Abstract
Hearing loss is the most common sensory disorder, affecting 1 in 650 newborns. Linkage analysis revealed linkage to locus DFNA22 in two Belgian families 1 and 2 with autosomal dominant sensorineural hearing loss. As MYO6 has previously been reported as responsible for the hearing loss at loci DFNA22 and DFNB37, respectively, DNA sequencing of the coding region and the promoter of MYO6 was performed but this analysis did not reveal any mutations. However, only in patients of family 2, an insertion of 108 bp was identified in the mRNA of the gene. The inserted fragment was part of intron 23 and sequencing of this intron revealed a new splice-site mutation c.IVS23+2321T>G, segregating with the hearing loss in the family. The mutation causes a frameshift and a premature termination codon, but real-time PCR revealed that only 15–20% of the mRNA is degraded by nonsense-mediated decay, while the other part may give rise to an aberrant protein. In family 1, a quantitative real-time PCR experiment revealed a 1.5–1.8-fold overexpression of MYO6 in patients compared to controls. The possible presence of a gene duplication could be excluded by real-time PCR on genomic level. Most likely, the overexpression is caused by a mutation in an unidentified regulatory region of the gene. This study indicates that the inner ear hair cells are sensitive to changes in expression levels of MYO6.
Similar content being viewed by others
Introduction
Hearing loss is the most common sensory disorder, affecting 1 in 650 newborns.1 In 30% of the cases, the hearing loss is part of a syndrome and is associated with other symptoms. In the remaining 70%, the hearing impairment is non-syndromic (NSHI) and almost exclusively due to cochlear defects. Prelingual hearing loss is mainly monogenic and in 70% of the cases inherited in an autosomal recessive way. Postlingual hearing impairment is in most cases multifactorial with presbycusis as the most common example. Monogenic postlingual hearing loss is uncommon and in most cases inherited in an autosomal dominant manner. Non-syndromic hearing loss is extremely genetically heterogeneous with 110 localizations and over 40 gene identifications (Guy Van Camp and Richard JH Smith: Hereditary Hearing Loss Homepage: http://webh01.ua.ac.be/hhh/). Mutations in one single gene, GJB2, account for up to 50% of all autosomal recessive cases. Among the loci for autosomal dominant hearing loss, there is no single locus that is substantially more frequent than the others. Despite the number of gene localizations already published for NSHI, new loci are still being regularly reported, and many loci probably remain undiscovered. In addition, for the known loci, the responsible gene has been identified in less than half of the cases. Therefore, the identification of new deafness genes is very important because it has proven to be an excellent way to elucidate the biology and (patho)physiology of the hearing process.
We performed linkage analysis on two large Belgian families 1 and 2 with non-syndromic autosomal dominant postlingual hearing loss. Linkage was found to the known locus DFNA22, with MYO6 as the disease-causing gene identified for this locus. MYO6 encodes myosin VI, an unconventional actin-based motor protein, expressed in a large variety of tissues. The protein is involved in endocytic trafficking and in maintaining the integrity of cellular organelles such as the Golgi complex.2
Mutations in MYO6 are responsible for non-syndromic autosomal dominant and recessive hearing loss. The gene is expressed in the hair cells of the inner ear and is necessary for maintaining their normal stereociliar structure. The initial evidence for the involvement of myosin VI in the hearing process was reported by Avraham et al.3 The unconventional myosin VI was identified in the Snell's waltzer mice, which exhibit deafness, typical circling behavior, head-tossing and hyperactivity. The mice's phenotype was found to be caused by a spontaneous mutation in Myo6. In 2001, a MYO6 mutation was found in humans in an Italian kindred with non-syndromic autosomal dominant hearing loss.4 In 2005, Mohiddin et al5 reported a second mutation in MYO6 causing a combination of autosomal dominant hearing loss and hypertrophic cardiomyopathy. In the meantime, mutations in MYO6 had also been found to cause non-syndromic autosomal recessive deafness at locus DFNB37 in three Pakistani families.6 Table 1 summarizes the mutations in MYO6 hitherto identified, as well as the clinical data for the families described.
In this study, we report the identification of a new splice-site mutation in MYO6 in family 2, and strong evidence that an increased expression of MYO6 is responsible for the hearing impairment in family 1.
Patients and methods
Family data
Figure 1 shows the ascertained pedigree of the Belgian family 1. The pedigree spans four generations, clearly showing an autosomal dominant mode of inheritance. In total, 68 family members (N=68) participated in the study. Figure 2 shows the pedigree of the second Belgian family 2 with autosomal dominant hearing loss, spanning five generations. Forty-three family members (N=43) participated in the study. This study was approved by the ethical committee of the University of Antwerp. All participants signed an informed consent and completed a questionnaire enabling us to obtain general and audiological data. An otoscopic examination was performed to identify other possible causes for the hearing impairment. In addition, air and bone conduction hearing threshold measurements were made. Family members were considered to be affected when three or more measurements exceeded the 90th percentile of the ISO 7029 normative values for specific age and sex.7 To be considered unaffected, members need to have thresholds better than 50th percentile over all frequencies measured.
Pedigree of the Belgian family 1 with non-syndromic autosomal dominant hearing loss. The haplotypes are shown below each symbol, with the linked haplotype indicated by a box. Black symbols indicate affected individuals, open symbols represent unaffected individuals and question marks indicate individuals with an uncertain diagnosis. The analyzed markers are listed from centromere (top) to telomere (bottom).
Pedigree of the Belgian family 2 with non-syndromic autosomal dominant hearing loss. The haplotypes are shown below each symbol, with the linked haplotype indicated by a box. Black symbols indicate affected individuals, open symbols represent unaffected individuals and question marks indicate individuals with an uncertain diagnosis. Individual VI:1 has congenital hearing loss and carries the splice-site mutation. The analyzed markers are listed from centromere (top) to telomere (bottom).
Genetic analysis
DNA was isolated from the peripheral blood samples of all participating family members by a standard salting out protocol. Fixed linkage parameters were used for all LOD score calculations, on the basis of an autosomal dominant mode of inheritance with an allele frequency of 0.001, a penetrance of 0% for wt/wt and 100% for wt/mt and mt/mt and a phenocopy rate of 0%. SLINK simulations were performed by the program Easylinkage (version 4.01, Berlin, Germany).8 If the SLINK score had a value of 3.3 or higher, the family was considered informative enough for genome-wide linkage analysis.9 A genome-wide search was performed by the company deCODE Genetics (Reykjavik, Iceland), using a polymorphic set of 500 microsatellite markers, covering the whole genome.
Linkage analysis was performed by calculating two-point and multipoint LOD scores with Easylinkage. Two-point LOD scores of 3.3 and higher were considered significant for linkage, and scores below −2 were seen as exclusion of a region. If the two-point LOD score was between 3.3 and −2, multipoint LOD scores were calculated to confirm or exclude linkage.9
All genotyping was performed by polymerase chain reaction (PCR) and fragment analysis on an ABI 3130 automated DNA sequencer (Applied Biosystems, CA, USA), using standard procedures.
The computer program Primer3 input was used for the design of most of the primers.10 Primers were designed for the amplification of the coding region (34 exons), the intron–exon boundaries, 1 non-coding exon and intron 23 of MYO6 (MIM 600970). Primers on cDNA level of MYO6 were constructed to generate overlapping fragments for DNA sequencing of the whole cDNA. Three primer sets were used to characterize the splice-site mutation (Table 2). PCR products were sequenced by standard procedures using an ABI 3130 automated DNA sequencer (Applied Biosystems).
Six different computer programs were used to predict the promoter region of MYO6, namely FirstEF,11 Promoter 2.0,12 Promoter Scan 2,13 DBTSS,14 McPromoter15 and Dragon Promoter Finder.16 The complete genomic sequence of MYO6 and 3000 bp upstream of the gene was analyzed by the programs to look for good promoter predictions. When two or more programs indicated the presence of a possible promoter in a certain region, this region was sequenced to look for possible disease-causing mutations.
Conservation of amino acids was evaluated using the ConSeq server.17 ConSeq scores vary from 1 (variable) to 9 (conserved).
To evaluate the expression of MYO6 in different human tissues, two Clontech broad-coverage multiple tissue cDNA panels of healthy individuals were used.
RNA isolation and cDNA synthesis
To isolate RNA, human peripheral blood lymphocytes were isolated, cultured and EBV-transformed by standard procedures. Total RNA was extracted using trizol RNA isolation protocol (Invitrogen, Merelbeke, Belgium). RNA was purified with Ambion's DNA-free™ Kit to remove contaminating DNA from the RNA samples. cDNA was synthesized using Invitrogen's SuperScript™ III First-Strand Synthesis System for RT-PCR with random hexamers. For each sample, RNA was isolated separately three times, and per RNA isolation, three independent cDNA syntheses were performed. Every cDNA sample was diluted 1:10 for use in real-time PCR experiments.
Gene expression analysis by real-time quantitative PCR
A quantitative real-time PCR was performed using gene-specific assays-on-demand (Applied Biosystems) containing two PCR primers (forward and reverse) and a TaqMan MGB probe (FAM dye labelled). For MYO6, two different assays Hs00192265_m1 and Hs01568230_m1 were used, each spanning an exon–exon boundary. Three housekeeping genes were used as an internal control: UBC (assay Hs00824723_m1), YWHAZ (assay Hs00237047_m1) and GAPDH (assay Hs99999905_m1). The detection was done using the LightCycler® 480 Instrument (Roche Applied Science, Vilvoorde, Belgium). Per sample and per assay, a mixture of 10 μl was prepared, containing 5 μl of 2 × Probes Master (Roche Applied Science, Vilvoorde, Belgium), 0.5 μl of the assay used, 2.5 μl of purified water and 2 μl of cDNA. Cycling conditions were as follows: preincubation: 10 min at 95°C; amplification: 40 cycles each consisting of 10 s at 95°C, 1 min at 60°C and 1 s at 72°C; cooling: 10 s at 40°C. For family 1, five patient samples and five unaffected family members were used in the experiments. For family 2, two patients and two unaffected family members were included. Data were analyzed using the LC480 software (Roche Applied Science) and subsequently exported for import into the relative quantification software qBase for further analysis.18 The coefficient of variance (CV) and M-value were calculated for the housekeeping genes as described19 to analyze the stability of expression of these genes. For both assays, the relative expression (RE) was calculated, as being the ratio of the geometric means of the normalized expression values of control persons versus patients. Statistical analysis was performed in SPSS by the non-parametric Mann–Whitney U-test.
Detection of gene copy number changes by real-time quantitative PCR
To investigate the possible presence of a gene duplication of MYO6 in patients from family 1, an assay-by-design MYO6ex11 was used, for which the primers and probes were located in exon 11 of the gene (Applied Biosystems). The RNase P kit was used as an internal control (Applied Biosystems). The effect of DNA concentration on PCR efficiency was determined using a dilution series from a genomic DNA sample, together with λ DNA as a carrier to stabilize the dilutions. The DNA dilution series was prepared containing 160, 80, 40, 20 and 10 ng DNA per reaction, including a control point with only carrier DNA. Each reaction was performed in quadruplicate. The threshold cycle was plotted against the log of the DNA amount in nanograms and efficiencies were calculated by using the formula 10−1/s−1 with s being the slope of the curve.
All samples were diluted in TE−4 buffer to a concentration of 20 ng/μl. A mix of 10 μl was prepared per well, containing 5 μl 2 × Probes Master (Roche Applied Science), 0.5 μl MYO6ex11 assay, 0.5 μl 20 × RNase P (VIC labelled), 1.5 μl of purified water and 2.5 μl of genomic DNA (50 ng). Thermal cycling conditions were identical to the gene expression assays. The gene copy number was calculated by the formula E−ΔΔCt, with E being the PCR efficiency and ΔΔCt=(Ct RNase Pcalibrator−Ct MYO6ex11calibrator)−(Ct RNase Psample−Ct MYO6ex11sample), in which a healthy control sample was used as a calibrating sample.
Results
Clinical data
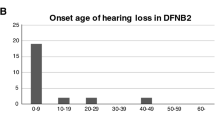
For family 1, 19 individuals were diagnosed as affected and were found to have a rather flat audiogram affecting all frequencies to a similar degree (Figure 3a). The hearing loss was mild at the onset, evolving to moderate to severe by the age of 50 years. Eleven individuals of family 2 were considered as affected, with a hearing loss mainly affecting the mid frequencies and ranging from mild to severe (Figure 3b). For both families, the hearing loss starts during the third decade for most family members and shows a slight progression.
Linkage analysis
After establishing the affection status of all participating family members, SLINK simulations confirmed that both families were informative enough for genome-wide linkage analysis. Maximum obtainable SLINK scores were 9.63 for family 1 and 5.05 for family 2. The samples from both families were sent to deCODE Genetics (Reykjavik, Iceland) for a genome-wide search and LOD scores were calculated for the 500 markers analyzed. For both families, linkage was found to locus DFNA22 on chromosome 6q with maximum two-point LOD scores of 6.06 for family 1 at θ=0.05 and 2.04 for family 2 at θ=0, both for marker D6S1557. For family 2, a maximum multipoint LOD score of 5.4 was obtained. Extra polymorphic markers in the region were analyzed to confirm linkage and to refine the candidate region. After constructing the haplotypes, the candidate regions for both families could be defined. The linked region of family 1 was established between markers D6S456 and D6S460, defining a region of 2.37 cM. For family 2, the candidate region was 9.08 cM between markers D6S1557 and D6S1652, a region completely comprising the linked region of family 1.
DNA sequencing of MYO6
MYO6 is located in the candidate region of both families and has been identified as a deafness gene for locus DFNA22, which makes it the best candidate gene. However, DNA sequencing of the coding and non-coding exons as well as the exon–intron boundaries and the UTRs of MYO6 did not reveal any possible mutations. In one patient from family 1, a new polymorphic 1 bp deletion was detected at genomic position 113.621 in intron 16 of the gene, but it did not segregate with the disease. In family 2, two new polymorphisms were identified. The first SNP was a nucleotide change g.107893A>G in intron 12, not segregating with the disease. The second nucleotide change was a C-T transition (c.2739C>T) in exon 25, causing the amino-acid change p.T845I. This polymorphism did segregate within the family but the ConSeq score of this amino acid was 6. In addition, this variant was also identified in 4 out of 192 independent Belgian control samples, which makes a pathogenic nature improbable.
As DNA sequencing of the exons of MYO6 did not reveal any pathogenic changes, we attempted to identify the position of the promoter region of the gene, using computer prediction programs. One region about 700 bp upstream of non-coding exon 1 was predicted to contain the promoter by four different prediction programs. This region was also confirmed to be the promoter region of MYO6 by Jung et al.2 The predicted promoter region was sequenced but no new sequence variants were identified.
Analysis of MYO6 on cDNA level
As no mutations were found by DNA sequencing of MYO6, we looked for splice-site mutations in the gene. For this purpose, cDNA was synthesized out of peripheral blood lymphocytes from a number of affected and unaffected family members of both families. Primers were designed to cover the whole cDNA in overlapping fragments. The lengths of the PCR fragments were estimated by loading the PCR products on a 1.5% agarose gel and were compared between patient and control samples of each family. For family 1, no length differences could be seen for any fragment between patients and healthy family members. Also DNA sequencing of the complete cDNA of MYO6 did not reveal any small deletions or insertions. The sequencing did reveal alternative splicing in MYO6. For two patients and one healthy family member, exons 29, 30 and 33 were not present in the cDNA, although these exons could be sequenced on genomic level. These alternative splicing forms could be found in the Alternative Splicing Database (ASD), and were considered not pathogenic.20
For family 2, a double band was seen on agarose gel for both patients but for none of the healthy family members, after performing a PCR using primer set 1 (Table 2). In addition to the wild-type 1141 bp fragment, a larger fragment of ∼1240 bp was observed, suggesting the presence of an insertion (Figure 4a). Subsequently, the fragment was split up into three smaller parts to localize the insertion more precisely and again a larger fragment was observed on agarose gel by use of primer set 2 (Table 2). A nested primer set 3 was used to intensify the fragment, with the forward primer lying even closer to the insertion point. Loading this PCR product on agarose gel revealed the wild-type fragment (161 bp) and a larger fragment (∼270 bp) (Figure 4b). Owing to the good separation of both fragments, the large fragment could be cut out from the agarose gel, purified and sequenced. The sequence showed an insertion of 108 bp located between the 3′ end of exon 23 and the 5′ end of exon 24 and corresponding to an intronic sequence in the gene at nucleotides 134 938–135 045, located in the middle of intron 23. To investigate the mutation leading to this insertion on genomic level, intron 23 was sequenced completely. A nucleotide change c.IVS23+2321T>G was identified in all affected family members but in none of the healthy individuals, creating a new splice donor site (Figure 5). The insertion of the 108 bp intronic fragment in the mRNA causes a premature termination codon (PTC) at the 16th nucleotide downstream of exon 23. Consensus values were calculated for the 3′ splice donor site and the 5′ splice acceptor site of the insertion with and without the mutation, according to Krawczak et al.21 The consensus value of the 3′ acceptor site was 0.86, which is a rather high score. The consensus value of the 5′ donor site changed from 0.83 in the normal sequence to a maximum value of 1.0 in the mutated sequence. The splice-site mutation could not be found in 139 independent, ethnically matched control samples.
RT-PCR of a fragment containing the cryptic exon in family 2. RNA was obtained from lymphoblasts. Two separate bands could be identified for both patients, but for none of the unaffected family members: the wild-type fragment and a fragment about 100 bp larger, containing the cryptic exon. (a) PCR obtained by using primer set 1, showing the wild-type fragment of 1141 bp and the larger fragment of ∼1240 bp. (b) Nested PCR by use of primer set 2 and nested primer set 3, resulting in a 161 bp wild-type fragment and a 269 bp fragment carrying the mutation. Above the 269 bp fragment, another fragment was visible, but we did not succeed in sequencing this fragment to obtain the exact sequence.
Quantitative real-time PCR on cDNA level
For family 2, we aimed to check the effect of the splice-site mutation IVS23+2321T>G on the expression level of MYO6. Two assays for MYO6 and three assays for different housekeeping genes were used. For all three housekeeping genes, the CV values were below 21%, indicating stable expression. The RE values were 1.25 and 1.17 for assays 1 and 2, respectively, indicating that the expression of MYO6 in patients has decreased to 80–85% of the expression level in unaffected individuals. Statistical analysis of these results with the Mann–Whitney U-test showed no statistically significant difference in expression between patient and control samples.
As no mutations were found by DNA sequencing of MYO6 in family 1, a quantitative real-time PCR was performed with the same MYO6 assays to look for changes in expression of the gene. The CV values for all housekeeping genes were below 18%, indicating stable expression. We found RE values of 0.55 and 0.65 for assays 1 and 2 respectively, meaning a 1.5–1.8-fold increase in MYO6 expression in the analyzed patients of the family (data not shown). The Mann-Whitney U-test gave a P-value of 0.008 and 0.002, respectively, indicating a statistically significant difference between the expression in patients and control samples for both assays.
Quantitative real-time PCR on genomic level
To check whether the overexpression of MYO6 in the patients from family 1 is caused by a gene duplication, a new MYO6 assay on genomic DNA was designed with primers and probes located in exon 11 of the gene. Firstly, we determined the efficiencies of this assay and of the RNase P assay, used as a control. PCR efficiencies were 96% for MYO6ex11 and 95% for RNase P. As these values are comparable, we could use the ΔΔCt method to compare the copy number of the gene in patient and control samples from the family. For the quantitative analysis, 18 control samples and 18 patient samples were run in quadruplicate.22 Taking the PCR efficiencies into account, the gene copy number of all patients compared to the controls was 1.01.
Expression profile of the cryptic exon
To determine whether the aberrant splicing in intron 23 of MYO6 occurred in other human tissues from healthy persons, an RT-PCR overspanning exons 23 and 24 was performed on two broad-coverage multiple tissue cDNA panels. Primer set 1 and nested primer set 3 were used to check the presence of the cryptic exon (Table 2). In none of the tissues, the larger fragment was detected (data not shown).
Discussion
In this study, we report two Belgian families with mild to severe sensorineural hearing loss, linked to DFNA22. Changes in splicing and expression of MYO6 are associated with hearing loss in both families, suggesting that MYO6 is the disease-causing gene.
Family 2
In family 2, a splice-site mutation c.IVS23+2321T>G could be identified on genomic level, segregating with the hearing loss in the family. The insertion of an extra fragment in the mRNA causes a premature termination codon (PTC) at the 16th nucleotide downstream of exon 23. The results of the real-time PCR experiments showed that the aberrant mRNA is probably degraded only partially by NMD because approximately 80–85% of the transcript is still present in family patients. The lower NMD efficiency could be explained by the influence of additional factors that are involved in the NMD machinery and its regulation.23
The cryptic exon of MYO6 has rather high consensus values (3′: 0.86; 5′: 0.83) although the exon is not included in the normal mRNA, indicating that it could be a pseudoexon. As the mutation causes an increase in consensus value for the splice donor site to a maximum value of 1, this seems to be a sufficient change for inclusion of the pseudoexon in the transcript of the gene.
Family 1
In family 1, we did not find any potentially pathogenic changes by DNA sequencing of MYO6 on genomic level and on cDNA level. However, by performing quantitative real-time PCR experiments on cDNA level, we observed a 1.5–1.8-fold increase in MYO6 expression in patients compared to unaffected family members. The possible presence of a gene duplication could be excluded by quantitative real-time PCR on genomic level, as the copy number for both patients and controls was very close to 1. Because the promoter region of the gene has already been identified and DNA sequencing did not reveal any possible pathogenic variants, we can assume that the overexpression is not due to a promoter mutation. An alternative explanation could be the presence of a mutation in a regulatory sequence of MYO6. In mice, a possible regulatory region of Myo6 was found in a second Snell's waltzer mutant sesv, showing deafness and circling due to an inverson causing a break 30–220 kb upstream of the gene.24 However, in humans, no regulatory regions have been identified so far, making it very difficult to identify the disease-causing variant in this family.
Possible genotype–phenotype correlation for dominant versus recessive mutations in MYO6
To date, MYO6 has been identified as the disease-causing gene in four dominant and two recessive families with non-syndromic hearing loss. When comparing the phenotype of the patients in all MYO6 families, we can put forward the hypothesis that a genotype–phenotype correlation is present for mutations in MYO6. Recessive mutations 36–37insT and 3496C>T both cause a PTC in the motor domain of the gene, leading to mRNA degradation by NMD. No normal protein product will be present and the mutations lead to profound, congenital hearing loss (Table 1). The third recessive mutation 647A>T does not cause a PTC but its pathogenic nature has not been confirmed. For the three dominant mutations that have been identified, haploinsufficiency is not a likely mechanism as heterozygous carriers of the recessive mutations have a normal hearing. A more plausible explanation is that all three mutations act in a dominant-negative way. This hypothesis is supported by the finding that myosin VI can form dimers in the presence of a cargo, established by an intermolecular interaction through the predicted high probability coiled-coil region.25 As the dominant mutations are not located in or very close to this coiled-coil region, they will not prevent dimerization. Binding of the aberrant protein to the wild-type protein may however exert a dominant-negative effect and inactivate the dimer. As part of the dimers consists of two wild-type proteins, there will be some residual myosin VI activity. This could explain the milder phenotype and later onset of the hearing loss in patients from the dominant families compared to the recessive families. For both dominant families described previously,4, 5 the characteristics of the hearing loss are comparable (Table 1). This could be explained by the fact that C442Y and H246R are both missense mutations, changing an important structural amino acid in the motor domain of the protein. The splice-site mutation causes hearing loss with a later age of onset and the audiograms have a different shape (Table 1 and Figure 3). However, this mutation does not change one single amino acid but leads to a protein in which 36 additional amino acids are inserted. This difference in mutation type could explain the distinct phenotypic outcome.
Influence of MYO6 expression level in the hair cells
The finding that overexpression of MYO6 may cause hearing loss may indicate that the hearing process is influenced by the level of MYO6 transcript. If the transcript level ranges between 50 and 100%, this seems sufficient for a normal protein function, which is supported by the finding that heterozygous carriers of recessive mutations do not have hearing loss. Patients of family 2 were found to have transcript levels of 80–85%, but in this case, the hearing loss is probably due to a dominant-negative effect. The absence of normal transcript leads to profound hearing loss as seen in homozygous carriers of recessive mutations. The effect of transcript levels between 0 and 50% is not known in humans. However, recessive sesv mutant mice, which have a deafness and circling phenotype, were found to have very low expression levels of Myo6 mRNA and total absence of protein expression. Furthermore, in recessive sesv mutant mice with a similar phenotype, expression of myosin VI protein in the inner ears was reduced to 11–16% of the levels found in control mice.24 These findings may indicate that expression levels between 0 and 50% may contribute to the hearing loss in both mice mutants. In addition, the dominant mutations with presumed dominant-negative effect suggest that these lower levels may lead to hearing loss with a milder phenotype due to the residual myosin VI activity. Our results for family 1 show that levels above 100% may also be deleterious for the hair cells.
It is remarkable that MYO6 is expressed in a wide variety of tissues while patients with MYO6 mutations only suffer from hearing loss. The specific phenotype for the patients could be explained by the hypothesis that mainly the hair cells in the inner ear, where expression is highest, are very sensitive to changes in the amount of MYO6 transcript below 50% and above 100%. Differences in NMD efficiencies in different cell types may partially explain the tissue-specific effect of the mutation.23 Another hypothesis is based on the fact that MYO6 is mainly expressed in epithelial cells and required for maintaining the structural integrity of cells and not for development.26 In epithelial cells such as the ones in the intestine or the kidney, MYO6 is not essential for maintenance because these cells have a very high turnover. On the contrary, the inner ear epithelial cells are postmitotic and do require MYO6 for structural maintenance as they cannot be renewed.
References
Mehl AL, Thomson V : The Colorado newborn hearing screening project, 1992–1999: on the threshold of effective population-based universal newborn hearing screening. Pediatrics 2002; 109: E7.
Jung EJ, Liu G, Zhou W, Chen X : Myosin VI is a mediator of the p53-dependent cell survival pathway. Mol Cell Biol 2006; 26: 2175–2186.
Avraham KB, Hasson T, Sobe T et al: Characterization of unconventional MYO6, the human homologue of the gene responsible for deafness in Snell's waltzer mice. Hum Mol Genet 1997; 6: 1225–1231.
Melchionda S, Ahituv N, Bisceglia L et al: MYO6, the human homologue of the gene responsible for deafness in Snell's waltzer mice, is mutated in autosomal dominant nonsyndromic hearing loss. Am J Hum Genet 2001; 69: 635–640.
Mohiddin SA, Ahmed ZM, Griffith AJ et al: Novel association of hypertrophic cardiomyopathy, sensorineural deafness, and a mutation in unconventional myosin VI (MYO6). J Med Genet 2004; 41: 309–314.
Ahmed ZM, Morell RJ, Riazuddin S et al: Mutations of MYO6 are associated with recessive deafness, DFNB37. Am J Hum Genet 2003; 72: 1315–1322.
International Organisation for Standardisation, 2001.
Lindner TH, Hoffmann K : easyLINKAGE: a PERL script for easy and automated two-/multi-point linkage analyses. Bioinformatics 2005; 21: 405–407.
Lander E, Kruglyak L : Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 1995; 11: 241–247.
Rozen S, Skaletsky H : Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 2000; 132: 365–386.
Davuluri RV, Grosse I, Zhang MQ : Computational identification of promoters and first exons in the human genome. Nat Genet 2001; 29: 412–417.
Knudsen S : Promoter2.0: for the recognition of PolII promoter sequences. Bioinformatics 1999; 15: 356–361.
Prestridge DS : Predicting Pol II promoter sequences using transcription factor binding sites. J Mol Biol 1995; 249: 923–932.
Suzuki Y, Yamashita R, Nakai K, Sugano S : DBTSS: database of human transcriptional start sites and full-length cDNAs. Nucleic Acids Res 2002; 30: 328–331.
Ohler U, Niemann H, Liao G, Rubin GM : Joint modeling of DNA sequence and physical properties to improve eukaryotic promoter recognition. Bioinformatics 2001; 17 (Suppl 1): S199–S206.
Bajic VB, Seah SH, Chong A, Zhang G, Koh JL, Brusic V : Dragon Promoter Finder: recognition of vertebrate RNA polymerase II promoters. Bioinformatics 2002; 18: 198–199.
Berezin C, Glaser F, Rosenberg J et al: ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics 2004; 20: 1322–1324.
Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J : qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 2007; 8: R19.
Vandesompele J, De Preter K, Pattyn F et al: Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002; 3 RESEARCH0034.
Stamm S, Riethoven JJ, Le Texier V et al: ASD: a bioinformatics resource on alternative splicing. Nucleic Acids Res 2006; 34: D46–D55.
Krawczak M, Reiss J, Cooper DN : The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet 1992; 90: 41–54.
Hoebeeck J, Speleman F, Vandesompele J : Real-time quantitative PCR as an alternative to Southern blot or fluorescence in situ hybridization for detection of gene copy number changes. Methods Mol Biol 2007; 353: 205–226.
Linde L, Boelz S, Neu-Yilik G, Kulozik AE, Kerem B : The efficiency of nonsense-mediated mRNA decay is an inherent character and varies among different cells. Eur J Hum Genet 2007; 15: 1156–1162.
Avraham KB, Hasson T, Steel KP et al: The mouse Snell's waltzer deafness gene encodes an unconventional myosin required for structural integrity of inner ear hair cells. Nat Genet 1995; 11: 369–375.
Park H, Ramamurthy B, Travaglia M et al: Full-length myosin VI dimerizes and moves processively along actin filaments upon monomer clustering. Mol Cell 2006; 21: 331–336.
Millo H, Leaper K, Lazou V, Bownes M : Myosin VI plays a role in cell–cell adhesion during epithelial morphogenesis. Mech Dev 2004; 121: 1335–1351.
Acknowledgements
This study was supported by grants from EUROHEAR (LSHG-CT-20054-512063) and by the Fund for Scientific Research Flanders (FWO-F, Grant G.0138.07). Nele Hilgert is a fellow of the Fund for Scientic Research Flanders (FWO-F). We thank Professor Karen Avraham for helpful advice.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hilgert, N., Topsakal, V., van Dinther, J. et al. A splice-site mutation and overexpression of MYO6 cause a similar phenotype in two families with autosomal dominant hearing loss. Eur J Hum Genet 16, 593–602 (2008). https://doi.org/10.1038/sj.ejhg.5202000
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5202000
Keywords
This article is cited by
-
Heterozygous missense variants of LMX1A lead to nonsyndromic hearing impairment and vestibular dysfunction
Human Genetics (2018)
-
Deep intronic mutations and human disease
Human Genetics (2017)
-
Can the ‘neuron theory’ be complemented by a universal mechanism for generic neuronal differentiation
Cell and Tissue Research (2015)