within a factor of 3 for all forms of DRONC. Interestingly, we obtained almost identical kcat/KM values for Ac-LALD-AFC and Ac-LALE-AFC, indicating that in the context of the optimal P4–P2 subsite occupancies, no forms of DRONC could distinguish between Asp or Glu in P1, at least by using these tetrapeptide substrates.
The kcat/KM values we determined were surprisingly low, with no form of DRONC exceeding 5.5 M−1 s−1(Table 1). These rates are of the orders of magnitude below the kcat/KM values of other caspases on their optimal peptidyl substrates. For example, caspase-3 has a kcat/KM of 1.3 × 106 M−1 s−1 on Ac-DEVD-pNA,20 and caspase-9 has a kcat/KM of 1.9 × 104 M−1 s−1 on Ac-LEHD-AFC.21 Clearly, we had to employ a better substrate with which to explore the specificity and activity of DRONC, and to this end, we chose to work with the natural substrate DRICE, an executioner caspase and downstream target of DRONC.16, 17
Kinetics and specificity of DRICE cleavage by DRONC and DRONC autoproteolysis
DRICE is cleaved at Asp230 during its activation by DRONC,14, 16 and DRONC cleaves itself at Glu352 during its autoactivation, and so clearly in the context of natural protein substrates, DRONC can cleave after Asp or Glu. To determine how efficient DRONC is in cleaving a natural (protein) versus synthetic (tetrapeptide) substrate, we incubated a constant concentration of the DRICE precursor with increasing concentrations of DRONC. The DRICE precursor contained a Cys211Ala catalytic mutation to prevent autoprocessing during expression in E. coli. We also replaced the natural Asp230 cleavage site with Glu in the background of the DRICE Cys211Ala mutant, and we compared the efficiency of DRICE processing of the wild type and the Glu mutant. The objective was to determine whether DRONC preferred Asp or Glu in P1 in the context of this natural substrate. Figure 5 reveals that a lower concentration of DRONC is required to cleave wt DRICE than the Asp230Glu mutant, indicating that Asp at position 230 is preferred over Glu. To quantitate the difference we determined the concentration of DRONC required to cleave 50% of the DRICE precursor, and applied this concentration to equation (2) described under Materials and Methods, allowing us to determine the kcat/KM value for each protein substrate as previously described.18 We were not able to determine cleavage of DRICE by NaCit-treated DRONC, since high concentrations of the salt tended to precipitate DRICE, but wt two-chain DRONC in caspase buffer cleaved wt DRICE with kcat/KM of 1.3 × 103 M−1 s−1 and the Asp230Glu mutant with kcat/KM of 1.6 × 102 M−1 s−1 (Table 1). Consequently, in the context of a natural substrate, DRONC prefers Asp to Glu by a factor of about 8.
DRONC prefers Asp over Glu in the context of a natural substrate. Catalytic Cys/Ala mutants of wt DRICE (a) and an Asp230Glu mutant (b) were expressed and purified from E. coli and used as substrates for cleavage by wt DRONC in caspase buffer. A standard concentration of DRICE wt or mutant (0.85 μM) was incubated with increasing concentrations (125 nM to12.5 μM) of DRONC (left to right) for 90 min at 37°C. The arrow indicates the concentration of DRONC required to cleave 50% of the respective DRICE precursor
In a related experiment, we analyzed the cleavage of a protein substrate that naturally contains a Glu at the DRONC cleavage site – DRONC itself. Because cleavage of DRONC by itself is very inefficient in vitro, we compared autoprocessing during expression in E. coli, where very high protein levels promote autoproteolysis. Figure 6 reveals that wt DRONC (Glu352) is processed to roughly the same extent as the Glu352Asp mutant. Thus, in the context of a natural, but rather poor substrate, there is no substantial preference of DRONC for Glu or Asp.
Specificity of DRONC autolytic processing. (a) Recombinant DRONC Glu352Asp and wt DRONC with a C-terminal His tag were expressed in E. coli. After induction with 0.1 mM IPTG, samples were taken from the culture at the indicated induction times. E. coli lysates from each time point were then analyzed by western blotting for the presence of a His tag. Cleavage of full-length DRONC (FL) to generate the small subunit (SS) at 11 kDa indicates cleavage at residue 352. (b) Quantitative image analysis of the same samples using a CCD camera from the developing PVDF membrane; •, wt and ▴, Glu352Asp mutant. Cleavage is expressed as fractional conversion of the immunoreactivity in the SS divided by the sum of total immunoreactivity in the FL plus SS
Discussion
The general mechanism of caspase activation requires transition of loops carrying the catalytic and specificity-determining residues from and exposed to a constrained conformation, allowing the enzyme to accept substrate and enhance catalysis.2, 3 This is achieved for the human initiator caspase-8 and caspase-9 by dimerization, and for the human executioner caspase-3 and caspase-7 by cleavage in the inter-chain linker of already assembled dimers.22 The distinction in mechanism is presumably due to the position of a caspase in a pathway, such that executioners are downstream of initiators, and therefore can be activated through proteolysis by the initiator, but initiators have no protease above them, and have to employ a non-proteolytic mechanism for their activation. Consequently, it came as somewhat of a surprise that the Drosophila initiator caspase DRONC, the ortholog of human caspase-9, was reported to be activated by proteolysis,14 since this would enforce a revision of the general mechanism of initiator caspase activation. We demonstrate that direct cleavage of single-chain DRONC, using endoprotease GluC, does not activate the enzyme. In contrast, incubation of the same protein, Glu352Ala DRONC, in 1.0 M NaCit activated the enzyme by more than two orders of magnitude. Since NaCit activates the human initiator caspase-8 and caspase-9 by dimerization,19 with a similar gain in activity, it is likely that the same mechanism is in play for DRONC. Our data support recent work questioning the role of autoproteolysis in DRONC activation.15
Cleavage in the inter-chain linker clearly has a role to play in the regulation of DRONC activity. In caspase buffer, single-chain DRONC has almost no activity on synthetic substrates when compared with the two-chain enzyme, and it was this observation that led to the suggestion that cleavage in the inter-chain linker was the mechanism of activation.14 However, the two-chain protein is dimeric, but the single-chain protein is monomeric, and it is this difference that accounts for the increase in activity. The simplest explanation is that cleavage in the inter-chain linker stabilizes dimerization of DRONC, very much as is seen with the human initiator caspase-8.23, 24 But first the protein must be dimerized, which can be achieved in vitro with high concentrations of NaCit, and presumably in vivo following recruitment to the Drosophila apoptosome.9 This scenario is fully consistent with the process that activates the human initiator caspase-9, occurring through apoptosome-driven dimerization of monomers to induce the active conformation of this ortholog of DRONC.13
A property of DRONC that distinguishes it from other caspases is its acceptance of Glu in the primary specificity pocket P1. This has been demonstrated previously on synthetic peptidyl substrates,16 and also in terms of its autolytic cleavage and cleavage of Drosophila IAP1.14, 16, 25 Interestingly, as shown in Figure 4a, caspase-9 also tolerates Glu in its primary specificity pocket, and can cleave its own inter-chain segment following a Glu residue, although this cleavage is subservient to the natural Asp cleavage sites in the caspase-9 linker.26, 27 We demonstrate, using a matched set of substrates, that DRONC may have almost equal activity on tetrapeptides containing Asp and Glu in P1, but this does not hold for a downstream substrate. DRONC cleaves itself with approximately equal preference for Asp and Glu, but for the natural substrate DRICE, Asp is substantially favored over Glu in P1. This apparent contradiction relates to the absolute values of the second-order rate of proteolysis, kcat/KM, for it appears that in the context of an optimal tetrapeptide substrate (Ac-LALD-AFC), or autoproteolysis, there is little discrimination at P1, but in the context of a good protein substrate (DRICE), discrimination between Asp and Glu is readily apparent.
The concept that DRONC is equally active on Asp and Glu substrates is inaccurate because in the context of a substrate that is cleaved more rapidly (DRICE), Asp dominates the P1 preference. But why is DRONC such a poor enzyme on itself, and also on tetrapeptides compared with a downstream natural protein substrate? After all, LALD is the optimal tetrapeptide sequence in our hands, very similar to the TATD sequence found previously by a similar combinatorial technique,16 in which study it was shown that DRONC had kcat/KM values of 2.73 M−1 s−1 for Ac-TQTE-AFC and 3.36 M−1 s−1 for Ac-TQTD-AFC. When another residue is added to the P5 position, DRONC catalysis increases but only by twofold,16 suggesting that the enhancement in catalysis on DRICE is largely due to interactions outside of the catalytic cleft. Such ‘exosite’ interactions are important in enhancing the catalysis of inherently poor enzymes, including metalloproteases28 and coagulation proteases.29, 30
Since DRONC has no intrinsic preference for Glu over Asp, why is the primary autolytic cleavage site at position 352 conserved as Glu in both D. melanogaster and Drosophila pseudoobscura? To answer this will require some sophisticated experiments, but it could simply be to prevent other caspases, such as DCP-1 and DRICE with the canonical Asp specificity,16 from adventitiously cleaving DRONC. Such cleavage may be associated with premature removal of DRONC via the ubiquitin system before its activation.31 Our profiling of DRONC's primary substrate specificity, and demonstration that cleavage of DRONC is neither necessary nor sufficient for its activation, are, of necessity, founded on in vitro observations. Assembling a Drosophila apoptosome that efficiently cleaves peptide substrates is not trivial. Indeed, in a recent paper,15 the only assay possible for ARK activated DRONC was to amplify the signal and measure downstream DEVD-ase activity. Unfortunately this cannot tell us about DRONC specificity on synthetic substrates because the signal needed executioner caspase activation.
On the basis of our observations, one could envision different scenarios for how DRONC activity and specificity are enhanced in vivo. Although cleavage does not activate DRONC, our data suggest that the stability of the active dimer is substantially enhanced by cleavage. This may be important if DRONC dissociates from the apoptosome following dimerization and cleavage, where it would be stable for long enough to cleave good targets such as DRICE. Indeed, stabilization may explain why, although cleavage of DRONC is not required for activity in vitro, it may be required for cell death in vivo.14 Association of DRONC and DARK may result in an improved ability to bind non-optimal substrates, such as DRONC itself and DCP-1 (the other proposed executioner caspase targeted by DRONC). When compared with DRICE, DCP-1 is reportedly a much poorer substrate of recombinant DRONC,16 and may require exosite interactions provided by the full apoptosome. Obviously, a combination of these options could be envisioned, but our data firmly suggest that the activity of DRONC is not only dependent on the formation of a correct dimer, but perhaps to a greater extent on interactions distant from the active site.
Materials and Methods
Materials
Ac-IETD-AFC was from MP Biomedicals (Irvine, CA, USA). The vector pET23b was from EMD Biosciences (San Diego, CA, USA). E. coli strain BL21pLysS(DE3) and QuickChange site-directed mutagenesis kit were from Stratagene (La Jolla, CA, USA). Caspase buffer contains 20 mM Pipes, 10% sucrose, 100 mM NaCl, 0.1% CHAPS, 1 mM EDTA and 10 mM dithiothreitol (DTT), pH 7.2. NaCit buffer contains 50 mM Tris, 100 mM sodium chloride, pH 7.4, containing NaCit in the range of 0.7 to 1.4 M, depending on experimental conditions.
Proteins
The plasmid pET23b encoding Drosophila wt DRONC and Glu352Ala DRONC plus a C-terminal His6 purification tag was a generous gift from Dr Rollie Clem. These plasmids were transformed into E. coli strain BL21pLysS(DE3) expressed and purified by a Ni-chelate affinity method.18 Expressed proteins were tested within 2 days of purification. QuickChange site-directed mutagenesis kits (Stratagene) were used to substitute Glu343 and Glu344 of DRONC with an Ala residue, and to create a Glu352Asp mutant.
The cDNA encoding Drosophila wt DRICE plus a C-terminal His6 purification tag was a generous gift from Dr Pascal Meier. This cDNA was inserted via NdeI/XhoI into pET23b, transformed into E. coli strain BL21pLysS(DE3) expressed and purified by a Ni-chelate affinity method.18 Expressed proteins were tested within 2 days of purification. QuickChange site-directed mutagenesis kit (Stratagene) was used to generate Glu230Asp and Cys211Ala mutants of DRICE. All plasmids were sequenced to verify the success of the mutagenesis and the absence of other mutations.
Positional-scanning substrate libraries
We employed two positional-scanning libraries to define the P1–P4 subsite preferences of DRONC. The first library was used to define the preferred P1, and contained 18 pools in which the P1 position is fixed as one of the 18 amino acids contained in proteins (with the exception of Met and Cys). The P2, P3 and P4 positions are evenly represented by these 18 amino acids in each pool.32 This fluorogenic 7-amino-4-methylcoumarin (AMC) peptide substrate library was synthesized by solid-phase chemistry on an Fmoc-AMC resin essentially as previously described.33 Selected peptide pools were subjected to amino-acid analysis to verify library composition and concentration, and the concentration of each remaining peptide pools was determined by absorbance at 320 nm. All reactions were carried out in 100 μl in a 96-well plate containing and substrate library concentration in the range 5–10 μM at 37°C, and analyzed with excitation at 320 nm and emission at 460 nm.
The second library was used to define the P4, P3 and P2 subsite preferences of DRONC, and we employed a positional-scanning substrate library with the general structure Ac-OXX-Asp-AMC for P4 scanning, Ac-XOX-Asp-AMC for P3 scanning and Ac-XXO-Asp-AMC for P2 scanning (where O is the positionally specified residue and X signifies the mixture of the 20 natural amino acids omitting Cys, and including D-Ala). Standard peptide synthesis was used to prepare the library, as previously described.19 The variable amino-acid pools were prepared as an isokinetic mixture with all 20 amino acids.
Synthesis of the Ac-LALD-AFC and Ac-LALE-AFC
Fmoc-Asp(OtBu)-AFC and Fmoc-Glu(OtBu)-AFC were synthesized as described34 and subsequently the Fmoc group was deprotected using four equivalents of diethylamine in N,N′-dimethylformamide for 1 h. Ac-LAL-COOH was synthesized by classic solution-phase peptide methods, and coupled to the deprotected Asp- or Glu-AFC.
Enzyme kinetics
Assays using fluorogenic AFC substrates (excitation wavelength 405 nm and emission wavelength 510 nm) were carried out on an fMax Fluorescence Microplate reader (Molecular Devices) operating in the kinetic mode. Reactions of 100 μl contained enzyme and substrate (final concentration 100 μM) in appropriate buffer. To determine the rates of synthetic substrate cleavage, we measured the release of the AFC fluorophore continuously using an fMax Fluorescence Microplate reader (Molecular Devices) operating in the kinetic mode. To determine the catalytic efficiency of the enzyme, initial velocities were measured as a function of substrate [S0]. When [S0]≪KM, the plot of velocity versus [S0] is a straight line with a slope of Vmax/KM. The kcat/KM values were calculated using the following expression.

To determine the rate of DRICE cleavage by DRONC, we calculated the enzyme concentration [E] required to decrease 50% (EC50) of precursor in time t from the respective SDS gels, and the catalytic efficiency was expressed as kobs. When the reaction is carried out below KM for protein substrate (which is probably true for the cases examined in our study), kobs becomes the kinetic constant kcat/KM.35

Enzyme concentrations [E] were calculated on the basis of total protein concentration estimated from the absorbance at 280 nm.36
SDS-PAGE and N-terminal sequencing
An 8–18% linear acrylamide gradient SDS gel run in a 2-amino-2-methyl-1,3-proanediol/glycine/HCL system was used for resolving proteins.37 Samples were boiled in SDS sample buffer containing 50 mM DTT for 5 min and then loaded on the stacking gel. For N-terminal sequencing, protein samples were resolved by SDS-PAGE and transferred to an Immobilon-P membrane (Millipore, Bedford, MA, USA) by electroblotting. The membrane was briefly stained with Coomassie Brilliant Blue R-250, destained and washed with water. Appropriate bands were excised and sequenced by Edman degradation on a 492 protein sequencer (Applied Biosystems, Foster City, CA, USA).38
Size-exclusion chromatography
Experiments were performed using a Pharmacia AKTA purifier system equipped with a Superdex 200 HR 10/30 column (Amersham Pharmacia Biotech) with a flow rate of 0.4 ml/min, and 0.4 ml fractions were collected for analysis. The buffer used for size-exclusion chromatography was 20 mM Tris (pH8.0)/100 mM NaCl. The column was calibrated using protein standards from Bio-Rad.
Mass spectrometry
Mass spectra of Endoproteinase Glu-C-digested fragments were collected using an Applied Biosystems Voyager DE Pro MALDI-TOF instrument with the help of the Burnham Institute Proteomics Facility.
Abbreviations
- AFC:
-
amino-4-trifluoromethylcoumarin
- AMC:
-
7-amino-4-methylcoumarin
- Apaf-1:
-
apoptosis protease-activating factor-1
- CARD:
-
caspase recruitment domain
- DARK:
-
Drosophila Apaf-1-related killer
- DCP-1:
-
death caspase-1
- DRICE:
-
Drosophila interleukin-1B-converting enzyme
- DRONC:
-
Drosophila Nedd2-like caspase
- NaCit:
-
sodium citrate
References
Kumar S . Caspase function in programmed cell death. Cell Death Differ 2007; 14: 32–43.
Fuentes-Prior P, Salvesen GS . The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J 2004; 384: 201–232.
Riedl SJ, Shi Y . Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol 2004; 5: 897–907.
Hay BA, Guo M . Caspase-dependent cell death in Drosophila. Annu Rev Cell Dev Biol 2006; 22: 623–650.
Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES et al. Cytochrome c and dATP-Dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 1997; 91: 479–489.
Rodriguez A, Oliver H, Zou H, Chen P, Wang X, Abrams JM . Dark is a Drosophila homologue of Apaf-1/CED-4 and functions in an evolutionarily conserved death pathway. Nat Cell Biol 1999; 1: 272–279.
Quinn LM, Dorstyn L, Mills K, Colussi PA, Chen P, Coombe M et al. An essential role for the caspase dronc in developmentally programmed cell death in Drosophila. J Biol Chem 2000; 275: 40416–40424.
Yu X, Acehan D, Ménétret J-F, Booth CR, Ludtke SJ, Riedl SJ et al. A structure of the human apoptosome at 12.8 Å resolution provides insights into this cell death platform. Structure 2005; 13: 1725–1735.
Yu X, Wang L, Acehan D, Wang X, Akey CW . Three-dimensional structure of a double apoptosome formed by the Drosophila Apaf-1 related killer. J Mol Biol 2006; 355: 577–589.
Dorstyn L, Read S, Cakouros D, Huh JR, Hay BA, Kumar S . The role of cytochrome c in caspase activation in Drosophila melanogaster cells. J Cell Biol 2002; 156: 1089–1098.
Zimmermann KC, Ricci JE, Droin NM, Green DR . The role of ARK in stress-induced apoptosis in Drosophila cells. J Cell Biol 2002; 156: 1077–1087.
Arama E, Agapite J, Steller H . Caspase activity and a specific cytochrome c are required for sperm differentiation in Drosophila. Dev Cell 2003; 4: 687–697.
Pop C, Timmer J, Sperandio S, Salvesen GS . The apoptosome activates caspase-9 by dimerization. Mol Cell 2006; 22: 269–275.
Muro I, Monser K, Clem RJ . Mechanism of Dronc activation in Drosophila cells. J Cell Sci 2004; 117: 5035–5041.
Dorstyn L, Kumar S . A biochemical analysis of the activation of the Drosophila caspase DRONC. Cell Death Differ 2007; e-pub ahead of print.
Hawkins CJ, Yoo SJ, Peterson EP, Wang SL, Vernooy SY, Hay BA . The Drosophila caspase DRONC cleaves following glutamate or aspartate and is regulated by DIAP1, HID, and GRIM. J Biol Chem 2000; 275: 27084–27093.
Fraser AG, McCarthy NJ, Evan GI . drICE is an essential caspase required for apoptotic activity in Drosophila cells. EMBO J 1997; 16: 6192–6199.
Stennicke HR, Salvesen GS . Caspases: preparation and characterization. Methods 1999; 17: 313–319.
Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen I et al. A unified model for apical caspase activation. Mol Cell 2003; 11: 529–541.
Stennicke HR, Renatus M, Meldal M, Salvesen GS . Internally quenched fluorescent peptide substrates disclose the subsite preferences of human caspases 1, 3, 6, 7 and 8. Biochem J 2000; 350: 563–568.
Renatus M, Stennicke HR, Scott FL, Liddington RC, Salvesen GS . Dimer formation drives the activation of the cell death protease caspase 9. Proc Natl Acad Sci USA 2001; 98: 14250–14255.
Boatright KM, Salvesen GS . Mechanisms of caspase activation. Curr Opin Cell Biol 2003; 15: 725–731.
Donepudi M, Mac Sweeney A, Briand C, Gruetter MG . Insights into the regulatory mechanism for caspase-8 activation. Mol Cell 2003; 11: 543–549.
Pop C, Fitzgerald P, Green DR, Salvesen GS . Role of proteolysis in caspase-8 activation and stabilization. Biochemistry 2007; 46: 4398–4407.
Muro I, Means JC, Clem RJ . Cleavage of the apoptosis inhibitor DIAP1 by the apical caspase DRONC in both normal and apoptotic Drosophila cells. J Biol Chem 2005; 280: 18683–18688.
Stennicke HR, Deveraux QL, Humke EW, Reed JC, Dixit VM, Salvesen GS . Caspase-9 can be activated without proteolytic processing. J Biol Chem 1999; 274: 8359–8362.
Sadhukhan R, Leone JW, Lull J, Wang Z, Kletzien RF, Heinrikson RL et al. An efficient method to express and refold a truncated human procaspase-9: a caspase with activity toward Glu–X bonds. Protein Expr Purif 2006; 46: 299–308.
Overall CM . Molecular determinants of metalloproteinase substrate specificity: matrix metalloproteinase substrate binding domains, modules, and exosites. Mol Biotechnol 2002; 22: 51–86.
Krishnaswamy S . Exosite-driven substrate specificity and function in coagulation. J Thromb Haemost 2005; 3: 54–67.
Larsen KS, Ostergaard H, Bjelke JR, Olsen OH, Rasmussen HB, Christensen L et al. Engineering the substrate and inhibitor specificities of human coagulation factor VIIa. Biochem J 2007; 405: 429–438.
Muro I, Hay BA, Clem RJ . The Drosophila DIAP1 protein is required to prevent accumulation of a continuously generated, processed form of the apical caspase DRONC. J Biol Chem 2002; 277: 49644–49650.
Snipas SJ, Wildfang E, Nazif T, Christensen L, Boatright KM, Bogyo M et al. Characteristics of the caspase-like catalytic domain of human paracaspase. Biol Chem 2004; 385: 1093–1098.
Maly DJ, Leonetti F, Backes BJ, Dauber DS, Harris JL, Craik CS et al. Expedient solid-phase synthesis of fluorogenic protease substrates using the 7-amino-4-carbamoylmethylcoumarin (ACC) fluorophore. J Org Chem 2002; 67: 910–915.
Alves LC, Almeida PC, Franzoni L, Juliano L, Juliano MA . Synthesis of N alpha-protected aminoacyl 7-amino-4-methyl-coumarin amide by phosphorous oxychloride and preparation of specific fluorogenic substrates for papain. Pept Res 1996; 9: 92–96.
Stennicke HR, Salvesen GS . Caspase assays. Methods Enzymol 2000; 322: 91–100.
Edelhoch H . Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry 1967; 6: 1948–1954.
Bury A . Analysis of protein and peptide mixtures: evaluation of three sodium dodecyl sulphate-polyacrylamide gel electrophoresis buffer systems. J Chromatogr 1981; 213: 491–500.
Matsudaira P . Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem 1987; 262: 10035–10038.
Acknowledgements
We thank Drs Rollie Clem and Pascal Meier for generous gifts of plasmids, and members of the Salvesen laboratory for helpful discussions. This work was supported by NIH grants CA-69381 and AG-15402.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by E Baehrecke
Rights and permissions
About this article
Cite this article
Snipas, S., Drag, M., Stennicke, H. et al. Activation mechanism and substrate specificity of the Drosophila initiator caspase DRONC. Cell Death Differ 15, 938–945 (2008). https://doi.org/10.1038/cdd.2008.23
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2008.23
Keywords
This article is cited by
-
Tango7 regulates cortical activity of caspases during reaper-triggered changes in tissue elasticity
Nature Communications (2017)
-
Highly sensitive and adaptable fluorescence-quenched pair discloses the substrate specificity profiles in diverse protease families
Scientific Reports (2017)
-
Cacidases: caspases can cleave after aspartate, glutamate and phosphoserine residues
Cell Death & Differentiation (2016)
-
A comprehensive characterization of the caspase gene family in insects from the order Lepidoptera
BMC Genomics (2011)
-
The role of IAP antagonist proteins in the core apoptosis pathway of the mosquito disease vector Aedes aegypti
Apoptosis (2011)