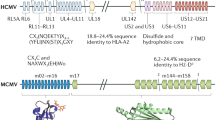
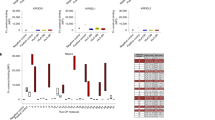
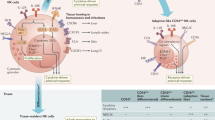
Abstract
Recognition and destruction of virus-infected cells by class I major histocompatibility complex (MHC) restricted cytotoxic T lymphocytes (CTL) is a central part of the immune system's attempts to control and eliminate virus infection. It is therefore not surprising that many viruses have evolved strategies to interfere with the processing and presentation of peptide antigen on class I MHC molecules (reviewed in ref. 1). These mechanisms act to prevent or reduce expression of MHC molecules at the cell surface. However, many natural killer (NK) cells are able to recognize and destroy host cells that no longer express class I MHC molecules (the 'missing self hypothesis2). Thus, any virus-infected cell that has lost cell-surface expression of MHC class I to avoid CTL attack should become susceptible to NK-cell-mediated destruction. We describe here the first example, to our knowledge, of a viral strategy to evade immune surveillance by NK cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Spriggs, M. K. One step ahead of the game: Viral immunomodulatory molecules. Annu. Rev. Immunol. 14, 101–130 (1996).
Ljunggren, H.-G. & Karre, K. In search of the ‘missing self’: MHC molecules and NK recognition. Immunol. Today 11, 237–242 (1990).
Beersma, M. F. C., Bijlmakers, M. J. E. & Ploegh, H. L. Human cytomegalovirus down-regulates HLA class I expression by reducing the stability of class I H chains. J. Immunol. 151, 4455–4464 (1993).
Jones, T. R. et al. Multiple independent loci within the human cytomegalovirus unique short region down-regulate expression of major histocompatibility complex class I heavy chains. J. Virol. 69, 4830–4841 (1995).
Wiertz, E. J. H. J. et al. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84, 769–779 (1996).
Wiertz, E. J. H. J. et al. Cytosolic destruction of a type I membrane protein by transfer via the Sec61p complex from the ER to the proteasome. Nature 384, 432–438 (1996).
Ahn, K. et al. Human cytomegalovirus inhibits antigen presentation by a sequential multistep process. Proc. Natl Acad. Sci. USA 93, 10990–10995 (1996).
Jones, T. R. et al. Human cytomegalovirus US3 impairs transport and maturation of major histocompatibility complex class I heavy chains. Proc. Natl Acad. Sci. USA 93, 11327–11333 (1996).
Hengel, H., Flohr, T., Hammerling, G. J. & Koszinowski, U. H. Human cytomegalovirus inhibits peptide translocation into the endoplasmic reticulum for MHC class I assembly. J. Gen. Virol. 77, 2287–2296 (1996).
Beck, S. & Barrell, B. G. Human cytomegalovirus encodes a glycoprotein homologous to MHC class-I antigens. Nature 331, 269–272 (1988).
Browne, H., Smith, G., Beck, S. & Minson, A. A complex between the MHC class I homologue encoded by human cytomegalovirus and β2-microglobulin. Nature 347, 770–772 (1990).
Fahnestock, M. L. et al. The MHC class I homologue encoded by human cytomegalovirus binds endogenous peptides. Immunity 3, 583–590 (1995).
Wiley, D. MHC gene in cytomegalovirus. Nature 331, 209–210 (1988).
Shimizu, Y. & DeMars, R. Production of human cells expressing individual transfered HLA-A, -B, -C genes using an HLA-A, -B, -C null human cell line. J. Immunol. 142, 3320 (1989).
Parham, P., Androlewicz, M. J., Holmes, N. J. & Rothenberg, B. E. Arginine-45 is a major part of the determinant of human β2-microglobulin recognised by mouse monoclonal antibody BBM.1. J. Biol. Chem. 258, 6179–6186 (1983).
Brocker, T., Peter, A., Traunecker, A. & Karjaleinen, K. New simplified molecular design for functional T cell receptor. Eur. J. Immunol. 23, 1435–1439 (1993).
Fuhrmann, U., Bause, E. & Ploegh, H. Inhibitors of oligosaccharide processing. Biochim. Biophys. Acta 825, 95–110 (1985).
Aramburu, J. et al. A novel functional cell surface dimer (Kp43) expressed by natural killer cells and T cell receptor-g/d+ T lymphocytes. I. Inhibition of the IL-2 dependent proliferation by anti-Kp43 monoclonal antibody. J. Immunol. 144, 3238–3247 (1990).
Aramburu, J., Balboa, M. A., Izquierdo, M. & Lopez-botet, M. A novel functional cell surface dimer (Kp43) expressed by natural killer cells and T cell receptor-g/d+ T lymphocytes. II. Modulation of natural killer cell cytoxocity by anti-Kp43 monoclonal antibody. J. Immunol. 147, 714–721 (1991).
Perez-Villar, J. J. et al. Functional ambivalence of the Kp43 (CD94) NK cell associated surface antigen. J. Immunol. 154, 5779–5788 (1995).
Phillips, J. H. et al. CD94 and a novel associated protein (94AP) form a NK cell receptor involved in the recognition of HLA-A, HLA-B and HLA-C allotypes. Immunity 5, 163–172 (1996).
Perez-Villar, J. J. et al. Biochemical and serologic evidence for the existence of functionally distinct forms of the CD94 NK cell receptor. J. Immunol. 157, 5367–5374 (1996).
Lazetic, S., Chang, C., Houchins, J. P., Lanier, L. L. & Phillips, J. H. Human natural killer cell receptors involved in MHC class I recognition are disulfide linked heterodimers of CD94 and NKG2 subunits. J. Immunol. 157, 4741–4745 (1996).
McGeoch, D. J., Cook, S., Dolan, A., Jamieson, F. E. & Telford, E. A. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J. Mol. Biol. 247, 443–458 (1995).
Mandelboim, O. et al. Protection from lysis by natural killer cells of group 1 and 2 specificity is mediated by residue 80 in human histocompatibility leukocyte antigen C alleles and also occurs with empty major histocompatibility complex molecules. J. Exp. Med. 184, 913–922 (1996).
Shimizu, Y., Geraghy, D. E., Koller, B. H., Orr, H. T. & DeMars, R. Transfer and expression of three cloned human non-HLA-A, B, C class I major histocompatibility genes in mutant lymphoblastoid cells. Proc. Natl Acad. Sci. USA 85, 227–231 (1988).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Reyburn, H., Mandelboim, O., Valés-Gómez, M. et al. The class I MHC homologue of human cytomegalovirus inhibits attack by natural killer cells. Nature 386, 514–517 (1997). https://doi.org/10.1038/386514a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/386514a0
This article is cited by
-
Cure My Virus: Hematemesis and Melena in a Transplant Recipient
Digestive Diseases and Sciences (2021)
-
Modulation of innate and adaptive immunity by cytomegaloviruses
Nature Reviews Immunology (2020)
-
Serum SAA1 and APOE are novel indicators for human cytomegalovirus infection
Scientific Reports (2017)
-
Classical and non-classical MHC I molecule manipulation by human cytomegalovirus: so many targets—but how many arrows in the quiver?
Cellular & Molecular Immunology (2015)
-
Role of HCMV miR-UL70-3p and miR-UL148D in overcoming the cellular apoptosis
Molecular and Cellular Biochemistry (2014)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.