Abstract
Background:
VEPH1 is amplified in several cancers including ovarian but its impact on tumour progression is unknown. Previous work has shown that VEPH1 inhibits TGFβ signalling while its Drosophila ortholog increases tissue growth, raising the possibility that VEPH1 could impact tumour growth or progression.
Methods:
A CRISPR approach was used to disrupt VEPH1 expression in ovarian cancer ES-2 cells, while VEPH1-negative SKOV3 cells were stably transfected with VEPH1 cDNA. The impact of altered VEPH1 expression was assessed using in vitro and in vivo assays and mechanistic studies were performed in vitro.
Results:
VEPH1 expression in SKOV3 cells resulted in a reduced tumour growth rate associated with increased necrotic area, and decreased microvessel density and VEGF-A levels relative to tumours formed by mock-transfected cells. VEPH1 expression also decreased VEGFA and IL8 expression in SKOV3 cells and was associated with decreased activated AKT levels. These effects were not observed in ES-2 cells, which bear a BRAFV600E activating mutation that leads to constitutively increased IL8 and VEGFA expression.
Conclusions:
VEPH1 expression in SKOV3 ovarian cancer cells inhibits AKT activation to decrease VEGFA and IL8 expression, which leads to decreased tumour vascularisation and progression.
Similar content being viewed by others
Main
The gene encoding Ventricular Zone Expressed Pleckstrin Homology Domain Containing 1 (VEPH1) was initially identified due to its expression in the neural tube ventricular zone of the mouse and zebrafish during neurogenesis. Downregulation of Veph1 expression in zebrafish embryos disrupted brain and otic vesicle development, suggesting a role in neural cell differentiation (Muto et al, 2004). VEPH1 is well conserved from flies to human, and selective overexpression of the Drosophila ortholog, melted, in the wing increased wing size, while ubiquitous overexpression increased overall body size (Teleman et al, 2005). This growth effect was attributed to decreased FOXO and increased TOR signalling through binding and plasma membrane sequestration of FOXO and Tsc1, the TOR inhibitor, by melted. These findings suggest that mammalian VEPH1 may also function to regulate growth.
Multiple studies indicate frequent amplification of the chromosome 3q region containing the VEPH1 gene locus in various cancers, including ovarian (Sjoblom et al, 2006; Hernandez et al, 2007; Qian and Massion, 2008; Bast et al, 2009; Ylipaa et al, 2011; Pharoah et al, 2013). Amplification of VEPH1 and increased mRNA levels were indicated in ∼40% of 68 primary human epithelial ovarian cancers in a genome-wide analysis study (Ramakrishna et al, 2010). VEPH1 copy number was also found to be increased in seven out of 12 human ovarian cancer cell lines, including ES-2 (Tan et al, 2009) and we recently demonstrated differential VEPH1 protein expression in a panel of ovarian cancer cell lines representing various histotypes. VEPH1 was detected by western blot analysis in ES-2, HEY and OVCA429 cells, but not in SKOV3, OVCAR3, CAOV3 or HOC7 cells (Shathasivam et al, 2015). Similarly, interrogation of the provisional high-grade serous ovarian cancer (HGSOC) data set in The Cancer Genome Atlas (TCGA) indicated amplification of VEPH1 in nearly 17% of cases (Shathasivam et al, 2015). Whether VEPH1 affects tumour growth is unknown; however, survival analysis of the HGSOC cases with amplified VEPH1 indicates a possible increase in overall survival (Supplementary Figure 1). Moreover, we recently demonstrated that VEPH1 inhibits canonical TGFβ signalling in ovarian cancer cells (Shathasivam et al, 2015). Thus, VEPH1 could act to alter the impact of TGFβ on tumour progression.
To establish whether VEPH1 impacts tumour progression, we altered the expression of VEPH1 in both ES-2 and SKOV3 cells and monitored their growth as mouse xenografts. ES-2 cells were originally derived from a tumour mass of a woman diagnosed with poorly differentiated clear cell carcinoma, whereas SKOV3 cells were isolated from the ascites of a woman initially diagnosed with ovarian adenocarcinoma. Both cell lines have mutated TP53, which is common in the HGSOC histotype (Beaufort et al, 2014). In addition, ES-2 cells have an activating V600E mutation in BRAF (Estep et al, 2007), whereas SKOV3 cells have mutations in ARID1A and PIK3CA (Kang et al, 2005; Domcke et al, 2013; Beaufort et al, 2014), which are more prevalent in endometrioid and clear cell histotypes.
Materials and methods
Cell culture
SKOV3 and ES-2 cells were purchased from American Type Culture Collection (ATCC; Manassas, VA, USA). SKOV3 cells were previously stably transfected with either a metallothionein-inducible vector encoding Flag-tagged VEPH1 (SKOV3-Ve) or the empty expression vector (SKOV3-M) (Shathasivam et al, 2015). Cells were maintained as monolayers in RPMI-1640 medium supplemented with 1% (v/v) penicillin (100 U ml−1) and streptomycin (100 μg ml−1) (Invitrogen; Burlington, ON, Canada) and 5% (v/v) fetal bovine serum (HyClone Laboratories Inc; Logan, UT, USA). Medium for SKOV3-Ve and SKOV3-M cells was further supplemented with G418 (100 μg ml−1; Invitrogen) and CdCl2 or ZnSO4 as indicated for each experiment. Cells were grown in a humidified 37 °C incubator in the presence of 5% CO2. All cell lines and derivatives were authenticated by short random repeat analysis just before their use. PLX4720 (B-RafV600E selective inhibitor; Selleckchem; Houston, TX, USA), AZD6244 (MEK1/2 inhibitor; Selleckchem), LY294002 (PI3 kinase inhibitor; Cell Signaling Technology; Danvers, MA, USA) and SB431542 (TGFβ type I receptor inhibitor; Tocris; Minneapolis, MN, USA) were reconstituted in DMSO and further diluted with culture medium just before use. TGFβ1 (R&D Systems, Minneapolis, MN, USA) was reconstituted and diluted in culture medium.
CRISPR-Cas9 genome editing of VEPH1
Oligonucleotide sequences targeting exon 3 of VEPH1 (5′-CACCGCAAAAAGATCTTTCACGAGC-3′ and 5′-AAACGCTCGTGAAAGATCTTTTTGC-3′) were designed using the website tool (http://crispr.mit.edu) and were annealed and inserted into the Bbs1 site of pSpCas9(BB)-2A-GFP (Addgene plasmid 48138) (Ran et al, 2013). This construct expresses both Cas9 and a chimeric guide RNA that combines the targeting guide sequence and tracrRNA. ES-2 cells were transfected with the sequence-verified construct and GFP-positive cells were clonally selected (ES-2ΔVe). Clones were screened for loss of VEPH1 expression by western blot analysis. The targeted region of the genome was then amplified using Turbo Pfu polymerase (Agilent; Mississauga, ON, Canada), cloned into pCR-BluntII-TOPO (Invitrogen) and sequenced. The ES-2ΔVe cells used were verified to have a single-base insertion at codon 16, resulting in a premature stop-codon substitution at codon 25.
Cell proliferation and colony formation
Proliferation was determined by MTT or XTT dye-reduction assays as described previously (Kollara and Brown, 2010). To assess colony formation, 50 or 100 cells were seeded into 24-well plates and maintained in culture for 8 days. SKOV3-Ve and SKOV3-M cells were treated with 1 μ M CdCl2. Cultures were fixed in 4% paraformaldehyde, stained with 0.05% crystal violet, and photographed. Colonies within each culture well were counted.
Tumour xenograft model
The study was approved by the University of Toronto Animal Care and Use Committee and was performed in accordance with the Ontario Animals for Research Act and the federal Canadian Council on Animal Care. For generation of xenografts, cells were harvested at 80% confluency and suspended in phosphate-buffered saline (PBS). Female 5–8-week-old BALB/c nude mice were purchased from Charles River Laboratories (Sherbrooke, QC, Canada). Following a 7-day acclimatisation period, 3 × 106 cells were injected subcutaneously (four mice/treatment group in each of two independent runs for ES-2 and three independent runs for SKOV3 cells) in a volume of 300 μl PBS. The drinking water for mice injected with SKOV3-Ve or SKOV3-M cells contained 25 mM ZnSO4. Animals were monitored every 24–48 h and the size of resulting tumours was determined by caliper measurements. Tumour volume was calculated as: (π/6)(length × width × depth) (Tomayko and Reynolds, 1989). Animals were killed once total tumour volume reached 520 mm3, as directed by the university veterinarian, or at 9 weeks post injection if tumours did not form or attain end point volume. Other end points included a shift in body weight of >20% or ulceration at injection site. At the time of killing, tumours were excised, weighed and divided, with one-half snap-frozen and the other half fixed in 10% formalin (Sigma; Oakville, ON, Canada) and paraffin-embedded.
Real-time quantitative RT-PCR (qPCR)
Total RNA was extracted using Trizol and DNase-treated using a Turbo DNA-free kit (Invitrogen), or RNeasy Plus Universal kit (Qiagen, Toronto, ON, Canada) and reverse-transcribed using random hexamers (Invitrogen). Real-time qPCR was performed using SYBR green in an ABI PRISM 7900HT Sequence Detection System using primers (for VEPH1, Interleukin 8 (IL8), glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or TATA binding protein (TBP)) and conditions previously described (Backman et al, 2014; Shathasivam et al, 2015). VEGFA primers used were forward: 5′-GGGCAGAATCATCACGAAGT-3′ and reverse: 5′-CACACAGGATGGCTTGAAGA-3′. A relative standard curve method with TBP or GAPDH transcripts as calibrator was used to normalise VEPH1, VEGFA and IL8 transcript levels.
Western blot analysis
Western blot analysis was performed as previously described (Shathasivam et al, 2015). Membranes were incubated with anti-VEPH1 (1 : 1000 or 1 : 2000, SDI Newark, DE, USA; or 1 : 250, Sigma), anti-Flag (1 : 500, Sigma), anti-AKT (1 : 1000, Cell Signaling Technology), anti-phospho(Ser473)-AKT (1 : 1000, Cell Signaling Technology), anti-p44/42 MAP Kinase (1 : 500, Cell Signaling Technology), anti-phospho(Thr202/Tyr204)-p44/42 MAP Kinase (1 : 500, Cell Signaling Technology), anti-GAPDH (1 : 1500, Santa Cruz Biotechnology, Dallas, TX, USA) or anti-α-tubulin (1 : 3000, Sigma) antibodies. Blots were visualised by enhanced chemiluminescence (Santa Cruz Biotechnology).
Immunohistochemistry
Sections (5 μm thick) cut from the centre of formalin-fixed paraffin-embedded tumours were deparaffinised and rehydrated. Antigen retrieval was performed by heating sections in 10 mM citrate buffer and processed for immunohistochemistry as previously described (Puy et al, 1995). Sections were incubated overnight at 4 °C with either primary antibody or non-immune IgG (Santa Cruz Biotechnology). Primary antibodies used include: Ki-67 (1 : 200, Immunotech, Westbrook, ME, USA), proliferating cell nuclear antigen (PCNA; 1 : 2000, Dako, Mississauga, ON, Canada), CD34 (1 : 250, Abcam, Toronto, ON, Canada), CD31 (1 : 50, Abcam), and VEGF (1 : 100, ThermoScientific, Waltham, MA, USA). Sections were incubated with biotinylated secondary antibody (1 : 200, Vector Laboratories, Burlington, ON, Canada) then incubated with streptavidin-peroxidase complex (1 : 400; Vector Laboratories). The immunocomplex was visualised by diaminobenzidine chromogen (DAB; Sigma). Sections were counterstained with Harris Hematoxylin solution (Sigma).
Double terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL) and cleaved caspase-3 staining
Sections were stained for TUNEL using the TACS2 TdT-DAB In Situ Apoptosis Detection Kit (Trevigen, Gaithersburg, MD, USA) following the manufacturer’s protocol. Upon completion of DAB staining, sections were washed with PBS for 20 min, stained with cleaved caspase-3 antibody (1 : 300, Cell Signaling Technology) using the Cell and Tissue HRP-AEC Staining Kit (R&D Systems) following the manufacturer’s protocol and counterstained with Gill No.1 haematoxylin (Sigma). A positive TUNEL control was included for each tumour by treating a section with TACs-Nuclease to generate DNA breaks in every cell. Fixed paraffin-embedded Jurkat cells treated with apoptosis-inducing etopiside (Sigma) were included as a positive control for cleaved caspase-3.
Immunohistochemistry imaging and quantitation
Digital images were captured using a Hamamatsu NanoZoomer 2.0-RS Digital Slide Scanner (Meyer Instruments, Houston, TX, USA). Ki-67 and PCNA nuclear labelling index (LI) were determined using the ImmunoRatio quantitative image analysis program (Tuominen et al, 2010). Each image was analysed for percentage of DAB-stained nuclei. Three randomly selected 20 × magnification field images per tumour, with areas of necrosis excluded, were analysed and average PCNA and Ki-67 nuclear LI were calculated. Data were compiled to create an average PCNA and Ki-67 nuclear LI for SKOV3-M and SKOV3-Ve tumours.
Necrotic tumour area was determined by annotating haematoxylin and eosin (H&E) stained whole sections of tumours using the Hamamatsu NanoZoomer Digital Pathology viewer. Areas of necrotic tissue, based on cell and tissue morphology, were outlined using a freehand area annotation tool and necrotic area was divided by total tumour area to determine the percentage of necrotic area. Necrosis morphology and necrotic tumour annotations were verified by a gynaecological pathologist (BC).
Double TUNEL/cleaved caspase-3 staining was quantified by counting the number of TUNEL and cleaved caspase-3-positive cells per mm2 necrotic and non-necrotic area of each tumour at 40x magnification.
Microvessel density (MVD), defined as the number of microvessels per mm2, was determined using guidelines described by the International Consensus (Vermeulen et al, 2002). Microvessels were defined as a distinct CD34+ or CD31+ cell or cell cluster regardless of the presence of a lumen. Large vessels with thick muscular walls and areas of necrosis were excluded from the count. Microvessels were counted at 40 × magnification in the three most vascularised areas of the tumour (0.74 mm2 per field). Images were analysed with the operator blinded to treatment condition. An average CD34+ and CD31+ MVD was calculated for each tumour.
Staining for VEGF was quantified by calculating the area of positive staining relative to total area within five independent randomly selected fields per slide using Northern Eclipse (version 8) software (Empix Imaging, Mississauga, ON, Canada).
Tube formation assay
An in vitro tube formation assay was used as described by Arnaoutova and Kleinman (2010). Briefly, 120 μl growth factor-reduced Matrigel (8 mg ml−1; Corning, Bedford, MA, USA) were added to 48-well culture plates and allowed to solidify at 37 °C for 30 min. Cells (25 000 per well) in 250 μl culture medium were seeded and images were taken at 6 h for SKOV3-Ve and SKOV3-M cells and at 17 h for ES-2 and ES-2ΔVe cells. The number of junctions and total tube length were determined using the Angiogenesis Analyzer plugin for Image J (v1.47, https://imagej.nih.gov/ij/).
Statistical analyses
Data were analysed using Student’s unpaired two-tailed t-test, Fisher’s Exact test, or ANOVA, followed by a Student–Newman–Keuls post hoc test. Tumour progression data are presented as a Kaplan–Meier survival plot created using GraphPad Prism v5 (GraphPad Software, La Jolla, CA, USA) and were analysed using a Gehan–Breslow–Wilcoxon Test. A growth rate plot indicating tumour volume (mm3) at each time point, with averaged exponential lines of best-fit, was created using GraphPad Prism. Extra sum of squares F-test was used to compare the exponential nonlinear regression lines generated. Statistical significance was accepted at P<0.05.
Results
VEPH1 expression does not affect cell proliferation or colony formation in vitro
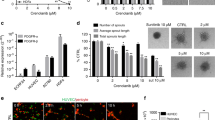
To determine if VEPH1 expression impacts cell proliferation or colony formation in vitro, we introduced a frameshift mutation near the translational ATG start site of VEPH1 in ES-2 cells using a CRISPR-Cas9 system. Loss of VEPH1 expression in these cells (ES-2ΔVe) was verified by western blot analysis (Figure 1A). Comparison of ES-2 to ES-2ΔVe cells indicated loss of VEPH1 expression did not affect cell proliferation (Figure 1B) or colony formation (Figure 1C). We previously showed that SKOV3 cells lack endogenous VEPH1 expression and generated cells stably transfected with full-length human VEPH1 cDNA (SKOV3-Ve) under regulation by a metallothionein promoter. These cells express Flag-tagged VEPH1 in the absence of promoter stimulation; however, CdCl2 or ZnSO4 induction further increased VEPH1 levels (Figure 1D). Comparison of SKOV3-Ve and mock-transfected SKOV3 (SKOV3-M) cells after CdCl2-induction further indicated that VEPH1 expression had no impact on cell proliferation (Figure 1E) or colony formation (Figure 1F).
VEPH1 expression does not alter in vitro cell growth or colony formation.(A) VEPH1 protein expression in ES-2 cells and loss of expression following introduction of a genomic frameshift mutation (ES-2ΔVe cells). Western blot analysis was performed with VEPH1 antibody and tubulin was used as loading control. (B) Growth of ES-2 and ES-2ΔVe cultures determined by XTT dye-reduction assay. Bars represent mean±s.e.m. of three independent experiments. (C) Number of colonies formed by ES-2 and ES-2ΔVe cells. Bars represent mean±s.e.m. of three independent experiments. (D) VEPH1 expression in SKOV3-Ve or SKOV3-M cells following treatment with CdCl2 or ZnSO4 for 24 h. Western blot analysis was performed using VEPH1 or Flag antibody as indicated with immunoblotting for tubulin used as a loading control. (E) Growth of SKOV3-M and SKOV3-Ve cultures maintained in media with CdCl2 as determined by MTT dye-reduction assay. Bars represent mean±s.e.m. of three independent experiments. (F) Number of colonies formed by SKOV3-M and SKOV3-Ve cells. Bars represent mean±s.e.m. of three independent experiments. Bars with different letters are statistically different from one another (P<0.05) as determined by ANOVA followed by SNK post hoc test.
Targeted disruption of VEPH1 expression does not impact ES-2 tumour growth
To determine if loss of VEPH1 expression affects tumour formation or growth in vivo, female BALB/c nude mice were injected subcutaneously with ES-2 (n=8) or ES-2ΔVe (n=8) cells and tumour volume was determined every 24 h. Tumours formed (Figure 2A) in all injected mice and animals were killed once tumours reached end point volume, or because of excessive ulceration (two mice) or a weight gain ⩾20% due to ascites accumulation (one ES-2ΔVe-injected mouse). Body weight changes were similar in both groups (Figure 2B) and no differences were observed in tumour progression (Figure 2C). Kaplan–Meier survival analysis using the date of killing as a surrogate for survival further demonstrated no significant impact of loss of VEPH1 expression in these cells (Figure 2D). Differential VEPH1 expression in tumours was verified by qPCR and western blot analysis. As expected, tumours derived from both ES-2 and ES-2ΔVe cells expressed similar levels of VEPH1 transcripts (Figure 2E) with ES-2ΔVe cells showing complete absence of VEPH1 protein (Figure 2F).
Loss of VEPH1 expression in ES-2 cells does not impact tumour progression in vivo.(A) Image of a representative tumour formed by ES-2 cell inoculation. (B, left) Body weight of individual mice at time of cell inoculation and at study end point. Two-way ANOVA indicated an effect of time (P=0.0001) but not cell type injected or interaction. (right) Percent change in final body weight from cell injection (P=0.7708). Line indicates group median. (C) ES-2 and ES-2ΔVe cell xenografts exhibited similar exponential growth trajectories. Extra sum of squares F-test was used to compare the exponential nonlinear regression lines generated for tumour growth as a function of time (P=0.5896). (D) Kaplan–Meier survival curve using attainment of end point as a surrogate for death (n=8/group; P=0.4724). (E) Relative VEPH1 transcript levels normalised to GAPDH measured in ES-2 and ES-2ΔVe cell tumours as determined by qPCR (n=8; P=0.6171). Line indicates group mean. (F) Western blots verifying loss of VEPH1 protein in ES-2ΔVe cell tumours. Immunoblotting for GAPDH was included as a loading control.
VEPH1 expression delays SKOV3 tumour growth
To determine if VEPH1 expression affects tumour formation or growth in vivo, mice (n=12 per group) were injected subcutaneously with SKOV3-Ve or SKOV3-M cells and tumour volume was determined every 24–48 h. Animals were killed when tumour volumes reached end point volume or 9 weeks post injection. Tumours (Figure 3A) appeared less vascularised compared with those formed by ES-2 cells (Figure 2A). Changes in body weight were similar in both SKOV3 groups (Figure 3B). VEPH1 expression had no impact on the number of tumours formed (SKOV3-Ve: 10/12; SKOV3-M: 11/12; P=1.00, Fisher’s exact test) or on those that achieved end point volume during the study period (SKOV3-Ve: 8/12; SKOV3-M: 9/12; P=1.00), indicating that VEPH1 expression did not alter engraftment. However, the tumour growth rate was significantly decreased by VEPH1 expression. SKOV3-Ve tumours advanced more slowly to end point volume than SKOV3-M tumours (45.8±2.2 vs 31.6±2.2 days, respectively; mean±s.e.m.; P<0.001). Plotting the growth of each tumour revealed an exponential growth pattern for both SKOV3-M and SKOV3-Ve tumours, with delayed progression for VEPH1-expressing tumours (Figure 3C). Substituting attainment of end point tumour volume for survival in a Kaplan–Meier survival analysis indicated a significant impact of VEPH1 expression on tumour progression (P<0.01, Figure 3D).
VEPH1 expression in SKOV3 cells delays tumour progression.(A) Image of a representative tumour formed by SKOV3 cell inoculation. (B, left) Body weight of individual mice at time of cell inoculation and at study end point. Two-way ANOVA indicated an effect of time (P<0.0001) but not cell type injected or interaction. (right) Comparison of percent change in body weight (P=0.9952). Line indicates group median. (C) SKOV3 xenografts exhibited exponential growth that was delayed in cells expressing VEPH1. Data shown are from animals in which tumours attained end point volume (n=9, SKOV3-M and n=8, SKOV3-Ve). Extra sum of squares F-test was used to compare the exponential nonlinear regression lines generated for tumour growth as a function of time (P<0.0001). (D) Kaplan–Meier survival curve using attainment of end point tumour volume as a surrogate for death. Only animals that exhibited tumour formation were included in the analysis (n=11, SKOV3-M and n=10, SKOV3-Ve). A Gehan–Breslow–Wilcoxon Test indicated a significant difference in survival between tumour groups (P=0.0097). (E) Relative VEPH1 transcript levels normalised to TBP measured in end point SKOV3-M (n=11) and SKOV3-Ve (n=10) tumours as determined by qPCR (*P<0.0001). Line indicates group mean. (F) Western blot verifying expression of VEPH1 protein in end point SKOV3-Ve cell tumours using Flag antibody. (G) VEPH1-expressing SKOV3 tumours have increased necrotic area. (top left) Mean necrotic area (necrotic area as a percentage of total tumour section area) for SKOV3-M (n=11) and SKOV3-Ve (n=10) tumours. Bars represent mean±s.e.m. (*P=0.034). (bottom left) Percentage of SKOV3-M (n=11) and SKOV3-Ve (n=10) tumours filled with liquid (*P=0.004). (right) Representative H&E sections show areas of necrosis (indicated by asterisks). Magnification bar, 100 μm. (H) SKOV3-Ve tumours exhibit increased levels of TUNEL staining but not cleaved caspase-3. (left) Number of TUNEL and cleaved caspase-3-positive cells per necrotic tumour area for SKOV3-M (n=11) and SKOV3-Ve (n=8) tumours. Bars represent mean±s.e.m. Bars with different letters are statistically significant from one another as determined by ANOVA followed by SNK post hoc test. (right) Number of TUNEL-positive cells per non-necrotic tumour area for SKOV3-M (n=11) and SKOV3-Ve (n=8) tumours. Bars represent mean±s.e.m. (*P=0.001).
To verify sustained VEPH1 expression in SKOV3-Ve tumours, qPCR and western blot analyses were conducted on tumours at end point. As expected, VEPH1 mRNA and protein expression were detected in all SKOV3-Ve tumours with little or no expression detected in SKOV3-M tumours (Figure 3E and F). An immunoreactive band was detected for one SKOV3-M tumour; however, this was at a lower molecular weight than Flag-VEPH1 with no measureable transcripts.
VEPH1 expression is associated with increased necrosis in SKOV3 cell tumours
To determine whether the delayed progression of SKOV3-Ve tumours reflects a decrease in proliferation, tumours were assessed for expression of PCNA and Ki-67 proliferation markers. Immunohistochemical analysis revealed no significant difference in either PCNA or Ki-67 staining between SKOV3-Ve and SKOV3-M tumours (Supplementary Figure 2).
Examination of H&E stained tumour sections indicated morphology consistent with HGSOC but with areas also consistent with clear cell carcinoma further evidenced by positive periodic acid-Schiff staining (not shown). Necrotic areas, characterised by cells with pyknotic nuclei, eosinophilia and residual ghost outlines of cells, were observed in all tumours (Figure 3G). However, SKOV3-Ve tumours exhibited more than twofold higher average area of necrosis compared with SKOV3-M tumours (Figure 3G). Upon tumour excision it was noted that 60% of SKOV3-Ve tumours were partially fluid-filled, a feature often associated with necrosis, whereas none of the SKOV3-M tumours contained fluid (Figure 3G).
To verify necrotic death, tumour sections were stained for DNA fragmentation (TUNEL), indicative of either apoptosis or necrosis, and cleaved caspase-3, indicative of apoptosis. TUNEL staining revealed a 3.2-fold greater number of cells with DNA fragmentation per necrotic area in SKOV3-Ve compared with SKOV3-M tumours (Figure 3H). Consistent with the predominance of necrotic cell death, fewer cleaved caspase-3 stained cells were identified in necrotic areas of both SKOV3-M and SKOV3-Ve tumours than TUNEL-positive cells (Figure 3H). No difference was observed in cleaved caspase-3 staining between SKOV3-Ve and SKOV3-M tumours (Figure 3H).
A 4.2-fold greater number of TUNEL-positive cells per non-necrotic area was also identified in SKOV3-Ve tumours compared with SKOV3-M tumours (Figure 3H). Cleaved caspase-3 staining was not detected in non-necrotic areas of tumours from either cell subline.
VEPH1 expression decreases microvessel density (MVD) in vivo and tube formation in vitro
Tumour necrosis may result from insufficient angiogenesis; therefore, the impact of VEPH1 expression on tumour MVD was examined by staining for endothelial cell markers CD31 and CD34. CD31 staining revealed a 35% decrease in MVD in SKOV3-Ve relative to SKOV3-M tumours (Figure 4A and B). Similar results were obtained for CD34 (Figure 4A). Consistent with our findings of differences in MVD, a 34% decrease in VEGF staining was observed in non-necrotic areas of SKOV3-Ve tumours compared with that of SKOV3-M tumours (Figure 4C). Altogether, these data indicate VEPH1 expression compromised tumour neovascularisation.
VEPH1 expression decreases MVD and VEGFA and IL8 expression.(A, left) CD31+ MVD for SKOV3-M (n=11) and SKOV3-Ve (n=10) tumours. Bars represent mean±s.e.m. (*P=0.041). (right) CD34+ MVD for SKOV3-M (n=11) and SKOV3-Ve (n=10) tumours. Bars represent mean±s.e.m. (*P=0.004). (B) Representative sections stained for CD31. Red arrows indicate microvessels. Scale bar, 25 μm. (C) Levels of VEGF staining in SKOV3-M (n=11) and SKOV3-Ve (n=10) tumours. Bars represent mean±s.e.m. (*P<0.05, one-tailed Student’s unpaired t-test). (D) Representative images of in vitro tubule formation. Scale bar, 200 μm. (E) Quantification of total tubule length in ES-2ΔVe vs ES-2 (P=0.4436) and SKOV3-M vs SKOV3-Ve (*P=0.0089) cells. Quantification of number of junctions in ES-2ΔVe vs ES-2 (P=0.1396) and SKOV3-M vs SKOV3-Ve (*P=0.0114) cells. Bars represent mean±s.e.m. (F) Levels of VEGFA transcripts measured in ES-2ΔVe, ES-2, SKOV3-M, and SKOV3-Ve cells by qPCR. Bars represent mean±s.e.m. of three independent experiments. Two-way ANOVA indicated a difference between cell lines and VEPH expression with a positive interaction. Bars with different letters are statistically significant from one another. (G) Levels of IL8 transcripts measured in ES-2 and SKOV3 cells by qPCR. Bars represent mean±s.e.m. of three independent experiments (*P<0.05). (H, left) IL8 transcript levels in ES-2ΔVe relative to ES-2 cells. (right) IL8 transcript levels in SKOV3-Ve relative to SKOV3-M cells (*P<0.0001).
Previous studies have reported SKOV3 cells are capable of vascular mimicry and readily form tubular structures in a 3D basement membrane matrix (Millimaggi et al, 2009), consistent with vessel formation. We compared the impact of VEPH1 expression on the ability of ES-2 and SKOV3 cells to form tubular structures on Matrigel. Both ES-2 and SKOV3 cells formed tubules (Figure 4D) with SKOV3 cells forming structures more rapidly than ES-2 cells (6 vs 17 h). Disrupted VEPH1 expression had no impact on tubule formation in ES-2 cells, whereas VEPH1 expression in SKOV3 cells led to a slight decrease in tubule length and number of junctions (Figure 4E).
ES-2 cells have an activating BRAF mutation (Estep et al, 2007) that leads to constitutive downstream MEK1/2 and ERK1/2 activation. This mutation has been shown in melanoma cells to increase VEGF-A and IL-8, a chemokine with potent angiogenic activity (Li et al, 2003), through ERK1/2 activation (Bottos et al, 2012; Khalili et al, 2012; Whipple and Brinckerhoff, 2014). We compared ES-2 and SKOV3 cells for VEGFA and IL8 expression using qPCR. ES-2 cells express eightfold more IL8 transcripts than SKOV3 cells, whereas SKOV3 cells express threefold more VEGFA transcripts than ES-2 cells (Figure 4F and G).
VEPH1 expression decreases VEGFA and IL8 expression in SKOV3 but not ES-2 cells
To determine why VEPH1 might alter vascularisation of SKOV3 but not ES-2 cell xenografts, we examined the impact of altered VEPH1 expression on VEGFA and IL8 transcript levels. Loss of VEPH1 in ES-2 cells did not impact either VEGFA or IL8 transcript levels, whereas VEPH1 expression in SKOV3 cells decreased both VEGFA and IL8 expression (Figure 4F and H). Treatment of ES-2 or ES-2ΔVe cells with either a MEK1/2 inhibitor, AZD6244 or a BRAFV600E-specific inhibitor, PLX4720, markedly decreased both VEGFA and IL8 expression (Figure 5A and B), consistent with the impact of the activated MAPK-ERK1/2 pathway on expression of these pro-angiogenic factors.
Activated MAPK-ERK1/2 regulates VEGFA and IL8 expression in ES-2 cells, whereas PI3K-AKT, MAPK-ERK1/2 and TGF β signalling regulate VEGFA and IL8 expression in SKOV3 cells.(A and B) Levels of VEGFA (A) and IL8 (B) transcripts measured in ES-2 and ES-2ΔVe cells by qPCR following treatment with the MEK1/2 inhibitor AZD6244 (1 μM) or the BRAFV600E inhibitor PLX4720 (10 μM). Bars represent mean±s.e.m. of two independent experiments with triplicates. Values are relative to vehicle-treated controls within each cell line. Bars with different letters are statistically significant from one another P<0.05). (C and D) Levels of VEGFA (C) and IL8 (D) transcripts measured in SKOV3-M and SKOV3-Ve cells by qPCR following treatment with AZD6244. Bars represent mean±s.e.m. of four (C) or three (D) independent experiments with triplicates. Values are relative to vehicle-treated controls within each cell line. *P<0.001. (E and F) Levels of VEGFA (E) and IL8 (F) transcripts measured in SKOV3-M and SKOV3-Ve cells by qPCR following treatment with TGFβ (100 pM). Bars represent mean±s.e.m. of three (E) or one (F) independent experiments with triplicates. Values are relative to vehicle-treated controls within each cell line. *P<0.01. (G and H) Levels of VEGFA (G) and IL8 (H) transcripts measured in SKOV3-M and SKOV3-Ve cells by qPCR following treatment with the ALK5 serine/threonine kinase inhibitor SB431542 (10 μM). Bars represent mean±s.e.m. of three (G) or five (H) independent experiments with triplicates. Values are relative to vehicle-treated controls within each cell line. *P<0.001.
In comparison, MEK1/2 inhibition in SKOV3 cells decreased IL8 expression by ⩾75% independent of VEPH1 expression, but did not alter VEGFA expression (Figure 5C and D), indicating that other signalling pathways affect VEGFA expression in these cells. We have previously shown that VEPH1 inhibits TGFβ signalling (Shathasivam et al, 2015). Consistent with this, TGFβ treatment increased VEGFA transcripts 1.4-fold in SKOV3-M cells but less than 1.2-fold in SKOV3-Ve cells (Figure 5E), and decreased IL8 expression by 60% in SKOV3-M but not in SKOV3-Ve cells (Figure 5F). Moreover, inhibition of TGFβ receptor type I (ALK5) with the serine/threonine kinase inhibitor SB431542 increased IL8 transcripts by sixfold (Figure 5H), indicating a potent effect of endogenous TGFβ in these cells. However, SB431542 treatment increased, rather than decreased, VEGFA expression (Figure 5G), which suggests non-canonical TGFβ signalling.
TGFβ activates PI3K/AKT through a non-canonical pathway (Zhang, 2009). In addition, SKOV3 cells have an activating PIK3CA mutation (Kang et al, 2005; Beaufort et al, 2014); therefore, we tested the impact of LY294002, a PI3K inhibitor, on VEGFA and IL8 expression in these cells. LY294002 treatment decreased VEGFA expression in both SKOV3-M and SKOV3-Ve cells by 80% (Figure 6A), and decreased IL8 expression by ∼75% in SKOV3-M cells; however, in SKOV3-Ve cells, this treatment increased IL8 expression by approximately eightfold (Figure 6B). Western blot analysis confirmed SKOV3, but not ES-2, cells have high levels of phosphorylated AKT (pAKT), whereas ES-2 cells have higher levels of phosphorylated ERK1/2 (pERK1/2; Figure 6C). VEPH1 expression in SKOV3 cells resulted in lower pAKT (Figure 6D). SB431542 increased pERK1/2 independent of VEPH1 expression (Figure 6D), which is consistent with the increase in IL8 transcripts following this treatment (Figure 6B). LY294002 increased pERK1/2 only in the presence of VEPH1 (Figure 6D), which is consistent with the increase in IL8 transcripts seen in these cells following this treatment (Figure 6B).
Differential effect of VEPH1 on AKT blockade of VEGFA and IL8 transcript levels in SKOV3 cells.(A and B) Levels of VEGFA (A) and IL8 (B) transcripts measured in SKOV3-M and SKOV3-Ve cells by qPCR following treatment with the PI3K inhibitor LY294002 (100 μM). Bars represent mean±s.e.m. of two independent experiments with triplicates. Values are relative to vehicle-treated controls within each cell line. *P<0.05, **P<0.0001. (C) Western blot showing pERK1/2 and pAKT levels in ES-2 and SKOV3 cells. A representative of three independent blots is shown. (D) Western blot showing pERK1/2 and pAKT levels in SKOV3-M and SKOV3-Ve cells following treatment with an inhibitor to MEK1/2, PI3K or ALK5 serine/threonine kinase. (E) Model of how VEPH1 affects VEGFA and IL8 expression in SKOV3 but not ES-2 cells. * indicates the presence of an oncogenic mutation.
Discussion
This study assessed the impact of VEPH1 expression in two ovarian cancer cell lines with divergent endogenous expression. The VEPH1 gene locus is amplified in ES-2 cells, which results in high levels of VEPH1 protein expression, whereas VEPH1 transcript and protein expression is undetectable in SKOV3 cells (Tan et al, 2009; Shathasivam et al, 2015). Disruption of the VEPH1 locus in ES-2 cells eliminated endogenous VEPH1 protein levels and we have previously shown that SKOV3-Ve cells express equivalent or less VEPH1 protein than ES-2 cells (Shathasivam et al, 2015). In contrast to what was initially expected based upon the impact of Drosophila melted on growth, lack of VEPH1 expression was not associated with decreased in vitro cell growth or colony formation and did not result in slowed expansion of tumours formed from xenografted cells. Rather, ectopic VEPH1 expression in SKOV3 cells delayed xenograft tumour expansion. This finding is consistent with a trend toward increased survival among TCGA HGSOC patient data set with tumour amplification of the VEPH1 locus (Supplementary Figure 1).
SKOV3-Ve and SKOV3-M cells formed tumours with equivalent proliferation indices, consistent with our in vitro data. However, examination of SKOV3-Ve tumours indicated increased necrotic area, consistent with decreased MVD, compared with SKOV3-M tumours. Thus, our data indicate that VEPH1 expression led to impaired neovascularisation resulting in tumour necrosis and ultimately delayed overall tumour growth.
Tumour neovascularisation is accomplished through multiple processes. These include the recruitment of bone marrow-derived endothelial precursor cells to the tumour or differentiation of cancer cells into bona fide endothelial cells (Alvero et al, 2009) for de novo microvessel formation, expansion of existing vasculature into the tumour through angiogenesis, and formation of acellular matrix-derived microvessels through vascular mimicry (Seftor et al, 2012). These latter microvessels are externally lined by tumour cells and are devoid of endothelial cells. SKOV3 cells undergo vascular mimicry (Millimaggi et al, 2009) and we found PAS+ vessel-like structures in both SKOV3 and ES-2 cell-generated tumours (data not shown). Both ES-2 and SKOV3 cells exhibited tube-forming capacity in vitro. Although VEPH1 expression resulted in slightly decreased tubule formation by SKOV3 cells, loss of expression in ES-2 cells was without effect.
Multiple cytokines and growth factors contribute to neovascularisation, most prominent among these is VEGF-A. In addition, IL-8/CXCL8 is a potent chemokine that promotes endothelial cell proliferation and migration (Waugh and Wilson, 2008; Martin et al, 2009) and is highly expressed by ES-2 cells which form highly vascularised tumours. Both VEGFA and IL8 transcript levels were decreased by VEPH1 expression in SKOV3 cells, but not in ES-2 cells. This finding with VEGFA is consistent with our previous gene profiling study showing decreased VEGFA transcripts in SKOV3-Ve relative to SKOV3-M cells (Shathasivam et al, 2015). Moreover, VEGFA transcript levels were decreased in SKOV3-Ve generated tumours compared with SKOV3-M cell tumours.
Multiple signalling systems regulate VEGF-A and IL-8, many of which act through activation of the MAPK-ERK1/2 and PI3K-AKT pathways. We attribute our inability to detect an impact of VEPH1 loss on ES-2 cell tumour neovascularisation to the presence of an oncogenic BRAFV600E mutation in these cells (Estep et al, 2007). This mutation results in constitutively activated BRAF that triggers high levels of IL8 and VEGFA expression through the MEK-ERK pathway to stimulate angiogenesis (Figure 6E; Bottos et al, 2012). Using a specific BRAFV600E inhibitor and a MEK1/2 specific inhibitor, we demonstrated that both VEGFA and IL8 expression in ES-2 cells is dependent upon the constitutive activation of this pathway. Our finding that VEPH1 did not impact the expression of these pro-angiogenic factors in ES-2 cells indicates that VEPH1 does not act upon this pathway downstream of BRAF.
The activating PIK3CA mutation in SKOV3 cells results in constitutive AKT activation. Inhibition of PI3K in SKOV3-M cells resulted in markedly decreased levels of both VEGFA and IL8 transcripts. Expression of VEPH1 in SKOV3 cells resulted in decreased pAKT, indicating that VEPH1 may act to inhibit IL8 and VEGFA expression by interfering with AKT activation. However, inhibition of PI3K in the presence of VEPH1 resulted in increased IL8 expression, whereas VEGFA transcript levels were decreased. Thus, while decreased pAKT by VEPH1 is consistent with an impact on VEGFA expression, its impact on IL8 expression involves an additional signalling pathway. It is interesting to note that inhibition of PI3K in SKOV3-Ve, but not SKOV3-M, cells resulted in increased pERK1/2, which could explain the increase in IL8 expression.
We recently reported that VEPH1 affects multiple signalling pathways and inhibits canonical TGFβ signalling (Shathasivam et al, 2015). Our current studies demonstrate that SKOV3 cells have endogenous TGFβ signalling and that this signalling represses IL8 expression through an ALK5 serine/threonine kinase-dependent pathway. It is thus likely that VEPH1 acts to diminish TGFβ-induced suppression of IL8 expression, while simultaneously suppressing IL8 expression through its effect on another pathway.
TGFβ from multiple cellular sources is present in the tumour microenvironment and promotes angiogenesis in part by increasing VEGFA expression (Liao et al, 2011). TGFβ signalling contributes to increased VEGFA expression in SKOV3 cells, likely through a non-canonical pathway since SB431542 also increased VEGFA transcript levels and did not block exogenous TGFβ-induced increased VEGFA transcript levels (data not shown). The increase observed with TGFβ was diminished in SKOV3-Ve cells relative to SKOV3-M cells, suggesting that VEPH1 may inhibit non-canonical TGFβ signalling.
Inhibition of MEK1/2 in SKOV3 cells resulted in decreased IL8 but not VEGFA transcript levels. We propose that this effect on IL8 is mediated through an impact on the TGFβ effector SMADs (model outlined in Figure 6E). ERK1/2-dependent phosphorylation of the linker region of SMAD2/3 inhibits their ability to accumulate in the cell nucleus (Kretzschmar et al, 1999). Inhibition of MEK1/2 would thus increase SMAD2/3 activity to result in increased IL8 inhibition.
The mechanism by which VEPH1 inhibits AKT activation, which is a central effector of multiple signalling pathways, remains to be determined. Drosophila melted has been shown to bind to TSC1 to prevent its stabilisation of TSC2, thereby disinhibiting TOR (Mikeladze-Dvali et al, 2005; Teleman et al, 2005). However, while TSC2 inhibits mTORC1, it activates mTORC2, which stimulates AKT phosphorylation at S473 (Sarbassov et al, 2005). Thus, if TSC1 binding is conserved in human VEPH1, this could be a mechanism by which VEPH1 decreases AKT activation. In addition, TGFβ binding results in tyrosine phosphorylation of ALK5 to create a docking site for the adaptor protein ShcA (Lee et al, 2007; Ursini-Siegel et al, 2012). This docking creates a platform for the subsequent binding of the regulatory unit of PI3K that results in activation of the catalytic subunit. Moreover, activated ShcA binds Grb2-SOS to activate RAS and the MAPK-ERK1/2 pathway. These pathways are dependent upon tyrosine phosphorylation of ALK5 rather than ALK5 serine-threonine kinase activity and may not be directly affected by SB431542. The increase in VEGFA transcripts in SKOV3 cells observed in the presence of either TGFβ or SB431542 likely reflects increased non-canonical signalling. Whether VEPH1 affects ShcA docking to ALK5 remains to be determined.
This study provides the first evidence of a role for VEPH1 on tumour progression and indicates a function of VEPH1 in regulating tumour vascularisation and necrosis in a xenograft model. This effect suggests a late acting tumour suppressor function; however, VEPH1 modulates multiple signalling pathways, including TGFβ, which has pleiotropic effects on cancer progression. Thus, it remains possible that VEPH1 may have effects that promote tumour initiation. It is also likely that VEPH1 has a role in other cancers as an increase in VEPH1 copy number and expression has been identified in breast (Hernandez et al, 2007) and gastrointestinal stromal tumours (Ylipaa et al, 2011). Although our findings suggest that the effect of VEPH1 is context-dependent, a recognised limitation of the present study is the inclusion of only two cell lines. Thus, in addition to determining the mechanisms by which VEPH1 affects AKT activation, it will be important to examine additional cell line models with different driver mutations to establish the generalisability of these findings.
Change history
11 April 2017
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Alvero AB, Fu HH, Holmberg J, Visintin I, Mor L, Marquina CC, Oidtman J, Silasi DAMor G (2009) Stem-like ovarian cancer cells can serve as tumor vascular progenitors. Stem Cells 27: 2405–2413.
Arnaoutova I, Kleinman HK (2010) In vitro angiogenesis: endothelial cell tube formation on gelled basement membrane extract. Nat Protoc 5: 628–635.
Backman S, Kollara A, Haw R, Stein L, Brown TJ (2014) Glucocorticoid-induced reversal of interleukin-1beta-stimulated inflammatory gene expression in human oviductal cells. PLoS One 9: e97997.
Bast RC Jr., Hennessy B, Mills GB (2009) The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer 9: 415–428.
Beaufort CM, Helmijr JC, Piskorz AM, Hoogstraat M, Ruigrok-Ritstier K, Besselink N, Murtaza M, Van IWF, Heine AA, Smid M, Koudijs MJ, Brenton JD, Berns EM, Helleman J (2014) Ovarian cancer cell line panel (OCCP): clinical importance of in vitro morphological subtypes. PLoS One 9: e103988.
Bottos A, Martini M, Di Nicolantonio F, Comunanza V, Maione F, Minassi A, Appendino G, Bussolino F, Bardelli A (2012) Targeting oncogenic serine/threonine-protein kinase BRAF in cancer cells inhibits angiogenesis and abrogates hypoxia. Proc Natl Acad Sci USA 109: E353–E359.
Domcke S, Sinha R, Levine DA, Sander C, Schultz N (2013) Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun 4: 2126.
Estep AL, Palmer C, Mccormick F, Rauen KA (2007) Mutation analysis of BRAF, MEK1 and MEK2 in 15 ovarian cancer cell lines: implications for therapy. PLoS One 2: e1279.
Hernandez P, Sole X, Valls J, Moreno V, Capella G, Urruticoechea A, Pujana MA (2007) Integrative analysis of a cancer somatic mutome. Mol Cancer 6: 13.
Kang S, Bader AG, Vogt PK (2005) Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci USA 102: 802–807.
Khalili JS, Liu S, Rodriguez-Cruz TG, Whittington M, Wardell S, Liu C, Zhang M, Cooper ZA, Frederick DT, Li Y, Zhang M, Joseph RW, Bernatchez C, Ekmekcioglu S, Grimm E, Radvanyi LG, Davis RE, Davies MA, Wargo JA, Hwu P, Lizee G (2012) Oncogenic BRAF(V600E) promotes stromal cell-mediated immunosuppression via induction of interleukin-1 in melanoma. Clin Cancer Res 18: 5329–5340.
Kollara A, Brown TJ (2010) Four and a half LIM domain 2 alters the impact of aryl hydrocarbon receptor on androgen receptor transcriptional activity. J Steroid Biochem Mol Biol 118: 51–58.
Kretzschmar M, Doody J, Timokhina I, Massague J (1999) A mechanism of repression of TGFbeta/ Smad signaling by oncogenic Ras. Genes Dev 13: 804–816.
Lee MK, Pardoux C, Hall MC, Lee PS, Warburton D, Qing J, Smith SM, Derynck R (2007) TGF-beta activates Erk MAP kinase signalling through direct phosphorylation of ShcA. EMBO J 26: 3957–3967.
Li A, Dubey S, Varney ML, Dave BJ, Singh RK (2003) IL-8 directly enhanced endothelial cell survival, proliferation, and matrix metalloproteinases production and regulated angiogenesis. J Immunol 170: 3369–3376.
Liao S, Liu J, Lin P, Shi T, Jain RK, Xu L (2011) TGF-beta blockade controls ascites by preventing abnormalization of lymphatic vessels in orthotopic human ovarian carcinoma models. Clin Cancer Res 17: 1415–1424.
Martin D, Galisteo R, Gutkind JS (2009) CXCL8/IL8 stimulates vascular endothelial growth factor (VEGF) expression and the autocrine activation of VEGFR2 in endothelial cells by activating NFkappaB through the CBM (Carma3/Bcl10/Malt1) complex. J Biol Chem 284: 6038–6042.
Mikeladze-Dvali T, Wernet MF, Pistillo D, Mazzoni EO, Teleman AA, Chen YW, Cohen S, Desplan C (2005) The growth regulators warts/lats and melted interact in a bistable loop to specify opposite fates in Drosophila R8 photoreceptors. Cell 122: 775–787.
Millimaggi D, Mari M, D' Ascenzo S, Giusti I, Pavan A, Dolo V (2009) Vasculogenic mimicry of human ovarian cancer cells: role of CD147. Int J Oncol 35: 1423–1428.
Muto E, Tabata Y, Taneda T, Aoki Y, Muto A, Arai K, Watanabe S (2004) Identification and characterization of Veph, a novel gene encoding a PH domain-containing protein expressed in the developing central nervous system of vertebrates. Biochimie 86: 523–531.
Pharoah PD, Tsai YY, Ramus SJ, Phelan CM, Goode EL, Lawrenson K, Buckley M, Fridley BL, Tyrer JP, Shen H, Weber R, Karevan R, Larson MC, Song H, Tessier DC, Bacot F, Vincent D, Cunningham JM, Dennis J, Dicks E, Aben KK, Anton-Culver H, Antonenkova N, Armasu SM, Baglietto L, Bandera EV, Beckmann MW, Birrer MJ, Bloom G, Bogdanova N, Brenton JD, Brinton LA, Brooks-Wilson A, Brown R, Butzow R, Campbell I, Carney ME, Carvalho RS, Chang-Claude J, Chen YA, Chen Z, Chow WH, Cicek MS, Coetzee G, Cook LS, Cramer DW, Cybulski C, Dansonka-Mieszkowska A, Despierre E, Doherty JA, Dork T, Du Bois A, Durst M, Eccles D, Edwards R, Ekici AB, Fasching PA, Fenstermacher D, Flanagan J, Gao YT, Garcia-Closas M, Gentry-Maharaj A, Giles G, Gjyshi A, Gore M, Gronwald J, Guo Q, Halle MK, Harter P, Hein A, Heitz F, Hillemanns P, Hoatlin M, Hogdall E, Hogdall CK, Hosono S, Jakubowska A, Jensen A, Kalli KR, Karlan BY, Kelemen LE, Kiemeney LA, Kjaer SK, Konecny GE, Krakstad C, Kupryjanczyk J, Lambrechts D, Lambrechts S, Le ND, Lee N, Lee J, Leminen A, Lim BK, Lissowska J, Lubinski J, Lundvall L, Lurie G, Massuger LF, Matsuo K, Mcguire V, McLaughlin JR, Menon U, Modugno F, Moysich KB, Nakanishi T, Narod SA, Ness RB, Nevanlinna H, Nickels S, Noushmehr H, Odunsi K, Olson S, Orlow I, Paul J, Pejovic T, Pelttari LM, Permuth-Wey J, Pike MC, Poole EM, Qu X, Risch HA, Rodriguez-Rodriguez L, Rossing MA, Rudolph A, Runnebaum I, Rzepecka IK, Salvesen HB, Schwaab I, Severi G, Shen H, Shridhar V, Shu XO, Sieh W, Southey MC, Spellman P, Tajima K, Teo SH, Terry KL, Thompson PJ, Timorek A, Tworoger SS, van Altena AM, van den Berg D, Vergote I, Vierkant RA, Vitonis AF, Wang-Gohrke S, Wentzensen N, Whittemore AS, Wik E, Winterhoff B, Woo YL, Wu AH, Yang HP, Zheng W, Ziogas A, Zulkifli F, Goodman MT, Hall P, Easton DF, Pearce CL, Berchuck A, Chenevix-Trench G, Iversen E, Monteiro AN, Gayther SA, Schildkraut JM, Sellers TA (2013) GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat Genet 45: 362–370,, 370e361-362.
Puy L, Maclusky NJ, Becker L, Karsan N, Trachtenberg J, Brown TJ (1995) Immunocytochemical detection of androgen receptor in human temporal cortex characterization and application of polyclonal androgen receptor antibodies in frozen and paraffin-embedded tissues. J Steroid Biochem Mol Biol 55: 197–209.
Qian J, Massion PP (2008) Role of chromosome 3q amplification in lung cancer. J Thorac Oncol 3: 212–215.
Ramakrishna M, Williams LH, Boyle SE, Bearfoot JL, Sridhar A, Speed TP, Gorringe KL, Campbell IG (2010) Identification of candidate growth promoting genes in ovarian cancer through integrated copy number and expression analysis. PLoS One 5: e9983.
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8: 2281–2308.
Sarbassov DD, Guertin DA, Ali SM, Sabatini DM (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307: 1098–1101.
Seftor RE, Hess AR, Seftor EA, Kirschmann DA, Hardy KM, Margaryan NV, Hendrix MJ (2012) Tumor cell vasculogenic mimicry: from controversy to therapeutic promise. Am J Pathol 181: 1115–1125.
Shathasivam P, Kollara A, Ringuette MJ, Virtanen C, Wrana JL, Brown TJ (2015) Human ortholog of Drosophila Melted impedes SMAD2 release from TGF-beta receptor I to inhibit TGF-beta signaling. Proc Natl Acad Sci USA 112: E3000–E3009.
Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE (2006) The consensus coding sequences of human breast and colorectal cancers. Science 314: 268–274.
Tan DS, Lambros MB, Rayter S, Natrajan R, Vatcheva R, Gao Q, Marchio C, Geyer FC, Savage K, Parry S, Fenwick K, Tamber N, Mackay A, Dexter T, Jameson C, Mccluggage WG, Williams A, Graham A, Faratian D, El-Bahrawy M, Paige AJ, Gabra H, Gore ME, Zvelebil M, Lord CJ, Kaye SB, Ashworth A, Reis-Filho JS (2009) PPM1D is a potential therapeutic target in ovarian clear cell carcinomas. Clin Cancer Res 15: 2269–2280.
Teleman AA, Chen YW, Cohen SM (2005) Drosophila melted modulates FOXO and TOR activity. Dev Cell 9: 271–281.
Tomayko MM, Reynolds CP (1989) Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother Pharmacol 24: 148–154.
Tuominen VJ, Ruotoistenmaki S, Viitanen A, Jumppanen M, Isola J (2010) ImmunoRatio: a publicly available web application for quantitative image analysis of estrogen receptor (ER), progesterone receptor (PR), and Ki-67. Breast Cancer Res 12: R56.
Ursini-Siegel J, Hardy WR, Zheng Y, Ling C, Zuo D, Zhang C, Podmore L, Pawson T, Muller WJ (2012) The ShcA SH2 domain engages a 14-3-3/PI3'K signaling complex and promotes breast cancer cell survival. Oncogene 31: 5038–5044.
Vermeulen PB, Gasparini G, Fox SB, Colpaert C, Marson LP, Gion M, Belien JA, De Waal RM, Van Marck E, Magnani E, Weidner N, Harris AL, Dirix LY (2002) Second international consensus on the methodology and criteria of evaluation of angiogenesis quantification in solid human tumours. Eur J Cancer 38: 1564–1579.
Waugh DJ, Wilson C (2008) The interleukin-8 pathway in cancer. Clin Cancer Res 14: 6735–6741.
Whipple CA, Brinckerhoff CE (2014) BRAF(V600E) melanoma cells secrete factors that activate stromal fibroblasts and enhance tumourigenicity. Br J Cancer 111: 1625–1633.
Ylipaa A, Hunt KK, Yang J, Lazar AJ, Torres KE, Lev DC, Nykter M, Pollock RE, Trent J, Zhang W (2011) Integrative genomic characterization and a genomic staging system for gastrointestinal stromal tumors. Cancer 117: 380–389.
Zhang YE (2009) Non-Smad pathways in TGF-beta signaling. Cell Res 19: 128–139.
Acknowledgements
Funded by CIHR grants MOP74726 and MOP106679 and NSERC grant RGPIN312475-11. We thank Suan Chin Yeo for lively discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Shathasivam, P., Kollara, A., Spybey, T. et al. VEPH1 expression decreases vascularisation in ovarian cancer xenografts and inhibits VEGFA and IL8 expression through inhibition of AKT activation. Br J Cancer 116, 1065–1076 (2017). https://doi.org/10.1038/bjc.2017.51
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2017.51
Keywords
This article is cited by
-
A 38-gene model comprised of key TET2-associated genes shows additive utility to high-risk prostate cancer cases in the prognostication of biochemical recurrence
BMC Cancer (2020)
-
Ventricular Zone Expressed PH Domain Containing 1 (VEPH1): an adaptor protein capable of modulating multiple signaling transduction pathways during normal and pathological development
Cell Communication and Signaling (2019)