ABSTRACT
We cloned cDNAs for Xenopus aldolases A, B and C. These three aldolase genes are localized on different chromosomes as a single copy gene. In the adult, the aldolase A gene is expressed extensively in muscle tissues, whereas the aldolase B gene is expressed strongly in kidney, liver, stomach and intestine, while the aldolase C gene is expressed in brain, heart and ovary. In oocytes aldolase A and C mRNAs, but not aldolase B mRNA, are extensively transcribed. Thus, aldolase A and C mRNAs, but not B mRNA, occur abundantly in eggs as maternal mRNAs, and strong expression of aldolase B mRNA is seen only after the late neurula stage. We conclude that aldolase A and C mRNAs are major aldolase mRNAs in early stages of Xenopus embryogenesis which proceeds utilizing yolk as the only energy source. aldolase B mRNA, on the other hand, is expressed only later in development in tissues which are required for dietary fructose metabolism. We also isolated the Xenopus aldolase C genomic gene (ca. 12 kb) and found that its promoter (ca. 2 kb) contains regions necessary for tissue-specific expression and also a GC rich region which is essential for basal transcriptional activity.
Similar content being viewed by others
INTRODUCTION
Studies on the temporally- and spacially-regu- lated expression of multilocus-type isozyme genes are important for understanding control mechanisms underlying growth and differentiation of cells in development of the animal embryo1. In Xenopus, L-lactate dehydrogenase (LDH) consists of two major [LDHA (muscle-type) and LDHB (liver-type)] and one minor [LDHC (oocyte-type)] subunits, and these subunits, coded for by different genes, are expressed in a tissue-specific manner during embryogenesis2. Creatine kinase consists of two (in tetrapods) or four (in amphibia and teleost fish) different subunits, and expression of these subunits also occurs in tissue-specific and developmentally-regulated manners3. Here, we review our recent studies on properties and spatial and temporal regulation of Xenopus aldolase genes in developing embryos and tissues.
aldolases as materials for studies on differential gene expression
aldolase, or fructose-1, 6-bisphosphate aldolase (EC 4, 1, 2, 13), provides a typical example of developmentally-controlled multilocus-type isozymes. Its functional form is either homo- or hetero-tetramer composed of two of the three different ca. 40,000-dalton proteins (A, B and C), each coded for by different genes4, 5. Holoenzymes catalyse different reactions depending on the nature of the constituent subunits. Basically, aldolases A and C catalyze the cleavage of fructose-1, 6-bisphosphate in the glycolytic pathway, giving rise to glyceraldehyde-3-phosphate and dihydroacetate phosphate4, 6, whereas aldolase B catalyses cleavage of fructose-1-phosphate derived from dietary carbohydrates to give rise to glyceraldehyde and dihydroacetate phosphate6, 7, 8. In addition, aldolase B catalyses gluconeogenesis or the hexose formation7, 8.
Interestingly, expression properties of aldolase subunit genes (A, B and C) differ according to the specialized roles which they perform. Thus, aldolase A is expressed mainly in muscles and other tissues with a high rate of glycolysis9, and aldolase C is expressed specifically in brain and nervous tissues10. By contrast, aldolase B, adapted to dietary fructose metabolism and gluconeogenesis, is expressed in liver, kidney and intestine which are active in these pathways4, 7, 8. As a result, mutation of this gene in humans causes the severe hereditary congenital disease fructose intolerance11, 12.
We have been interested in control mechanisms of the expression of the three aldolase genes in Xenopus embryogenesis and we asked which of the three aldolase genes (so-called muscle-type A, liver-type B and brain-type C) are utilized in early embryonic development, during which an unicellular and totipotent egg cell cleaves rapidly and becomes a multicellular embryo composed of many differentiated cells. We also cloned a genomic DNA of the aldolase C gene and characterized its promoter function.
Cloning of Xenopu s aldolases A, B and C cDNAs
We first attempted cloning and expression studies of aldolase A, B and C cDNAs, and we obtained XALDA, XALDB and XALDC. Fig 1 shows amino acid sequences of XALDA, XALDB and XALDC (respectively 1.5, 1.8 and 1.8 kb in length). As for the number of amino acids deduced from cDNAs there was no difference in all the three Xenopus aldolases (A, B and C) (Fig 1, left, top). The overall identities at the amino acid sequence level between Xenopus aldolases A and B and between B and C were lower (67 and 68% respectively) than that between aldolases A and C (left, top) as in rats10, 13 and humans14. In Fig 1 (left, bottom), we marked 4 isozyme group-specific regions (IGS I to IGS IV), which are conserved in each group of aldolases (A, B and C). We differently marked two of these (IGS I and IGS IV) (Fig 1, left, top), since these are particularly important for aldolase B-specific function (utilization of fructose-1-phosphate)15. Furthermore, we marked 7 regions which are highly or relatively highly conserved in all three types of aldolases5. Sequences in 4 IGSs were considerably different among the three Xenopus aldolases (A, B and C) (less than 50% identity) (Fig 1, left, top), yet those in 7 conserved regions were similar (between A and B, 85.0%; B and C, 85.0%; A and C, 93.8% in amino acid identity). Xenopus aldolases A, B and C, contained Lysine at amino acid number 230 (K; orange-colored in Fig 1, left, top) as an active center4. The three aldolases have a similar three dimensional structure (Fig 1, right).
Structure of Xenopus aldolase A, B and C. (left, top): Amino acid sequences. {from Kajita et al26}. (left, bottom): Charasteristic features of the three aldolases. The regions(1–7) highly conserved among all the three types of aldolases are indicated gray. Isozyme group-specific sequences (IGS) are indicated three colours; A-type specific as red, B-type as yellow, C-type as blue. Sequences which are not conserved are not coloured. Δ(right): Steric structure of aldolase A. Red regions are IGSs. The highly conserved regions among all types of aldolases are indicated by green. Gray-coloured regions are low conserved sequences. The orange portion is the active center. N-terminus and C-terminus are shown.
The phylogenetic tree constructed by the neighbor-joining method showed that the ancestral gene of aldolase B genes segregated from the ancestral gene of aldolase A and C genes before segregation of aldolase A and C genes in Xenopus as in other animal species10, 16. We obtained a cell line from Xenopus adult kidney and Fluorescence in situ hybridization (FISH) studies showed that the three aldolase genes are located on different chromosomes17.
Expression of three aldolase mRNAs in adult frogs
Northern blot analysis was done using RNAs extracted from various Xenopus adult tissues. We obtained a very strong aldolase A mRNA signal of ca. 1.8 kb in muscle, a moderately strong signal in heart, brain and intestine, and low levels of expression in all the other tissues, probably due to the transcription from the minor “pan-expression promoter”18. aldolase B mRNA appeared as a signal of ca. 1.8 kb in kidney, liver, intestine, stomach and interestingly, in skin, as noted for humans14, rat19, sheep20, sea bream (Sparus aurata)21 and salmon 22. The expression in skin is as in salmon22. We obtained a strong signal of aldolase C mRNA of ca. 2 kb in heart, brain and ovary, and a weak signal in intestine, testis, lung and other tissues.
Using in vitro transcribed aldolase A, B and C mRNAs as references, northern blot analyses were done and calibration curves for determinations of absolute amounts of aldolase A, B and C mRNAs in each adult tissue (we estimated the amounts permg of total RNA). We then calculated the percent composition of the three aldolase mRNAs (A, B and C) for each tissue and expressed the results as circle graphs (Fig 2). The total amount of three aldolase mRNAs (A plus B plus C) per unit amount (1mg) of the total RNA is largest in muscle (270 pg/mg total RNA), and next largest in heart (120 pg/mg total RNA), due to the occurrence of an extremely large amount of aldolase A mRNA in muscle and aldolase C mRNA in heart. This result is reasonable in view of the fact that these two tissues are most active in energy consumption. It is interesting here that skeletal muscles express almost exclusively aldolase A but the heart (cardiac muscles) expresses much more aldolase C mRNA than aldolase A mRNA. aldolase A mRNA constituted as much as 99% of the total aldolase mRNAs in muscle, whereas aldolase C mRNA constituted 70% and 60%, respectively, of the total aldolase mRNAs in heart and brain. Thus, the percent composition of three aldolase mRNAs in brain, a typical ectodermal tissue, is similar to that in heart, a typical mesodermal tissue. aldolase B mRNA, on the other hand, constitutes the major aldolase mRNA in liver and kidney, and also in stomach and intestine, again reflecting the fact that aldolase B is actively expressed in tissues which are necessary for metabolism of dietary fructose7.
aldolase mRNA expression in various adult organs {from Kajita et al17. Sizes of circles indicate the relative amount of aldolase mRNAs which is comparable among tissues.
It is worth pointing out that the kidney, which is mesodermal in origin, has an aldolase mRNA composition almost identical to that of the liver, which is endodermal in origin: In both of these tissues aldolase B mRNA constituted ca. 85% of the total aldolase mRNAs, the rest being aldolase A mRNA with only a trace amount of aldolase C mRNA. In intestine and stomach, which are both endodermal, the percent composition of aldolase B mRNA exceeded 50%, and skin which is ectodermal also strongly expressed aldolase B mRNA albeit the amount being slightly lower than 50% of the total aldolase mRNAs. Thus, each tissue has its characteristic aldolase mRNA composition, thereby reflecting functional differences between the three aldolases, rather than differences in developmental origin of each tissue.
Expression of three aldolase mRNAs in oocytes and embryos
Northern blot analyses were made using RNAs from oocytes. aldolase A mRNA occurred as a major aldolase mRNA and the level increased greatly as the oocyte grew, and the increased level was maintained during maturation. We detected no signal of aldolase B mRNA over the background level throughout oogenesis and oocyte maturation. We obtained a substantial amount of aldolase C mRNA, the level increased throughout oogenesis and remained constant during maturation.
Northern blot analysis were made using RNAs from embryos at different stages and in vitro transcribed three aldolase mRNAs. Using the calibration curves to correlate strengths of northern blot signals to the amounts of aldolase RNAs, we here again estimated the absolute amount of each of the three aldolase mRNAs in embryos. A Xenopus fertilized egg contained approximately 60 pg of aldola se A mRNA, 45 pg of aldolase C mRNA but only 1.5 pg or less aldolase B mRNA. Thus, aldolase A and C mRNAs together constitute 99% of the total egg aldolase mRNAs (A plus B plus C), and aldolase mRNAs constitutes altogether ca. 0.0025% of the total egg RNA, since a Xenopus egg contains ca. 4pg of RNA a major portion of which is rRNA23, 24. During cleavage the amount of aldolase A mRNA per embryo decreases to a level as low as 15 pg/embryo (at the late gastrula stage). From the early neurula stage, however, the level increased sharply to a level as high as 600 pg/embryo at the tadpole stage (stage 35). The period of this great increase in the amount of aldolase A mRNA corresponds to the period of somitegenesis. The amount of aldolase C mRNA per embryo also decreased sharply until the gastrula stage (ca. 3 pg/ μmbryo). Unlike that of aldolase A mRNA, however, the post-gastrular increase in the amount of aldolase C mRNA was not large and the level remained at 50 pg/embryo even at stage 35 (tadpole stage). The very low level of aldolase B mRNA per embryo was maintained until the midgastrula stage (stage 11). At the late gastrula stage (stage 13), however, a slight increase was detected, and from the late neurula stage (stage 22) on, the amount continuously increased and eventually reached the level as high as 140 pg/embryo at the tadpole stage (stage 35). Based on the quantitative data, we calculated the percent composition of three aldolase mRNAs for embryos at different stages (Fig 3). It is apparent that the A plus C-type aldolase composition is “egg-specific” or “early embryo-specific”. Probably, this aldolase composition is suitable for fructose metabolism during the yolk-dependent developmental stages, during which aldolase B is probably not necessary. aldolase B mRNA, on the other hand, is expressed only in later stages so that embryos get ready to metabolize dietary fructose at the feeding stages.
Expression of three aldolase mRNAs in developing embryos from Kijita et al17. Size of the circle is comparable among different stages. The total amount ofaldolase mRNAs in an egg is 106.5 pg.
Tissue-specific expression of aldolase genes in embryos
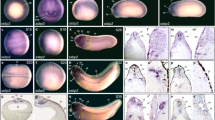
According to whole mount in situ hybridization, the earliest stage at which we detected localized signals for aldolase A, B and C mRNA was stage 16 (early neurula stage), 25 (late-neurula stage), and 21 (neurula stage), respectively25, 26. Throughout later stages, the aldolase A mRNA signal was noted in somites. At the tailbud stage the aldolase B mRNA signal appeared in regions where liver rudiments and proctodeum are formed. After hatching, aldolase B mRNA was strongly detected in pronephros, liver rudiments and hindgut, and at this stage, the tail fin (epidermis) was also stained. At stage 23, the aldolase C mRNA signal was in the neural tube, and at stage 33, it was in brain, spinal code, heart anlage, and pronephros. Interestingly, aldolase A mRNA was detected in pronephros at stage 36.
In the dorsal view of hatched embryos, the regionally-different expression of three aldolase mRNAs was clearly seen (Fig 4). Thus, the aldolase A mRNA signal was detected exclusively in somites. aldolase B mRNA signals occurred in pronephros and in epidermis. aldolase C mRNA signals occurred in retina, mesencephalic optic tectum, and rhombencephalon. aldolase C signals occurred also in the spinal cord which is elongated toward the tail tip. We obtained essentially the same results in sectioned materials of tailbud embryos.
Differental expression of three aldolase mRNAs detected by whole mount in situ hybridization. A, B and C indicate expression pattern of aldolase A, B and C mRNA, respectively {from Kajita et al26}.
Genomic DNA structure of Xenopus aldolase C
Genomic DNA cloning of aldolase C was also done27. We obtained a clone named XACl3-1, containing a 12.2 kb DNA insert. XACl3-1 contains the entire sequence for 5′-non-coding, coding, and 3′ -non-coding regions of Xenopus aldolase C gene (Fig 5A). From comparison of this genomic gene with Xenopus aldolase C cDNA10, the Xenopus gene was found to consist of nine exons and spanned a total 9.6 kb from the beginning of exon-1 to the end of exon-9 (Fig 5A). Fig 5A shows a comparison of this Xenopus gene with rat and human counterparts. Xenopus aldolase C gene closely resembled human and rats aldolase genes28, 29, 30. Thus, the number of exons was the same, and furthermore, lengths of all the exons, except for those including 5′-UTR and 3′-UTR, were quite similar between Xenopus and mammalian genes. However, size of the gene as a whole was quite different between Xenopus and mammals, due to the fact that all the introns in the Xenopus gene are several-fold larger than human and rat counterparts.
Structure of the Xenopus aldolase C genomic gene. A: The overall structure. B: various elements found in the promoter region. {from Yatsuki et al27}.
Primer extension analyses was done using oocyte poly(A)+ RNA as a template, and we determined the transcription initiation site (+1) (G). We made a sequence analysis of the 5′-flanking region (in total 1596 bp), conventionally dividing it into six different regions (A-F) (Fig 5B). The proximal part of the 5′-flanking region (regions D, E and F) contained a number of potentially cis-acting elements, as compared with mammalian genes. Like rat and human genes31, the Xenopus aldolase C gene contained a GC box (−47 bp in region E) and an aldolase C-specific element (ACSE) (from −14 to +2). Furthermore, in the Xenopus gene a CCAAT box (at −62 bp in region E) and a TATA-like element (at −28 bp in region F) were found, and these not found in rat and human genes31 (Fig 5B).
Expression studies with the variously-modified promoters were done using Xenopus embryos by DNA microinjection and a kidney cell line by DNA transfection. The GC box is the crucial element in the basal promoter, although the TATA-like element appeared to have a slightly stimulative effect on GC box functioning. We showed by gel retardation and foot-printing assays the occurrence in A6 cells of a nuclear facter(s) that binds specifically to the GC box27.
Expression characteristics of genomic Xenopus aldolase C gene
To test if the 5′-upstream region obtained here directs temporally and regionally regulated expression of the aldolase C gene in developing Xenopus embryos, we first injected an aldolase C promoter connected to bacterial CAT gene (pXAC-CAT) into Xenopus embryos and tested the CAT gene expression at different stages of development. pXAC-CAT was not expressed during cleavage to the blastula stage, expressed only weakly at gastrula and early neurula (neural plate) stages, but was strongly expressed at the late neurula (neural tube) stage (22 h). This time course was reproducible and roughly coincided with that of mRNA expressed from the endogenous aldolase C gene10. Strong expression of the aldolase C gene in the neurula stage is reasonable, since this gene is expressed extensively in neural tissues (Figs 2 and 4).
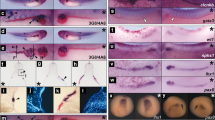
We then injected pXAC-lacZ, consisting of the Xenopus aldolase C gene promoter (1596 bp) and lacZ gene, at the animal side of both of the blastomeres at the two-cell stage. We examined X-gal expression at late neurula and tail-bud stages (Fig 6). Most of the injected embryos showed a blue colour which reflects lacZ gene expression mainly at the dorsal and central portion of the embryo axis, where the neural tube is formed. Fig 6A shows an example of an embryo with such an expresson pattern. At the tail-bud and swimming tadpole stages, lacZ gene expression was found mainly in the head part. Fig 6B shows an example of the late tail-bud stage embryo which showed the signal at the hind brain. Fig 6C shows a swimming tadpole with the signal apparently seen in the midbrain region. From these observations, we concluded that the 1596 bp of the 5′-flanking sequence supported the temporally and spacially regulated expression pattern characteristic of the aldolase C gene in developing embryos.
Expression in brain and spinal cord regions of injected aldolase C genomic gene promoter connected to Lac Z gene {Yatsuki et al, 27}.
Conclusion
In amphibian embryogenesis, it is a general phenomenon that a large amount of maternal stockpiles support early development and these are replaced by materials which are newly formed, based on zygotic gene expression during development32, 33. Such a switch from the maternal to zygotic mRNA was also observed for aldolases A and C. The maternal aldolase B mRNA occurs only at a residual level and extensive transcription takes place only after the late neurula stage. This agrees with the the zymogram analysis: Predominant enzymes present in early embryos were aldolase A and C25. We confirmed that the tissue-specific expression pattern in embryos was mostly as in adults. It should be emphasized here that aldolase A and C genes are quite actively expressed in oocytes and are preserved abundantly in early embryos which develop as a closed system depending on the yolk as the only energy source. By contrast, active aldolase B gene expression beings only late during embryoenesis, but before embryos reach the feeding tadpole stage. It follows then that the zygotic expression of aldolase B gene is to prepare for food intake. In mammals, there are data which parallel our present results.
Finally, one may ask why the aldolase B gene which is needed for metabolism of dietary fructose is expressed so early (at the gastrula stage, albeit at a low level) during embryogenesis. In embryos, however, such a precocious activation also occurs in other genes. For instance, transcription of rRNA genes begins as early as at the late blastula stage34, despite the fact that embryos contain ribosomes of an amount sufficient support development up to stage 41 (feeding tadpole stage)35.
References
Markert CL, Moller F . Multiple forms of enzymes: Tissues, ontogenetic, and species specific patterns. Proc Natl Acad Sci USA 1959; 45:753–63.
Tsuji S, Qureshi MA, Hou EW, Fitch WM, LiS S-L . Evolutionary relationships of lactate dehydrogenases (LDHs) from mammals, birds, an amphibian, fish, barley, and bacteria: LDH cDNA sequences from Xenopus, pig, and rat. Proc Natl Acad Sci USA 1994; 91:9392–6.
Robert J, Pasquier DL, Kobel HR . Differential expression of creatine kinase isozymes during development of Xenopus laevis: an unusual heterodimeric isozyme appears at metamorphosis. Differentiation 1991; 46:23–34.
Horecker BL, Tsola O, Lai CY . aldolases. In: Boyer PD. Ed. The Enzymes Vol. 7, Academic Press, N. Y. 1972:pp.213–58.
Hori K, Mukai T, Joh K, Arai Y, Sakakibara M, Yatsuki H . Structure and expression of human and rat aldolase isozymes Genes: multiple mRNA species of aldolase A produced from a single gene, in: Isozymes. Cur leftics Biol Med Res Vol. 14, 1987; 153–75.
Hers HG, Kusaka T . Le metabolisme du fructose 1-phosphate dans le foie, Biochem. Biophys Acta 1953; 11:427–30.
Munnich A, Besmond C, Darguy S, Reach G, Valmont S, Dreyfus JC, Kahn A . Dietary and hormonal regulation of aldolase B gene expression. J Clin Invest 1985; 75:1045–52.
Decaux JF, Marcillat O, Pichard AL, Henry J, Kahn A . Glucose-dependent and – independent effect of insulin on gene expression. J Biol Chem 1991; 266:3432–8.
Izzo P, Costanzo P, Lupo A, Rippa E, Paolella G, Salavatore F . Human aldolase A gene. Structural organization and tissue-specific expression by multiple promoters and alternate mRNA processing. Eur J Biochem 1988; 174:569–78.
Atsuchi Y, Yamana K, Yatsuki H, Hori K, Ueda S, Shiokawa K . Cloning of a brain-type aldolase cDNA and changes in its mRNA level during oogenesis and early embryogenesis in Xenopus laevis. Biochem Biophys Acta 1994; 1218:153–7.
Shiokawa O, Nunoi K, Kikuchi M, Matsuo E, Iino K, Oryoji Y . The first adult patient with fructose intolerance found in Japan. Diabetes J 1981; 9:77–81 (In Japanese).
Tolan D R . Molecular basis of hereditary fructose intolerance: mutations and polymorphisms in the human aldolase B gene. Hum Mutat 1995; 6:210–8.
Tsutsumi K, Mukai T, Tsutsumi R, Mori M, Daimon M, Tanaka T, Yatsuki H, Hori K, Ishikawa K . Nucleotide sequence of rat liver aldolase B messenger RNA. J Biol Chem 1984; 259:14572–75.
Rottmann WH, Tolan DR, Penhoet EE . Complete amino acid sequence for human aldolase B derived from cDNA and genomic clones. Proc Natl Acad Sci USA 1984; 81:2738–42.
Kusakabe T, Motoki K, Sugimoto Y, Takasaki Y, Hori K . Human aldolase B: liver-specific properties of the isozyme depend on type B isozyme group-specific sequences. Protein Eng 1994; 7:1387–93.
Burgess D G, Penhoet E E . Characterization of the chicken aldolase B gene. J Biol Chem 1985; 260:4604–14.
Kajita E, Moriwaki J, Yatsuki H, Hori K, Miura K, Hirai M, Shiokawa K . Quantitative expression studies of aldolase A, B and C genes in developing embryos and adult tissues of Xenopus laevis. Mech Dev 2001; 102:283–7.
Mukai T, Joh K, Arai Y, Yatsuki H, Hori K . Tissue-specific expression of rat aldolase A mRNAs. Three molecular species differing only in the 5′-terminal sequences, J Biol Chem 1986; 261:3347–54.
Simon M-P, Besmond C, Cottreau D, Weber A, Chaument-Riffaud P, Dreyfus J-C . Trepat J S, Marie J, Kahn A . Molecular cloning of cDNA for rat L-type pyruvate kinase and aldolase B. J Biol Chem 1983; 258:14576–84.
Gianquinto L, Pailhoux EA, Bezard J, Servel N, Kirszenbaum M, Cotinot C . Cloning and characterization of a full-length cDNA coding for ovine aldolase B from fetal mesonephros. Biochem Biophys Acta 1994; 1219:223–7.
Llewellyn L, Ramsurn VP, Sweeney GE, Wigham T, Santos CR, Power DM . Cloning and characterisation of a fish aldolase B gene. Biochem Biophys Acta 1995; 1263:75–8.
Llewellyn L, Sweeney GE, Ramsurn VP, Rogers SA, Wigham T Cloning and unusual expression profile of the aldolase B gene from Atlantic salmon. Biochem Biophys Acta 1998; 1443:375–80.
Brown DD, Littna E . RNA synthesis during the development of Xenopus laevis, the South African clawed toad. J Mol Biol 1964; 8:669–87.
Shiokawa K, Yamana K . Pattern of RNA synthesis in isolated cells of Xenopus laevis embryos. Dev Biol 1967; 16:368–88.
Hikasa H, Hori K, Shiokawa K . Structure of aldolase A (muscle-type) cDNA and its regulated expression in oocytes, embryos and adult tissues of Xenopus laevis. Biochem Biophys Acta 1997; 1354:189–203.
Kajita E, Wakiyama M, Miura K, Mizumoto K, Oka T, Komuro I, Miyata T, Yatsuki H, Hori K, Shiokawa K . Isolation and characterization of Xenopus laevis aldolase B cDNA and expression patterns of aldolase A, B and C genes in adult tissues, oocytes and embryos of Xenopus laevis. Biochem Biophys Acta 2000; 1493:101–18.
Yatsuki H, Outida M, Atsuchi Y, Mukai T, Shiokawa K, Hori K . Cloning of the Xenopus laevis aldolase C gene and analysis of its promoter function in developing Xenopus embryos and A6 cells. Biochem Biophys Acta 1998; 1442:199–217.
Mukai T, Yatsuki H, Masuko S, Arai Y, Joh K . Hori K . The structure of the brain-specific rat aldolase C gene and its regional expression. Biochem Biophys Res Comm 1991; 174:1035–42.
Tolan DR . Penhoet EE . Characterization of the human aldolase B gene. Mol Biol Med 1986; 3:245–64.
Joh K, Arai Y, Mukai T . Hori K . Expression of three mRNA species from a single rat aldolase A gene, differing in their 5′ non-coding regions. J Mol Biol 1986; 190:401–10.
Mukai T, Arai Y, Yatsuki H, Joh K, Hori K . An additional promoter functions in the human aldolase A gene, but not in rat. Eur J Biochem 1991; 195:781–7.
Shiokawa K . Gene expression from endogenous and exogenously-introduced DNAs in early embryogenesis of Xenopus laevis. Develop Growth Differ 1991; 33:1–8.
Shiokawa K, Kurashima R, Shinga J . Temporal control of gene expression from endogenous and exogenously-introduced DNAs in early embryogenesis of Xenopus laevis. Int J Dev Biol 1994; 38:249–55.
Shiokawa K, Misumi Y, Yamana K . Demonstration of rRNA synthesis in pre-gastrula smbryos of Xenopus laevis, Develop. Growth Differ 1981; 23:579–87.
Brown DD, Gurdon JB . Absence of ribosomal RNA synthesis in the anucleolate mutant of Xenopus laevis. Proc Natl Acad Sci USA 1964; 51:139–46.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
SHIOKAWA, K., KAJITA, E., HARA, H. et al. A developmental biological study of aldolase gene expression in Xenopus laevis. Cell Res 12, 85–96 (2002). https://doi.org/10.1038/sj.cr.7290114
Issue Date:
DOI: https://doi.org/10.1038/sj.cr.7290114
Keywords
This article is cited by
-
The role of glycolytic metabolic pathways in cardiovascular disease and potential therapeutic approaches
Basic Research in Cardiology (2023)
-
Identification of a fructose-1,6-bisphosphate aldolase gene and association of the single nucleotide polymorphisms with growth traits in the clam Meretrix meretrix
Molecular Biology Reports (2012)
-
ExprAlign - the identification of ESTs in non-model species by alignment of cDNA microarray expression profiles
BMC Genomics (2009)
-
Expressed sequence tag analysis of adult Clonorchis sinensis, the Chinese liver fluke
Parasitology Research (2006)