ABSTRACT
Mouse sp56 is considered as one of the candidates for mouse zona pellucida 3 (mZP3) receptor. Up to date, its homologue has only been cloned from guinea pig, namely AM67. Based on the cDNA sequence of mouse sp56, we designed a pair of primer to amplify its homologue from rat testis cDNA. Using RT-PCR, two fragments of 743 bp and 938 bp were amplified. The PCR products show very high homology to mouse sp56. However, the 743 bp product completely lacks one of the seven Sushi domains of mouse sp56. Using the 743 bp product as the probe to detect the expression profile of rat sp56 in rat tissues, Northern blot shows that a ∼ 2.0 kb mRNA expresses specifically in testis. Employed the RACE method, two full cDNA sequences of rat sp56 were obtained. A Mr ∼ 42 KD band was detected in denatured and non-reducing protein sample of rat testis and sperm with anti-mouse sp56 monoclonal antibody by Western blot method. Rat sp56 was localized on rat sperm head by the indirect immunofluorescence method. Rat sp56 immunoreactivity was detected from the early pachytene spermatocytes and throughout the spermatogenesis. Its cloning will further our understanding of the mechanism of the sperm-egg recognition and binding.
Similar content being viewed by others
INTRODUCTION
To fertilize an egg, mammalian sperm must bind tightly to the zona pellucida. The sperm-egg binding process, in a species -specific manner, is mediated by ZP3 and its receptor on the sperm1. Upon binding to its receptor, ZP3 induces the acrosome reaction, a membrane fusion event in which plasma membrane of the sperm head fuses with the acrosome2.
Up to date, several candidates for mZP3 receptor have been proposed. These candidates include β-1,4 galactosyltransferase3, a 95-KD phosphotyrosine-bearing protein with hexokinase activity4, and mouse sp565, 6. Mouse sp56 was first identified to be responsible for recognition of and binding to mZP3 by crosslinking and affinity chromatography5. The affinity purified sp56 has coordinates of 40 KD under non-reducing conditions and 56 KD under reducing conditions when subjected to 2-D SDS-PAGE6. The subsequent cDNA cloning demonstrated that this protein belonged to the complement 4B-binding protein superfamily7. As initially reported, mouse sp56 was localized to the plasma membrane of acrosome-intact sperm heads6. The latest evidence shows that the molecular weight of mouse sp56 in cauda epididymal sperm approximates 67 KD, and it's a component of the acrosomal matrix but not of the plasma membrane8, 9. The previously reported Mr ∼ 56 KD may be a C-terminal truncation form of mouse sp56 because of the proteolysis during the protein extraction process. During the spermatogenesis, mouse sp56 did decrease in size because of the differential oligosaccharide processing with polypeptide backbone conserved8.
Sperm-egg binding is inhibited by the membrane vesicles from the acrosome-intact sperm, which is relieved by the monoclonal antibody against mouse sp56. In addition, the evidence that mouse sp56 and mZP3 coimmunoprecipatate with each other indicates their specific association or interaction2.
The presence or absence of sp56 on sperm from different species accounts for species specificity of sperm-egg recognition7. Up to date, the only cloned orthologue of mouse sp56 was AM67 from guinea pig9. The present study is to clone its homologue from rat testis. Unlike the mouse sp56 or guinea pig AM67, rat sp56 has two transcripts, one of which completely lacks one of the seven Sushi domains of mouse sp56. And it expresses in the early stage of spermatogenesis and localizes on rat sperm head.
MATERIALS AND METHODS
Materials
The following chemicals and reagents were purchased from Sigma Chemical Co.: Bovine Serum albumin (BSA, Fraction V), EDTA, Tween-20, PMSF, FITC-conjugated goat anti-mouse IgG secondary antibody, HRP-conjugated goat anti-mouse IgG secondary antibody, AEC substrate, PerfectHybTM Plus Hybridization Buffers, and rat Multiple Tissue Northern Blots. Trizol and Superscript II Reverse Transcriptase were purchased from GIBCO-BRL. Mouse anti-sp56 monoclonal antibody (clone mAb7C5) from Biodesign. The random prime labeling system was from Amersham Pharmacia Biotech. Smart RACE cDNA Amplification Kit was from Clontech. TaKaRa Ex Taq™ was from TaKaRa. pGEM T-Easy vector was from Promega.
Preparation of protein extracts from rat sperm, rat testis and mouse testis
Sperm from the caudal epididymis of 3-month old Wistar male were released into PBS prewarmed to 37°C. After washing the sperm with PBS twice, the sperm pellet was suspended in Lammli buffer without DTT and boiled for 5 min. After centrifugation at 12000 rpm for 5 min, the supernatant was collected as the total protein extracts of rat sperm. The rat and mouse testis were homogenized in 50 mM Tris.Cl (pH 7.5), 5 mM EDTA and 1 mM PMSF. Equal volume of 2 × Lammli buffer without DTT was added to each homogenization and boiled for 5 min. After centrifugation at 12000 rpm for 5 min, the supernatant was collected as the total protein extracts of rat and mouse testis.
RNA extraction and RT-PCR
Total RNA from rat testis was extracted using Trizol and isolated according to the procedure supplied by the manufacture. Reverse transcription was carried out according to the manufacture's instructions (GIBCO-BRL). The first strand of cDNA was generated from 5 μ g of total RNA prepared from rat testis using oligo-dT primer and Superscript II Reverse Transcriptase.
The rat sp56 primers were designed according to the cDNA sequence of mouse sp56. Forward primer: TGAGAATGGTTCATGGGTGC; reverse primer: CTCCTTCCATCCGTAGACATA. PCR was performed initially by denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, extension at 72°C for 1 min, and a final extension step at 72°C for 10 min. Amplified PCR products were electrophoretically fractioned on 1% agarose gels, cloned and sequenced.
Western blot
Protein extracts from rat sperm, rat testis and mouse testis were analyzed by electrophoresis on 10% SDS-PAGE gel, and transferred onto a NC membrane. The membrane was blocked for 2 h with TBS containing 0.1% Tween 20 (TBST) and 5% dehydrated skim milk. Blots was then incubated overnight at 4°C in the presence of monoclonal antibody to mouse sp56 (clone mAb7C5, Biodesign). The blots were washed 3 × 15 min with TBST, and incubated with HRP-conjugated goat anti-mouse IgG secondary antibody at room temperature for 1 h. After washing 4 × 15 min with TBST, the bound anti-sp56 antibody was detected by visualization using Supersignal West Pico Chemiluminescent Substrate (Pierce).
Indirect immunofluoresence staining
Both live, unfixed rat sperm and fixed rat sperm were used in indirect immunofluorescence. Rat sperm were collected from the caudal epididymis and fixed in 4% paraformaldehyde in PBS for 30 min at 4°C. After three washes with TBS, the cells were incubated with mAb7C5 in 1% BSA in TBS at a 100-dilution overnight at 4°C. After washing with TBS three times, the sperm were incubated with FITC-conjugated goat anti-mouse IgG secondary antibody (1:400 dilution) for 1 h at room temperature and examined by fluorescence microcope.
Northern blot
The rat multiple tissues Northern blots were purchased from Sigma. The probe was the 743 bp RT-PCR product that had been sequenced to be homologous to mouse sp56. The probe was labeled using the random prime labeling system (Amersham Pharmacia Biotech). Hybridization was carried out with PerfectHybTM Hybridization Buffers Plus (Sigma) at 65°C for 3 h. The blots were washed in 2 × SSC, 0.1% SDS at 25°C for 30 min followed by 0.5 × SSC, 0.1% SDS at 50°C for 10 min, and exposed to Kodak BioMax MR film at −80°C.
RACE-PCR
The full-length cDNA of rat sp56 was obtained using a Smart RACE cDNA Amplification Kit (Clontech). The 5′and 3′ RACE-ready cDNA was prepared using mRNA isolated from 3-month rat testis according to the procedure supplied by the manufacture. RACE reactions were done using 5′ or 3′ RACE-ready cDNA with a polymerase (TaKaRa Ex Taq™) suitable for amplification of long regions of DNA with high fidelity. The PCR conditions were set and performed according to the protocols given by Clontech. Four oligonucleotides were used, two for 3′ RACE and two for 5′ RACE using a nested PCR procedure to obtain an increased specificity. For 3′ RACE, the external primer was GSS1 (5′TGGCTCCTGTTCTTGGTGACTGTGGACC 3′), and the internal one was GSS2 (5′CTTGGTGACTGTGGACCTCCACCC 3′). For 5′ RACE, the external primer was GSA1 (5′CACATTCTGGGAGAGAATCACTCC 3′), and the internal one was GSA2 (5′GCGAACGGTAAACTGGGTGGAGGTCC 3′).
The amplification products of 3′ and 5′ RACE were cloned into pGEM T-Easy vector (Promega) and sequenced. Ten clones were sequenced both for 3′ and 5′ RACE amplification products.
Immunohistochemical localization
Immunohistochemical staining was performed by the indirect HRP-conjugate method on 10 μ m-thick frozen sections of rat testis tissue. The sections were treated with 10% methanol containing 0.3% hydrogen peroxide for 30 min at room temperature to reduce endogenous peroxidase activity. Following washes (3 × 10 min) in 0.1 M PBS, the sections were incubated for 1 h in 1% normal goat serum. Sections were then incubated with mAb7C5 for overnight at 4°C. Following washes, sections were incubated with HRP-conjugated goat anti-mouse IgG secondary antibody at room temperature for 2 h. After washes in 0.1 M PBS, bound antibodies were detected with AEC substrate.
RESULTS
Rat sp56 cloning
Based on mouse sp56 cDNA sequence, primers were designed to amplify rat sp56 from rat testis cDNA. With the use of primer, two PCR products of 938 bp and 743 bp were obtained (Fig 1). DNA sequencing analysis showed that both PCR products were homologous to mouse sp56. The 743 bp fragment differs only from the 938 bp fragment in that it lacks the third Sushi domain of the mouse sp56, which contains seven Sushi domains7.
Electrophoresis analysis of rat sp56 cDNAs prepared from rat testis RNAs by RT-PCR. Rat testis total RNA was reverse-transcribed and PCR was performed using primers designed according to the cDNA sequence of mouse sp56. The lengths of the products were 938 bp and 743 bp respectively. Te: testis. M: 100 bp ladder marker.
Based on the sequence of the two RT-PCR products, 5′ and 3′ primers were designed to amplify the full-length sequence by the 5′ RACE and 3′ RACE method. 3′ RACE generated two products of 1.6 kb and 1.8 kb. 5′ RACE generated a 390 bp product. Two full-length rat sp56 sequences of 2021 bp and 1826 bp, namely rat sp56 (L) and rat sp56 (S) respectively, were generated based on the 5′ RACE and 3′ RACE products (Fig 2). Like the difference between the two RT-PCR products, the analysis of the two full-length sequences showed that rat sp56 (S) lacks the third Sushi domain of mouse sp56. These results show that there are two rat sp56 transcripts in rat testis. Both full-length sequences encoded 88 nucleotides of 5′-untranslated region and 199 nucleotides of 3′-untranslated region. A possible poly (A) signal TATAAA was found at 1915-1921 positions of rat sp56 (L), but at 1810-1816 positions of rat sp56 (S) (Fig 2).
Cloning of cDNA encoding rat sp56 (L), sp56 (S). 2A (upper): rat sp56 (S); 2B (below): rat sp56 (L). The deduced amino acid is shown below the nucleotide sequence numbered in the 5′ to 3′ direction. The translation initial site (ATG) is underlined; the stop codon (TAA) is double underlined; the polyadenylation signal (TATAAA) is underlined. The putative Sushi domains I-VI in rat sp56 (S) or I-VII in rat sp56 (L) are indicated under the deduced amino acids. Potential sites of asparagine-linked glycosylation are indicated by asterisk at amino acids.
Rat sp56 (L) encodes 577 amino acids, and rat sp56 (S) encodes 512 amino acids. While mouse sp56 is composed of 579 amino acids. Among the 577 amino acids of rat sp56, 449 residues are identical to, and 506 residues are related to that of mouse sp56. The very carboxyl terminal of rat sp56 shows low homology to that of mouse sp56. Among the 26 carboxyl terminal amino acids, only 9 residues are identical (Fig 3). Rat sp56 (L) and rat sp56 (S) have twelve, ten consensus sites for asparagine-linked glycosylation respectively (Fig 2).
Like mouse sp56 and guinea pig AM67, rat sp56 contains the repetitive structure called Sushi domain or short consecus repeat (SCR)7, 9. However, unlike mouse sp56 and guinea pig AM67, both of which have four conservative cystine residues in each Sushi domain, only four out of seven Sushi domains of rat sp56 (L) have the four conservative cystine residues, and the other three Sushi domains, the first, fourth and the fifth Sushi domains have only three cystine residues. Among the three Sushi domains lacking one of the conservative cystine residues, the first and the fifth Sushi domain lack the fourth or the last cystine residue; the fourth Sushi domain lacks the first cystine residue. It must be noted that rat sp56 (S) completely lacks the third Sushi domain of mouse sp56 or rat sp56 (L) (Fig 2).
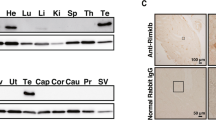
Using the 743 bp product as the probe to detect the expression profile of rat sp56 in rat tissues, Northern blot shows that a ∼ 2.0 kb mRNA expresses specifically in testis (Fig 4). This result didn't detect two expected bands of rat sp56 (L) and rat sp56 (S). The reason may be the small size difference between rat sp56 (L) and rat sp56 (S).
Northern Blot analysis of rat sp56 mRNA in rat mutiple tissues. The blot was hybridized with rat sp56 cDNA fragment (obtained from RT-PCR) radiolabeled by random priming. Lane 1. Brain; 2. Heart; 3. Liver; 4. Kidney; 5. Spleen; 6. Testis; 7. Lung; 8. Thymus; 9. Placenta; 10. Muscle. Numbers refer to molecular mass (Kb) based on RNA stand.
Western blot
Western blot of rat testis and sperm protein samples, using a monoclonal antibody directed against sp56 (mAb7C5), revealed that sp56 expressed in rat testis and sperm (Fig 5). And under non-reducing conditions, both mouse sp56 and rat sp56 have an apparent Mr of ∼ 42 KD. This result is consistent with that of Cohen and Wassarman2.
Western blot analysis of proteins prepared from mouse testis, rat testis and rat sperm detected with mAB7C5. Rat sperm protein (Sp) were extracted in Lammli sample buffer without DTT. Rat testis protein (rT) and mouse testis protein (mT) were prepared as described under “materials and methods”. Each lane contained 100 μ g of protein separated by SDS-PAGE. Note under denatured and non-reducing conditions, both mouse and rat sp56 have an apparent Mr of ∼42 KD. In addition, mouse sp56 can dimerize under denatured and non-reducing conditions. Numbers refer to molecular mass (KD) based on protein stand.
Localization of rat sp56 on the rat sperm
Using the indirect immunofluorescence technique, rat sp56 was located on the head of rat sperm (Fig 6). Almost all of the fixed sperm with intact acrosome exhibited fluorescence, but no staining was observed in live unfixed sperm (data not shown). Control sperm incubated with an equal concentration of nonspefic whole IgG showed no staining (data not shown). When the rat sperm were extracted with 50 mM sodium acetate, pH 5.2, containing 0.15 mM NaCl, 0.625% Triton X-100, 5% sucrose according to the method of Kye et al8. The native architecture of the intact acrosome is maintained after the detergent extractiooon, which made rat sp56 sp56 detectable in unfixed sperm head (data not shown). This result is in agreement with that of Kye et al, which shows that mouse sp56 is not located on the mouse sperm membrane but resides in the acrosomal matrix8.
Indirect immunofluorescence staining of paraformaldehyde-fixed rat sperm with mAB7C5. Shown are corresponding immunofluorescence (A) and phase-contrast (B) micrographs of mature rat sperm immunostained with anti-sp56 monoclonal antibody clone 7C5. Note localization of staining to sperm head. Magnification × 1000.
Immunohistochemical localization
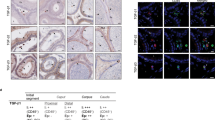
Rat sp56 immunoreactivity was detected from the early pachytene spermatocytes and throughout the spermatogenesis (Fig 7). It doesn't express uniformly in the spermatocytes but concentrates to some region of plasma, which may results in the absence of staining in some spermatocytes of the section. And its immunoreactivity is localized to the head of elongating spermatids. Control section incubated with an equal concentration of nonspefic whole IgG showed no staining (data not shown).
Immunohistochemical localization of rat sp56 in rat testis. Section was incubated with anti-sp56 antibody followed by overlaying with corresponding HRP-labeled secondary antibody. Note the absence of staining in some spermatocytes (indicated with arrowhead), whereas the staining is apparent in the head of elongating spermtids (indicated with arrow).
DISCUSSION
Mouse sp56 was first identified to be responsible for recognition of and binding to mZP35. Sperm-egg binding is inhibited in the presence of Anti-sp56 antibody2, 7. The inhibitory effect can be relieved by the acrosome vesicles containing mouse sp562. The coimmunoprecipatation of mouse sp56 and ZP3 directly demonstrates their specific association or interaction with each other2. These evidences demonstrate mouse sp56 plays an important role in sperm-egg binding process.
Previous research detected mouse sp56-like transcripts in hamster testis but not in guinea pig testis7. This is consistent with the fact that hamster sperm can bind to mZP10. Given the specificity of sperm-egg recognition and binding is mediated by ZP3 and its receptor, such as mouse sp56 in mouse, it is reasonable that other mouse sp56-like proteins exist in other species. AM67 from guinea pig is the first orthologue of mouse sp56 that have been cloned9. The present research was conducted to clone the orthologue of mouse sp56 from rat testis. It is very surprising that rat sp56 transcribes into two isoforms of rat sp56 (L) and rat sp56 (S), with the latter lacking the counterpart of Sushi domain 3 of mouse sp56.
Because mAb7C5 recognizes a sp56 epitope, the secondary or tertiary structure after denaturation in SDS, but only when intracellular disulfides remain intact6. There are may be two rat sp56 proteins due to two sp56 transcripts in rat testis, but mAb7C5 detected only one band of 42 KD in rat testis and sperm protein samples. The reason is either the epitope recognized by mAb7C5 lies in the third Sushi domain or related to the Sushi domain, the absence of which in rat sp56 (S) altered its secondary or tertiary structure otherwise recognized by mAb7C5; or rat sp56 (S) doesn't express in rat testis or expresses in a low level undetectable by western blot. The result of western blot showed that mouse sp56 dimerizes under non-reducing condition. But rat sp56 doesn't dimerize even under non-reducing condition. The cystine residues have been shown to be important in establishing and maintaining the three-dimensional structure of SCR -containing proteins, such as the complement regulatory protein superfamily11. The disulfide bonds are localized within the Sushi domain, with the first and third and the second and fourth half-cystines in each SCR being linked. The pattern of disulfide may confer to the SCR-containing protein a conformation that allows the assembly of the SCR units in a tandem fashion. Such an arrangement of the polypeptide chains may partially explain the aggregation of the SCR-containing proteins11. The absence of one cystine residue of the four cystine residues in rat sp56 Sushi domain 1, 4, 5 may account for the absence of dimerization or aggregation of rat sp56.
The expression profile of rat sp56 is different from that of mouse sp56, which expresses in late stages of spermatogenesis. Mouse sp56 is an acrosomal matrix component. In the late pachytene spermatocytes, it is first detectable in a cluster of small proacrosomal granule; in the haploid round spermatids, mouse sp56 was found with the developing acrosomal granule; in condensing spermatids and epididymal sperm, mouse sp56 was localized in the acrosome. And its polar localization pattern may be related to the structure alterations of the acrosome that occur during the spermatogenesis8.
It remained to determine whether rat sp56 (L) and sp56 (S) express in different spermatocytes (as seen in the result of Immunohistochemical staining, a small part of spermatocytes don't show rat sp56 immunoreactivity) or coexist in one spermatocyte. In addition, the temporal expression profile of rat sp56 needs further investigation.
Because of the very similarity of rat sp56 and mouse sp56, it's reasonable to deduce that rat sp56 is the receptor of rat ZP3, and that their interaction mediates the recognition and binding of rat sperm and egg. Nonetheless, the exact role of rat sp56 (L) and sp56 (S) needs to be determined. The question is raised whether their function overlap, are complement or independent of each other? And because our previous research focus on the molecules relevant to the acrosome reaction of rat sperm12, 13, we believe that with the cloning of rat sp56, we will have a further understanding of rat fertilization.
References
Wassarman PM, Jovine L, Litscher ES . A profile of fertilization in mammals. Nat Cell Biol 2001; 3(2):E59–64.
Cohen N, Wassarman PM . Association of egg zona pellucida glycoprotein mZP3 with sperm protein sp56 during fertilization in mice. Int J Dev Biol 2001; 45(3 Spec No)(3–Spec–No):569–76.
Miller DJ, Macek MB, Shur BD . Complementarity between sperm surface beta-1, 4-galactosyltransferase and egg-coat ZP3 mediates sperm-egg binding. Nature 1992; 357(6379):589–93.
Kalab P, Visconti P, Leclerc P, Kopf GS . p95, the major phosphotyrosine-containing protein in mouse spermatozoa, is a hexokinase with unique properties. J Biol Chem. 1994; 269(5):3810–7.
Bleil JD, Wassarman PM . Identification of a ZP3-binding protein on acrosome-intact mouse sperm by photoaffinity crosslinking. Proc Natl Acad Sci USA 1990; 87(14):5563–7.
Cheng A, Le T, Palacios M, et al. Sperm-egg recognition in the mouse: characterization of sp56, a sperm protein having specific affinity for ZP3. J Cell Biol 1994; 125(4):867–78.
Bookbinder LH, Cheng A, Bleil JD . Tissue- and species-specific expression of sp56, a mouse sperm fertilization protein. Science 1995; 269(5220):86–9.
Kim KS, Cha MC, Gerton GL . Mouse sperm protein sp56 is a component of the acrosomal matrix. Biol Reprod 2001; 64(1):36–43.
Foster JA, Friday BB, Maulit MT, et al. AM67, a secretory component of the guinea pig sperm acrosomal matrix, is related to mouse sperm protein sp56 and the complement component 4-binding proteins. J Biol Chem 1997; 272(19):12714–22.
Schmell ED, Gulyas BJ . Mammalian sperm-egg recognition and binding in vitro. I. Specificity of sperm interactions with live and fixed eggs in homologous and heterologous inseminations of hamster, mouse, and guinea pig oocytes. Biol Reprod 1980; 23(5):1075–85.
Janatova J, Reid KB, Willis AC . Disulfide bonds are localized within the short consensus repeat units of complement regulatory proteins: C4b-binding protein. Biochemistry 1989; 28(11):4754–61.
Hu JH, Yan YC . Identification of gamma1 subunit of GABA(A) receptor in rat testis. Cell Res 2002; 12(1):33–7.
Hu JH, He XB, Yan YC . Identification of gamma-aminobutyric acid transporter (GAT1) on the rat sperm. Cell Res 2000; 10(1):51–8.
Acknowledgements
This project was supported by the grant from National Key Basic Research Project, “973”, No. G199905592.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
HE, X., YAN, Y., LI, Y. et al. Cloning of rat sp56, the homologue of mouse sperm ZP3 receptor–sp56. Cell Res 13, 121–129 (2003). https://doi.org/10.1038/sj.cr.7290156
Issue Date:
DOI: https://doi.org/10.1038/sj.cr.7290156
Keywords
This article is cited by
-
Acrosome markers of human sperm
Anatomical Science International (2016)