Abstract
The Aconitum lycoctonum complex is a widespread yellow-flowered group of species found in central and southern Europe. Because of extreme morphological variability, the systematics of this group is confusing, and hybridizations among taxa are often hypothesized. To determine whether hybridization, realized mating system within populations or colonization from different Pleistocene refugia might explain some of the morphological variation, the genetic structure of 19 populations from central and southern Europe was studied using starch gel electrophoresis and 10 enzyme loci, of which eight were polymorphic. A pattern typical of an outcrossing species was expected because A. lycoctonum flowers are adapted to pollination by long-tongued bumblebees. However, heterozygosity was very low (between 0.031 and 0.150), which is atypical of either widespread or outcrossing species. The inbreeding coefficient, FIS, also suggested inbreeding in more than half the populations. Analysis of molecular variance showed that 31% of the genetic variation is found among populations, again suggesting inbreeding. A neighbour-joining tree based on Reynolds’s genetic distance showed a clear separation between the central/eastern European populations and the samples from the Iberian peninsula and Alpes Maritimes. These data are consistent with the hypothesized refugia and the phylogeographical histories of several European forest trees. The genetic identity of all populations was very high, suggesting that all investigated populations belong to the same species despite high morphological variability. Hybridization was not supported by the data.
Similar content being viewed by others
Introduction
The extent and structure of genetic variation within and among plant populations are known to be strongly influenced by mating systems. Comparisons of large numbers of species have shown that outbreeding populations have greater genetic diversity, with higher levels of heterozygosity and more polymorphic loci, than self-fertilized populations. Furthermore, the genetic structure of outcrossing populations is different. Outcrossing species have greater gene flow, leading to little differentiation among populations. Selfing species, on the other hand, show more differentiation among populations (e.g. Hamrick & Godt, 1989, 1997). Although interspecific studies provide information on general patterns, intraspecific studies allow a more direct investigation of the effects of breeding system on genetic variation and can, therefore, give valuable insights into the evolution of mating systems in plants (e.g. Waycott & Sampson, 1997; Fleming et al., 1998).
Mating systems are important, but the natural history of populations can also affect genetic diversity. A population that undergoes a rapid loss of individuals may lose many rare alleles as a result of genetic drift, and thus experience a reduction in average heterozygosity (a bottleneck effect). Once average heterozygosity is reduced, it can take a long time to rebuild to the original level, whereas the mean number of alleles is expected to increase much faster if population size increases again (Nei et al., 1975).
Ice ages have caused considerable reductions in the ranges of many plants and have thus strongly influenced the population histories of organisms in the northern hemisphere. Hewitt (1996) examined the effects of Pleistocene ice ages on population genetics in Europe. During the ice age, many species that are now distributed across Europe had their refugia in southern Europe. When the climate warmed, these species expanded north from these refugia. Because this colonization process involved a series of bottlenecks for the colonizing genome, it led to a loss of alleles and increased homozygosity. Taberlet et al’s. (1998) compared the phylogeographies of 10 European taxa, including mammals, amphibians, arthropods and trees. They showed that evidence for the three main refugia in southern Europe is found in the phylogeographical histories of many organisms. Taberlet et al (1998) review includes studies on the genetic variation and postglacial histories of European forest trees (Lagercrantz & Ryman, 1990; Konnert & Bergmann, 1995; Demesure et al., 1996; Dumolin-Lapègue et al., 1997), but little is known about the recolonization histories of herbaceous plants in central Europe.
The species complex of yellow-flowered Aconitum lycoctonum is distributed widely across southern and central Europe. These plants occur mainly in mountainous regions in shady habitats, often in forests. The complex is renowned for extreme morphological variability, which has led to the description of numerous taxa and confusing, inconsistent systematics (Warncke, 1964; Hegi, 1974; Hess et al., 1977; Tamura & Lauener, 1978; Tutin et al., 1993). Subspecies lycoctonum (nomenclature after Warncke, 1964) is described as typical for central Europe, and ssp. ranunculifolium for southern Europe. Hybrids are supposed to occur in contact areas, especially in the Alps (Warncke, 1964; Hegi, 1974; Hess et al., 1977; Tutin et al., 1993). Populations in the Alps with glandular hairs on flowers and peduncles are treated as A. penninum at the species or subspecies level (e.g. Hess et al., 1977), although Warncke (1964) mentioned that this character is typical for populations in both the Alps and the Balkan peninsula, and is therefore without any systematic value.
We used allozymes to investigate the breeding system, phylogeography and systematics of the yellow-flowering A. lycoctonum in central and southern Europe. Allozymes remain the most widely used technique for mating system estimation. They are also a useful tool for analysing hybrid zones (Cruzan, 1998).
Materials and methods
Study sites
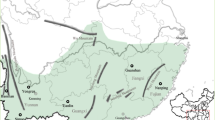
A total of 19 yellow-flowering A. lycoctonum s.l. populations from central, southern and eastern Europe were investigated (Fig. 1). The populations ranged from 360 to 2450 m above sea level (Table 1). From each population, 33 plants were collected (except population 13345, from which only 10 were collected) between May 1995 and September 1997. After collection, the plants were maintained in a greenhouse in Zürich.
Geographical locations of the sampling sites for Aconitum lycoctonum s.l. One population, no. 13345 in Romania, is not shown (see Table 1 for further details).
Enzyme electrophoresis
Fresh leaf material was homogenized in the phosphate grinding buffer–PVP solution described by Soltis et al. (1983). The extraction was then either immediately absorbed on paper wicks and loaded onto gels or stored at −80°C before electrophoresis. Horizontal starch gel (12.8%) electrophoresis was performed. The best results were obtained when fresh extractions were used.
After screening 24 enzymes, eight with scorable bands representing 10 loci were chosen for further investigations. Gel and electrode buffer system 1 (Soltis et al., 1983) was used for malate dehydrogenase (MDH; EC 1.1.1.37), system 5 (Soltis et al., 1983) for acid phosphatase (AcPH; EC 3.1.3.2), aspartate aminotransferase (AAT; EC 2.6.1.1), 6-phosphogluconate dehydrogenase (6PGD; EC 1.1.1.44) and malic enzyme (ME; EC 1.1.1.40). Phosphoglucomutase (PGM; EC 5.4.2.2, two loci) was resolved on system 1 and system 5. Two enzymes were resolved using system 6 (Soltis et al., 1983): phosphoglucoisomerase (PGI; EC 5.3.1.9, two loci) and triosephosphate isomerase (TPI; EC 5.3.1.1). Additional putative loci of 6PGD and MDH were occasionally detected but could not always be resolved or consistently scored, and therefore were not used in the analysis. Heterodimers coded by PGI-2c and PGI-2d and all other more cathodally located bands migrated towards the cathode on system 6 of Soltis et al. (1983). Enzymes were stained according to the recipes of Soltis et al. (1983).
Where more than one isozyme of a given enzyme was observed, they were numbered sequentially from the most anodal (no. 1) to the more cathodal isozyme. Alleles were also named sequentially from the most anodal (a) to the most cathodal.
Data analysis
The computer program BIOSYS-1 (Swofford & Selander, 1989) was used to calculate the mean number of alleles per locus, percentage polymorphic loci (95% criterion), observed and expected heterozygosities and allele frequencies. The correlations of these variability estimates with altitude were investigated, and the effect of population size on variability was tested by a one-way ANOVA. Population size was estimated as small (fewer than 100 reproductive individuals), medium (between 100 and 500) and large (more than 500 reproductive individuals). Nei’s (1972) genetic identity was calculated to facilitate comparisons with the literature.
For estimating and testing FIS values, FSTAT (Goudet, 1995) was used. FIS measures the heterozygote deficit within a population and thus indicates the probability that an individual is homozygous.
Deviations from Hardy–Weinberg equilibrium (HWE) were tested using the program ARLEQUIN (Schneider et al., 1997). It computed a test analogous to Fisher’s exact test using a Markov chain (chain length 100 000, dememorization 1000). Weir (1996) describes this procedure as the most powerful test for deviations from HWE.
An isolation by distance effect generates a positive correlation between geographical distance and FST/(1 − FST), where geographical distance and FST are calculated for each pair of populations. We tested for isolation by distance using the ‘Mantel test’ option of GENEPOP (Raymond & Rousset, 1995), which tests the correlation between the natural logarithm of geographical distances and estimates of FST/(1 − FST). Logarithm of distance is recommended if populations are spread in two dimensions (Rousset, 1997).
To estimate genetic distances among populations, the allele frequency table calculated in BIOSYS-1 was imported into the package PHYLIP (Felsenstein, 1993). The option SEQBOOT produced a file with 100 bootstrapped data sets. Genetic distance matrices for these data sets were calculated by running GENDIST. We chose Reynolds’s distance (Reynolds et al., 1983) and Cavalli-Sforza’s chord distance (Cavalli-Sforza & Edwards, 1967). Both these models assume no mutation and that all gene frequency changes are caused by genetic drift alone. They do not assume that population sizes have remained constant and equal in all populations. NEIGHBOUR was used to construct neighbour-joining and UPGMA trees for all 100 matrices of both distances. CONSENSE then found the consensus tree for each distance and clustering procedure, indicating how well each node was supported.
In addition, a maximum likelihood tree based on gene frequencies was calculated using the option CONTML. This tree is also built on the assumption that each locus evolves independently by pure genetic drift.
To test the hypothesis that there are hybrids in the Alps, principal components were computed for the 11 populations of central Europe using angular-transformed allele frequencies. Correlations between the first principal component axis and the geographical longitude and latitude were calculated.
Finally, we used ARLEQUIN (Schneider et al., 1997) to analyse genetic structure among and within populations and among and within geographical groups of populations by AMOVA (analysis of molecular variance). First, a partitioning of the variation among and within all populations was calculated. Then the populations were divided into three geographical regions (Iberian peninsula, Alpes Maritimes, and central and eastern Europe). This division was suggested by the cluster analysis. We calculated variation among regions, among populations within regions and among individuals within populations. In addition, the three groups were analysed separately to investigate possible differences among geographical regions.
Results
Loci and alleles scored
Eight of the 10 scored loci were polymorphic in at least one population. Up to five alleles were detected per locus. Allele frequencies are available from the corresponding author.
Significant deviations from Hardy–Weinberg equilibrium (P < 0.05) were found in four populations, at one locus each: nos 13042 (AcPH), 14411 (AcPH), 14424 (PGM-1) and 14445 (AcPH). All deviations were the result of heterozygote deficiencies.
Genetic variation within populations
Genetic variation differed strongly among populations. Table 1 summarizes the measures of genetic variability. The average number of alleles per locus (A) varies between 1.1 and 1.7 (mean 1.41) and the percentage of loci polymorphic (P) between 10% and 50% (mean 26.32%). Expected heterozygosity (unbiased estimate; see Nei, 1978) ranges from 0.028 to 0.147 (mean 0.08) and the inbreeding coefficient FIS from −0.279 to 0.342 (mean 0.03). The value of 0.342 in population no. 14411 is the only value significantly (P < 0.05) different from zero. Positive FIS values, indicating inbreeding, were found in 58% of the populations (11 out of 19); in one population, FIS was near zero, indicating random mating; and 37% of the populations (seven out of 19) had negative FIS values, indicating outcrossing.
No significant correlation could be found between altitude and number of alleles per locus (r = 0.317, P = 0.186), percentage of loci polymorphic (r = 0.378, P = 0.110), observed heterozygosity (r = 0.419, P = 0.075) or FIS (r = 0.126, P = 0.607). Nor did population size explain any differences among these variability measures (alleles per locus: F2,16 = 0.483, P = 0.626; percentage of loci polymorphic: F2,16 = 0.520, P = 0.604; observed heterozygosity: F2,16 = 0.509, P = 0.613; expected heterozygosity: F2,16 = 0.629, P = 0.546; inbreeding coefficient: F2,16 = 1.841, P= 0.191). The only significant correlation (r= 0.486, P< 0.05) was observed between altitude and expected heterozygosity.
Genetic distances and relationship among populations
The investigated populations are genetically very similar. The genetic identity of the 19 populations was large; the mean identity of Nei was 0.958 ± 0.032 SD (maximum 1, minimum 0.845). A value of one indicates that two populations are genetically identical. The calculated genetic distance values are small. Cavalli-Sforza’s chord distance values are between 0.002 and 0.256 (mean 0.099 ± 0.060), Reynolds’s between 0.002 and 0.650 (mean 0.283 ± 0.153).
The bootstrapped UPGMA and neighbour-joining trees, based on Reynolds’s and Cavalli-Sforza’s distance, respectively, gave very similar results. The neighbour-joining tree based on Reynolds’s genetic distance had the highest bootstrap values and is shown in Fig. 2. This tree shows an obvious clustering between the populations of the Iberian peninsula and the Alpes Maritimes, and between the populations of central and eastern Europe.
Neighbour-joining tree of 19 Aconitum lycoctonum populations based on Reynolds’s genetic distance (Reynolds et al., 1983). Populations originating from the Iberian peninsula are shown in italics; those from the Alpes Maritimes are shown in bold. Bootstrap values from 100 bootstraps are given at each fork. For population numbers, see Table 1 and Fig. 1.
The maximum likelihood tree also supports the three main groups, except that no. 14414 (Picos de Europa, northern Spain) is shown to be more closely related to nos 14438 and 14445 (Alpes Maritimes) than to the other four populations from the Iberian peninsula.
PCA analysis was used to test for the occurrence in the Alps of hybrids between more southern and northern taxa. The populations plotted on the first, second and third principal components did not show any clustering coherent with the geographical distribution (Fig. 3); thus, the hypothesis of hybridization is not supported. The correlation between the first component and the geographical latitude was not significant (r = −0.071, P = 0.835; Fig. 4a), but the correlation between the first component and longitude was marginally significant (r = −0.582, P = 0.060; Fig. 4b).
Scatter plot of (a) the first (PC 1) and second (PC 2) and (b) the first and third (PC 3) principal components calculated from allele frequencies of 11 populations of Aconitum lycoctonum in central Europe. The percentage of variation that is explained by each component is shown on each axis. Open circles indicate populations north of or in the northern part of the Alps; filled circles indicate populations in the main or southern part of the Alps. For population numbers, see Table 1 and Fig. 1.
Genetic differentiation
Mantel’s test indicated an isolation by geographical distance effect (r = 0.474, P = 0.001). Comparing levels of variation within and among populations, we found about 32% of the total genetic diversity among populations (FST = 0.317, P < 0.001; Table 2). Grouping the 19 populations within geographical regions (Iberian peninsula, Alpes Maritimes, central and eastern Europe), we found 24.01% of the variation among, and 14.78% within, these groups. There is a higher level of interpopulation variation in the Iberian peninsula (25.45%) than in the Alpes Maritimes (20.75%) and in central and eastern Europe (17.06%).
Discussion
Genetic variation
To interpret the causes of the level and pattern of genetic variation within and among populations of A. lycoctonum, two factors need to be considered. The first is the breeding system of the plant. The other is the natural history of the populations as influenced by glaciation, including possible refugia and migration during interglacials. These two factors acting alone, or in concert, are likely to have been the primary forces influencing the genetic structure of these populations.
The genus Aconitum is famous as ‘the bumblebee flower par excellence’ (Hegi, 1974). It is pollinated primarily by long-tongued bumblebees, which are the most efficient pollinators of these plants (Thøstesen & Olesen, 1996). The flowers of A. lycoctonum are protandrous. Within racemes, flowers open from the bottom to the top. The bees generally work the plant from the lower to the upper flowers. This should encourage outcrossing if flowers are completely protandrous. However, the fact that more than half of the FIS values found showed a deficiency of heterozygotes suggests that, at least in some populations, selfing or mating with close relatives is common. One likely cause of selfing in this species is geitonogamy. Plants typically have more than one inflorescence, and bees usually move between them. Because the plants are not self-incompatible (A.-B. Utelli & B. A. Roy, unpubl. data), these within-plant movements may lead to selfing. In addition, the small black seeds of A. lycoctonum are released from the open capsules by slight wind, but no long-distance dispersal is known. Thus, seeds are most likely to fall near the mother plant, further increasing the likelihood of mating with close relatives.
Hamrick & Godt (1989) reported a highly significant influence of the breeding system on the variation of allozymes among populations. The mean percentage of variation among populations of animal-pollinated outcrossers was 19.7%, and among populations with mixed-mating systems it was 21.6%, whereas the percentage of variation among complete selfers was significantly higher at 51.0%. In our data, the percentage of variation among all 19 populations is 31.7%. This overall value lies between those reported for species with mixed-mating systems and absolute selfers. However, when we separate the data by region, the percentages of variation among populations in the Alpes Maritimes and in central Europe are more similar to outcrossers or species with mixed-mating systems (20.8% and 17.0%, respectively), whereas the Iberian populations show a pattern suggesting more selfing with variation at 25.5% (see Table 2). These differences among the geographical regions might be caused by sampling bias, because the sampling density in central Europe was higher than in the Iberian peninsula, and Mantel’s test showed an isolation effect by geographical distance. Because we found a range of FIS values in all geographical regions, we conclude that the observed differences in interpopulation variation among the three regions are not caused by a regional change in the breeding system. Instead, the individual natural history of each population may have led to more or less inbreeding.
A low mean heterozygosity of 0.076 was estimated for endemics by Godt et al. (1996), presumably because genetic variability decreases as a result of bottlenecks and founder events, and a continual reduction of heterozygosity is found when populations remain small. Our mean value of observed heterozygosity was 0.08, close to that typical for endemic species, which A. lycoctonum with its wide range is not. The highest heterozygosity is found in Romania in a population that probably lies near to its ice age refugium. The Romanian population is also the population with the most polymorphic loci. Our data suggest, therefore, that post-glacial recolonization of central Europe by A. lycoctonum was characterized by founder events of small populations that remained small and isolated for a long time. There is, however, little relationship between current population size and genetic variability.
Phylogeography
Recently, Taberlet et al. (1998) showed some general trends concerning refugia during glaciation and subsequent recolonization in Europe. First, there are three main potential refugia: in the Iberian peninsula, in southern Italy and in the Balkans. But the Carpathian Mountains are also a potential refugium area (Lagercrantz & Ryman, 1990; King & Ferris, 1998). Secondly, Taberlet et al. (1998) stated that the northern part of Europe has been colonized primarily from the Iberian and Balkan refugia, whereas populations evolving in Italy were not able to spread northwards because of the Alpine barrier. However, some species also colonized central Europe from the Italian refugium (e.g. oaks; Demesure et al., 1996) or from a refugium in the Carpathian Mountains (e.g. black alder; King & Ferris, 1998). Our data are consistent with these results. First, central and eastern Europe were probably colonized from ancestors surviving in eastern Europe (Carpathian Mountains) or in south-eastern Europe (Balkans), because the population from Romania is clustering with the ones from central Europe, and an east–west gradient is shown by the correlation between the first principal component and the longitude of the populations in central Europe. Moreover, the population from Romania is the most variable, as is expected for populations near their ice age refugium. However, without any samples from the Balkans and southern Italy, we cannot completely exclude the possibility that the Carpathian Mountains and central Europe were colonized from either potential refugium. Our data also show that all populations from the Iberian peninsula are closely related, because they have common ancestors. To determine whether the Alpes Maritimes had been colonized from Italy, or whether migration from the Iberian peninsula had taken place as Küpfer (1974) has postulated for several plant species, populations from southern Italy would need to be examined.
Because A. lycoctonum grows mostly in forests or in shady places, it is not surprising that its postglacial history is similar to that found for some forest trees. Konnert & Bergmann (1995) postulated three refugia from which silver fir (Abies alba) recolonized central and eastern Europe: central Italy, central or eastern France and the Balkan peninsula. Silver fir in the Pyrenees must have originated from a further refugium. Silver fir from the Italian refugium crossed the Alps. Dumolin-Lapègue et al. (1997) and Ferris et al. (1998) found that oaks were present in refugia in the Iberian peninsula, southern Italy and the Balkans, and that the Alps were recolonized by white oaks from the Italian refugium. On the other hand, Demesure et al. (1996) showed that Fagus silvatica from the Balkan refugium colonized central and eastern Europe very rapidly, whereas the beeches from Italy seemed to be stopped south of the Alps, similar to our hypothesis for A. lycoctonum. Leonardi & Menozzi (1995) also demonstrated a tendency for an east–west recolonization by the beech in the northern part of Italy. Additionally, the central European populations of Norway spruce (Picea abies) are supposed to have originated from refugia in the Carpathians and the Dinaric Alps (Lagercrantz & Ryman, 1990), similar to Alnus glutinosa, which colonized northern and central Europe from a refugium in the Carpathian Mountains (King & Ferris, 1998).
Systematics
Our data do not support any of the suggested systematics of the A. lycoctonum group based on morphological characters (see Warncke, 1964; Hegi, 1974; Hess et al., 1977; Tamura & Lauener, 1978; Tutin et al., 1993). Instead, our data suggest that all investigated populations belong to the same species because genetic differences among populations are small. Crawford (1989) stated that the mean identity for populations belonging to the same taxon is often above 0.90. We calculated a mean identity of 0.96. One could argue that there is a subdivision between the south of Europe and the central and eastern part of Europe (see Fig. 2) and treat these populations as different subspecies. But the often-raised hypotheses that the Alps, especially the Swiss Alps, are the contact zone between a more northern and a more southern (sub)species (Warncke, 1964; Hegi, 1974; Hess et al., 1977; Tutin et al., 1993) is not supported by our data. We did not find any significant correlation between genetic variation and a north–south gradient in central Europe. Nor did Beul (1990) find any correlations between the presence or amount of secondary compounds and collection site or morphology in a study of the diterpenalkaloids of 24 A. lycoctonum populations in Switzerland.
If the Alpes Maritimes were colonized from Italy, and if there was a contact area between this Italian and a possible Balkan line, it might be somewhere between the Alpes Maritimes and south-west Switzerland, or in the Slovenian Alps, as has been reported for silver fir (Konnert & Bergmann, 1995). However, the lack of a contact hybrid zone in Switzerland, where one has been hypothesized, suggests that, instead of hybridizing, the species is simply morphologically variable.
Hess et al. (1977) call plants with glandular hairs on the flowers and peduncles A. penninum, at the species level. According to this character, populations 14438 and 14445, both of the Alpes Maritimes, and population 13042 of south-west Switzerland would belong to A. penninum. However, our cluster analysis does not show a closer relationship among these three populations than among any other central European populations.
The morphological differences that have caused all the systematic confusion may be correlated with population genetic structure. Inbreeding, either as a result of matings among close relatives in small populations or as a result of selfing, would tend to fix morphological differences among populations (Baker, 1959). To shed more light on the systematics of A. lycoctonum s.l., additional investigations, including DNA sequencing, crossing experiments, and morphological measurements are needed.
Conclusions
We have shown that the investigated A. lycoctonum populations are not as outcrossed as they were expected to be based on their pollination relationship with bees. Cruzan (1998) lists several factors causing intraspecific variation in outcrossing. He includes flower colour, degree of dichogamy (i.e. temporal separation of sexual function within flowers), plant density, population structure and variation in pollinator abundance. Further investigations will show whether any of these factors are causing the different inbreeding coefficients among A. lycoctonum populations. Because of the close relationship between specialized bumblebees and Aconitum, the question of pollinator availability is of special interest. It is known that pollination systems change with the available array of pollinators or revert to autogamy when pollen vectors are scarce or unreliable (Traveset et al., 1998). Because some populations of A. lycoctonum are often visited by nectar robbers (pers. obs.) and nectar robbers can influence reproduction (e.g. Roubik et al., 1985; Morris, 1996; Traveset et al., 1998), we will also test their influence on the reproductive success of A. lycoctonum.
The genetic distances found among 19 A. lycoctonum populations support the hypothesized ice age refugia in the south and east of the European continent. Central Europe was recolonized by plants from the Carpathians or the Balkan refugium. A second refugium was in the Iberian peninsula. If plants came from Italy towards the Alps, then they are restricted to the most south-west of the Alps.
Finally, allozyme data suggest strongly that all of the yellow-flowered A. lycoctonum in central and southern Europe belong to one single species.
References
Baker, H. G. (1959). Reproductive methods as factors in speciation in flowering plants. Cold Spring Harb Symp Quant Biol, 24: 177–191.
Beul, M. (1990). Diterpenalkaloide der in der Schweiz vorkommenden Subspecies von Aconitum lycoctonum L.. Inaugural Dissertation, University of Basle.
Cavalli-Sforza, L. L. and Edwards, A. W. F. (1967). Phylogenetic analysis: models and estimation procedures. Evolution, 21: 550–570.
Crawford, D. J. (1989). Enzyme electrophoresis and plant systematics. In: Soltis, D. E. and Soltis, P. S. (eds) Isozymes in Plant Biology,. pp. 146–164. Dioscorides Press, Portland, OR.
Cruzan, M. B. (1998). Genetic markers in plant evolutionary ecology. Ecology, 79: 400–412.
Demesure, B., Comps, B. and Petit, R. (1996). Chloroplast DNA phylogeography of the common beech (Fagus sylvatica L.) in Europe. Evolution, 50: 2515–2520.
Dumolin-Lapègue, S., Demesure, B., Fineschi, S., Lecorre, V. and Petit, R. J. (1997). Phylogeographic structure of white oaks throughout the European continent. Genetics, 146: 1475–1487.
Felsenstein, J. (1993). PHYLIP (Phylogeny Inference Package), Version 3.5. Department of Genetics, University of Washington, Seattle.
Ferris, C., King, R. A., Vainola, R. and Hewitt, G. M. (1998). Chloroplast DNA recognizes three refugial sources of European oaks and suggests independent eastern and western immigrations to Finland. Heredity, 80: 584–593.
Fleming, T. H., Maurice, S. and Hamrick, J. L. (1998). Geographic variation in the breeding system and the evolutionary stability of trioecy in Pachycereus pringlei (Cactaceae). Evol Ecol, 12: 279–289.
Godt, M. J. W., Johnson, B. R. and Hamrick, J. L. (1996). Genetic diversity and population size in four rare southern Appalachian plant species. Conserv Biol, 10: 796–805.
Goudet, J. (1995). FSTAT, version 1.2: a computer program to calculate F-statistics. J Hered, 86: 485–486.
Hamrick, J. L. and Godt, M. J. (1989). Allozyme diversity in plant species. In: Brown, A. D. H., Clegg, M. T., Kahler, A. L. and Weir, B. S. (eds) Plant Population Genetics Breeding, and Genetic Resources pp. 43–63. Sinauer, Sunderland, MA.
Hamrick, J. L. and Godt, M. J. (1997). Effects of life history traits on genetic diversity in plant species. In: Silvertown, J., Franco, M. and Harper, J. L. (eds) Plant Life Histories. pp. 102–118. Cambridge University Press, Cambridge.
Hegi, G. (1974). Illustrierte Flora Von Mitteleuropa. Parey, Berlin.
Hess, H. E., Landolt, E. and Hirzel, R. (1977). Flora der Schweiz und Angrenzender Gebiete. Birkhäuser-Verlag, Basel.
Hewitt, G. M. (1996). Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc, 58: 247–276.
King, R. A. and Ferris, C. (1998). Chloroplast DNA phylogeography of Alnus glutinosa (L.) Gaertn. Mol Ecol, 7: 1151–1161.
Konnert, M. and Bergmann, F. (1995). The geographical distribution of genetic variation of silver fir (Abies alba pinaceae) in relation to its migration history. Pl Syst Evol, 196: 19–30.
Küpfer, P. (1974). Recherches sur les liens de parenté entre la flore orophile des Alpes et celle des Pyrénées. Boissiera, 23: 1–322.
Lagercrantz, U. and Ryman, N. (1990). Genetic structure of Norway spruce (Picea abies): concordance of morphological and allozymic variation. Evolution, 44: 38–53.
Leonardi, S. and Menozzi, P. (1995). Genetic variability of Fagus silvatica L. in Italy: the role of postglacial recolonization. Heredity, 75: 35–44.
Morris, W. F. (1996). Mutualism denied? Nectar-robbing bumble bees do not reduce female or male success of bluebells. Ecology, 77: 1451–1462.
Nei, M. (1972). Genetic distances between populations. Am Nat, 106: 293–291.
Nei, M. (1978). Estimations of average heterozygosity and genetic distance from a small number of individuals. Genetics, 89: 583–590.
Nei, M., Maruyama, T. and Chakraborty, R. (1975). The bottleneck effect and genetic variability in populations. Evolution, 29: 1–10.
Raymond, M. and Rousset, F. (1995). GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered, 86: 248–249.
Reynolds, J., Weir, B. S. and Cockerham, C. C. (1983). Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics, 105: 767–779.
Roubik, D. W., Holbrook, N. M. and Parra, V. G. (1985). Roles of nectar robbers in reproduction of the tropical treelet Quassia amara (Simaroubaceae). Oecologia, 66: 161–167.
Rousset, F. (1997). Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics, 145: 1219–1228.
Schneider, S., Kueffer, J. -M., Roessli, D. and Excoffier, L. (1997). ARLEQUIN, Version 1.1: A Software for Population Genetic Data Analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland.
Soltis, D. E., Haufler, C. H., Darrow, D. C. and Gastony, G. J. (1983). Starch gel electrophoresis of ferns: a compilation of grinding buffers, gel and electrode buffers, and staining schedules. Am Fern J, 73: 9–27.
Swofford, D. L. and Selander, R. B. (1989). BIOSYS-1. A Computer Program for the Analysis of Allelic Variation in Population Genetics and Biochemical Systematics. Release 1.7 University of Illinois, Urbana, IL.
Taberlet, P., Fumagalli, L., Wust-Saucy, A. G. and Cosson, J. F. (1998). Comparative phylogeography and postglacial colonization routes in Europe. Mol Ecol, 7: 453–464.
Tamura, M. and Lauener, L. A. (1978). A synopsis of Aconitum subgenus lycoctonum: II. Notes R Bot Garden, Edinb, 37: 113–124.
Thøstesen, A. M. and Olesen, J. M. (1996). Pollen removal and deposition by specialist and generalist bumblebees in Aconitum septentrionale. Oikos, 77: 77–84.
Traveset, A., Willson, M. F. and Sabag, C. (1998). Effect of nectar-robbing birds on fruit set of Fuchsia magellanica in Tierra Del Fuego: a disrupted mutualism. Funct Ecol, 12: 459–464.
Tutin, T. G., Burges, N. A., Chater, A. O., Edmondson, J. R., Heywood, V. H. and Moore, D. M. et al. (1993). Flora Europaea. Cambridge University Press, Cambridge.
Warncke, K. (1964). Die europäischen Sippen der Aconitum lycoctonum -Gruppe Ph.D. Thesis, Ludwig-Maximilians-Universität, Munich.
Waycott, M. and Sampson, J. F. (1997). The mating system of an hydrophilous angiosperm Posidonia australis (Posidoniaceae). Am J Bot, 84: 621–625.
Weir, B. S. (1996). Intraspecific differentiation. In: Hillis, D. M., Moritz, C. and Mable, B. K. (eds) Molecular Systematics. pp. 385–405. Sinauer Associates, Sunderland, MA.
Acknowledgements
We thank B. Gautschi, M. A. Gautschi, S. Horat, P. Senn, M. Soliva and A. Widmer for help in collecting plant material, M. Fotsch for cultivating the plants in the greenhouse, and A. Leuchtmann for introducing A.-B.U. to laboratory methods. We also thank A. Widmer and an anonymous reviewer for helpful comments on the manuscript. The project was funded by the Swiss National Fund (no. 3100.49168.96).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Utelli, AB., Roy, B. & Baltisberger, M. History can be more important than ‘pollination syndrome’ in determining the genetic structure of plant populations: the case of Aconitum lycoctonum (Ranunculaceae). Heredity 82, 574–584 (1999). https://doi.org/10.1038/sj.hdy.6885070
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6885070
Keywords
This article is cited by
-
Genetic variability in a narrow endemic snapdragon (Antirrhinum subbaeticum, Scrophulariaceae) using RAPD markers
Heredity (2002)
-
Molecular and morphological analyses of EuropeanAconitum species (Ranunculaceae)
Plant Systematics and Evolution (2000)