Abstract
Both epidermal growth factor receptor (EGFR) and ErbB-2 play an important role in cancer biology and constitute promising molecular targets of therapy. EGFR and ErbB-2 expression has been observed in rhabdomyosarcoma cell lines but not analyzed systematically in rhabdomyosarcoma tumors. Tissue microarray sections representing 66 rhabdomyosarcoma tumors (34 embryonal rhabdomyosarcoma, 32 alveolar rhabdomyosarcoma) were surveyed by immunohistochemistry using antibodies specific for EGFR and ErbB-2. Immunostains were assessed for intensity (0: no immunostaining; 1: weak; 2: moderate; 3: strong) and percentage of at least 500 neoplastic cells exhibiting membranous or membranous and cytoplasmic immunostaining. EGFR and ErbB-2 expression was considered positive if the product of intensity and percentage was greater than 10. Patients had a median age of 5.7 years (range 8 months–19.1 years), and of 65/66 patients, 38 were males and 27 were females. Expression of ErbB-2 was identified in 22/66 (33%) cases and tended to be more frequent in the alveolar subtype (13/32, 41%, vs 9/34, 26%, P=0.30). Expression of EGFR was identified in 31/66 (47%) cases and correlated with the embryonal subtype (26/34, 76%, vs 5/32, 16%, P<0.0001) independent of stage, age, and gender. Coexpression of EGFR and ErbB-2 was identified in eight tumors, of which six were embryonal rhabdomyosarcoma. None of the cases exhibited EGFR or ErbB-2 gene amplification, as assessed using fluorescence in situ hybridization. Furthermore, analysis of 11 additional rhabdomyosarcoma tumors (six alveolar; five embryonal) revealed no evidence of mutations in EGFR exons 18, 19, 20, and 21. In summary, expression of EGFR and/or ErbB-2 is detected in a sizeable subset of rhabdomyosarcoma tumors without evidence of EGFR or ErbB-2 amplification or mutations in the EGFR tyrosine kinase domain. Notably, expression of EGFR correlates with the embryonal subtype, which is also more likely to coexpress EGFR and ErbB-2.
Similar content being viewed by others
Main
Rhabdomyosarcoma is the most common malignant soft tissue neoplasm in individuals under 19 years of age, with an annual incidence in the United States of approximately 4.6 cases per million.1 Despite a higher prevalence among younger individuals, rhabdomyosarcoma also affects adults.2, 3 Based on morphology and molecular biology, rhabdomyosarcoma tumors are currently classified using the modified International Classification of Rhabdomyosarcoma into three main categories: alveolar rhabdomyosarcoma, embryonal rhabdomyosarcoma, and pleomorphic rhabdomyosarcoma.4
Although both alveolar and embryonal rhabdomyosarcoma tumors exhibit features indicative of rhabdomyoblastic differentiation, the two groups diverge in several aspects that include morphology, molecular genetics, and clinical outcome. Alveolar rhabdomyosarcoma is comprised of small neoplastic cells with scant cytoplasm and harbors nonrandom chromosomal translocations t(2;13)(q35;q14) or t(1;13)(p36;q14) that lead to the fusion of PAX3 and PAX7, respectively, to FOXO1a (FKHR).5, 6, 7 On the other hand, embryonal rhabdomyosarcoma, which also includes spindle cell, botryoid, and anaplastic rhabdomyosarcoma, is generally comprised of neoplastic cells recapitulating various stages of rhabdomyogenesis and lacks the presence of recurrent chromosomal translocations.8 Notably, outcome data consistently indicate that patients with alveolar rhabdomyosarcoma have a worse prognosis than those with embryonal rhabdomyosarcoma.9, 10
The epidermal growth factor receptor (EGFR) family constitutes subclass I of the receptor tyrosine kinase superfamily, and it consists of four genes encoding receptors that play an important role in coordinating many normal cellular processes.11 Two of the EGFR family members—EGFR (ErbB-1) and ErbB-2 (HER2)—have been the subject of intense investigation in view of their involvement in numerous types of human cancers and their potential as molecular targets of therapy.12 Amplification of EGFR and ErbB-2 leading to overexpression of their respective gene products has been demonstrated to be of clinical utility in a variety of human cancers, particularly in those of neuronal and epithelial derivation.13, 14, 15, 16 In addition, the presence of acquired somatic mutations of the EGFR tyrosine kinase domain has been associated with susceptibility to EGFR tyrosine kinase inhibitors (gefitinib or erlotinib).17, 18, 19 Inhibitors of EGFR and ErbB-2 are currently in clinical use or in advanced clinical trials,12, 20 including a recent phase I trial of gefitinib in children with refractory solid tumors.21
Although expression of EGFR and ErbB-2 has been demonstrated22, 23 in rhabdomyosarcoma cell lines, and in vitro evidence suggests that inhibition of EGFR adversely affects the proliferation and survival of rhabdomyosarcoma cells;23, 24 no systematic analysis of EGFR and ErbB-2 expression in rhabdomyosarcoma tumors has been undertaken. In this study, we assessed the expression and copy number status of EGFR and ErbB2 in a large group of rhabdomyosarcoma tumors and correlated our findings with histologic subtype and clinical features. We also surveyed a group of rhabdomyosarcoma tumors for the presence of mutations in the EGFR tyrosine kinase domain.
Materials and methods
Paraffin-Embedded Rhabdomyosarcoma Tumors and Tissue Microarray Construction
Sections from two tissue microarray blocks containing tissue cores from 66 formalin-fixed paraffin-embedded rhabdomyosarcoma tumors from 66 patients who were enrolled on Children's Oncology Group protocols at various participating sites were obtained from The Biopathology Center, Columbus, OH, USA. All tumors were obtained from patients enrolled on the Intergroup Rhabdomyosarcoma Study Group IV protocol25, 26, 27 or later studies. Cases were selected on the basis of submission by the treating institution of a paraffin block or formalin-fixed tissue for banking at The Biopathology Center. The criteria for submitting blocks for banking were not based on any clinical or pathological factors. An average of 3.1 (range, 1 to 6 cores) 1 mm cores of tumor tissue from each case were used to construct the tissue microarray blocks using a manual arrayer (Beecher Instruments, Sun Prairie, WI, USA). Cores from normal tissues were scattered in each of the tissue microarray blocks as controls.
All tumors fulfilled the diagnostic criteria of rhabdomyosarcoma as defined in the World Health Organization classification.8 Tumors included 32 alveolar rhabdomyosarcoma and 34 embryonal rhabdomyosarcoma, all of which expressed myogenin and/or Myo-D1. Alveolar rhabdomyosarcoma tumors were comprised of small neoplastic cells with scant cytoplasm and minimal morphologic evidence of differentiation. Embryonal rhabdomyosarcoma tumors were comprised of spindle cells with variable recapitulation of skeletal muscle differentiation stages. For the purpose of statistical analysis, one tumor with mixed alveolar and embryonal morphologic features was included with the alveolar rhabdomyosarcoma group.8, 28 Two alveolar rhabdomyosarcoma tumors had notable nuclear pleomorphism. Samples included three metastatic tumors, and four samples were procured after initiation of therapy.
This study was conducted with the approval of the St Jude Children's Research Hospital's Institutional Review Board.
Immunohistochemistry
Monoclonal antibodies against EGFR (31G7, 1:10, Zymed, San Francisco, CA, USA), ErbB-2 (1:50, Dako, Carpinteria, CA, USA), myogenin (1:1600, Dako), and MyoD1 (1:25, Dako) were utilized for immunohistochemical analysis. Immunostaining using the EGFR antibody was carried out on the automated Ventana Benchmark XT immunostainer (Ventana Medical Systems, Tucson, AZ, USA) using the manufacturer's deparaffinization, antigen retrieval, and detection reagents. Briefly, for EGFR immunostaining, tissue microarray sections were deparaffinized using EZ Prep followed by 4-min incubation in Protease-1 solution according the manufacturer's recommended protocol. Visualization of EGFR immunostaining was by a biotin-avidin immunoperoxidase assay (IVIEW DAB Detection Kit). Immunostaining using the ErbB-2, myogenin, and MyoD1 antibodies was carried out on a Dako autostainer (Dako) with off-line deparaffinization and pretreatment. Briefly, for ErbB-2, myogenin and MyoD1 immunostaining, tissue microarray sections were deparaffinized using xylene and hydrated using alcohols to distilled water followed by antigen retrieval in a steamer with Target Retrieval (Dako) at pH 6.0. Visualization of ErbB-2, myogenin, and MyoD1 immunostaining was by biotin-avidin immunoperoxidase assay with staining performed on the Dako autostainer using LSAB2 (Dako) detection kit.
Immunostains for EGFR and ErbB-2 were interpreted as described previously.29 The intensity of immunostaining was estimated as follows: 1, weak; 2, moderate; or 3, strong. The percentage of tumor cells positive for EGFR and ErbB-2 in a membranous staining pattern (with or without significant cytoplasmic staining) was estimated by counting at least 500 tumor cells in each core. Results were scored by multiplying the percentage of positive cells by the estimated intensity of immunostaining (score=percentage × intensity; maximum=300). The core with the highest score determined the final value for each case. For the purposes of statistical analysis, a score of 20 or higher was used to define positivity based on data distribution plot (Figure 1). For myogenin and Myo-D1, any unequivocal nuclear immunostaining by tumor cells was considered positive, irrespective of intensity.
Fluorescence In Situ Hybridization
Fluorescence in situ hybridization (FISH) was performed to estimate the copy number of the EGFR and ErbB-2 genes. Four bacterial artificial chromosome-derived probes were used for this purpose. A fluorescein-tagged probe targeting the EGFR locus on chromosome 7p11.2 (Invitrogen, Carlsbad, CA, USA; clone RPCI-11 148P17, 155 kb) was paired with a rhodamine-tagged control probe targeting the CFTR locus on 7q31.2 (Invitrogen, clone RPCI-11 460J21, 167 kb). Similarly, a rhodamine-labeled probe targeting the ErbB-2 locus on chromosome 17q12 (Invitrogen, clone CTD-2019C10, 351 kb) was paired with a fluorescein-labeled control probe targeting the RPA1 locus on 17p13.3 (Invitrogen, clone RPCI-11 4F24, 140 kb). All probes were paired for dual target hybridization in DenHyb buffer (Insitus Laboratories, Albuquerque, NM, USA) at a dilution of 1:50. Hybridization was carried out as described previously.30 DAPI (1 μl/ml) (Insitus Laboratories) was used as a nuclear counterstain, and sections were visualized under a Nikon E800 fluorescence microscope with appropriate filters (Nikon Instruments, Melville, NY, USA) and AI Cytovision software (Applied Imaging, Santa Clara, CA, USA).
Cores in which >90% of nuclei showed hybridization signals were considered informative, and those were scored for the number of fluorescent signals by counting 100 non-overlapping intact nuclei. Samples were considered amplified for EGFR or ErbB-2 if they demonstrated nuclei with EGFR:CFTR or ErbB-2:RPA1 signal ratio >2, respectively.
Mutation Analysis
Eleven frozen rhabdomyosarcoma tumor samples were obtained from the SJCRH Tissue Resources Core Facility. A small fragment of each sample was fixed in formalin and embedded in formalin for quality assurance. All samples represented viable rhabdomyosarcoma tumors (six alveolar; five embryonal), as determined by reviewing hematoxylin and eosin-stained slides, and were positive for myogenin. EGFR expression was detected by immunohistochemistry in 5 samples (one alveolar; four embryonal).
Extracted tumor DNA was used to amplify EGFR exons 18, 19, 20, and 21 using primer sequences and PCR conditions kindly provided by Dr M Meyerson (Dana-Farber Cancer Institute, Boston, MA, USA)17 and available upon request. Following amplification, PCR products were purified using the Qiaex II gel extraction kit (Qiagen, Valencia, CA, USA) or Microcon-100 purification columns (Millipore, Billerica, MA, USA). Purified DNA was then sequenced in both sense and antisense directions by Big Dye® Terminator (v.3) Chemistry using PCR primers on Applied Biosystem 3700 DNA Analyzers.
Statistical Analysis
Associations between EGFR/ErbB-2 immunostaining and stage were examined using the exact Kruskal–Wallis test. Fisher's exact test was used to examine the association between EGFR/ErbB-2 immunostaining and histologic subtype. The exact Wilcoxon rank sum test was used to examine the association between age at diagnosis and histologic subtype. Multiple logistic regression models were used to explore the correlation between ErbB-2 (or EGFR) expression (positive vs negative) and the predictor variables including histologic subtype, stage, age and gender.
Results
Patient Characteristics
The demographic and clinical characteristics of the study group cases are summarized in Table 1. Gender data were available on 65/66 patients, of whom 38 were males (14 alveolar, 24 embryonal) and 27 were females (18 alveolar, 9 embryonal). The patients had a median age of 5.7 years (range, 8 months–19.1 years). As expected, patients with alveolar rhabdomyosarcoma tended to be older than patients with embryonal rhabdomyosarcoma (median age 10 years, range 10 months–19.1 years, vs 4.7 years, range 8 months–17.2 years, respectively; P=0.06).
Patients were staged according to the Intergroup Rhabdomyosarcoma Study Group criteria,28 and staging data were available on 65/66 patients. There was no correlation between histologic subtype and stage (P=0.81) in this study group. Stage was as follows: 12 stage I patients (5 alveolar, 7 embryonal); 9 stage II patients (5 alveolar, 4 embryonal); 30 stage III patients (15 alveolar, 15 embryonal); and, 14 stage IV patients (7 alveolar, 7 embryonal). No follow up data was available.
ErbB-2 and EGFR Expression in Rhabdomyosarcoma Tumors
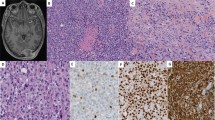
Expression of ErbB-2 (Figure 2b, d) was identified in 22 (33%) of 66 rhabdomyosarcoma tumors. ErbB-2 expression tended to be more frequent in the alveolar subtype, with 13 (41%) of 32 alveolar rhabdomyosarcoma tumors compared to 9 (26%) of 34 embryonal rhabdomyosarcoma tumors positive for ErbB-2, although this difference did not attain statistical significance (P=0.30) (Table 2a). There was no correlation between ErbB-2 expression and stage or between ErbB-2 expression and localized (stages I–III) vs metastatic (stage IV) disease (P=0.27 and P=0.35, respectively). Using a multiple logistic regression model, we further examined whether histologic subtype in rhabdomyosarcoma was significantly correlated with ErbB-2 expression level after adjusting for other variables such as stage, age, and gender and found no evidence of such a correlation.
Expression of EGFR (Figure 2a, c) was identified in 31 (47%) of 66 rhabdomyosarcoma tumors. Notably, there was a significant correlation between EGFR expression and the embryonal subtype, with 26 (76%) of 34 embryonal rhabdomyosarcoma tumors compared to 5 (16%) of 32 alveolar rhabdomyosarcoma tumors positive for EGFR (P<0.0001) (Table 2b). Immunostaining intensity was moderate or strong in 25 of 31 EGFR-positive tumors. There was no correlation between EGFR expression and stage or between EGFR expression and localized (stages I–III) vs metastatic (stage IV) disease (P=0.96 and P=1.0, respectively). A multiple logistic regression model demonstrated that EGFR expression is significantly correlated with the histologic subtype of rhabdomyosarcoma after adjusting for other variables such as stage, age, and gender.
The above analyses were also performed using a lower cutoff score of 10 or higher for both the ErbB-2 and EGFR immunostains and yielded identical conclusions.
Coexpression of EGFR and ErbB-2 was identified in eight (12%) of 66 rhabdomyosarcoma tumors, six of which were of the embryonal subtype. The six embryonal rhabdomyosarcoma tumors with EGFR and ErbB-2 co-expression had advanced stage disease (stage III or IV), while the two alveolar rhabdomyosarcoma tumors had low stage disease (stage I and II). Conversely, alveolar rhabdomyosarcoma tumors were significantly more likely to lack expression of both EGFR and ErbB-2 compared to embryonal rhabdomyosarcoma tumors (16/32, 50%, vs 5/34, 15%, P=0.003). The tumor with mixed alveolar and embryonal histology exhibited strong immunostaining with ErbB-2 and was negative for EGFR. Of the two cases with notable nuclear pleomorphism, one case was positive for ErbB-2 and negative for EGFR, while the second was negative for both.
ErbB-2 and EGFR Gene Copy Number Analysis
Fluorescence in situ hybridization was performed using probes specific for the EGFR gene at 7p11.2 and the ErbB-2 gene at 17q12. In order to control for potential aneuploidy involving chromosomes 7 and 17, both test probes (EGFR and ErbB-2) were paired with control probes targeting loci on opposing chromosomal arms, specifically 7q31.2 and 17p13.3, respectively. Of the 66 cases analyzed, none had nuclei with EGFR:CFTR or ErbB-2:RPA1 signal ratio > 2, thereby indicating no evidence of genomic amplification at either of the test loci (Figure 3).
EGFR Tyrosine Kinase Domain Mutation Analysis
The EGFR exons 18, 19, 20, and 21 were amplifiable using polymerase chain reaction (PCR) in all 11 rhabdomyosarcoma tumor samples analyzed, which included six alveolar and five embryonal rhabdomyosarcoma tumors. Sequencing revealed no detectable mutations, suggesting that none of these tumors harbored mutations in the EGFR tyrosine kinase domain.
Discussion
Proteins of the EGFR family are expressed in various tissues of epithelial, mesenchymal and neuronal origin.12 EGFR expression in the developing human fetus has been detected in a wide variety of tissues,31 including skeletal muscle tissue.32 The expression of EGFR has been also demonstrated in rhabdomyosarcoma cell lines, in which abrogation of EGFR expression was found to impair proliferative ability.23 Likewise, ErbB-2 is required for myoblast cell survival,33 and ectopic expression of ErbB-2 in a transgenic mouse that lacks the p53 tumor suppressor gene has been shown to promote rhabdomyosarcoma development34 (although p53 mutation frequency has been found to be extremely low in rhabdomyosarcoma tumors35). Therefore, EGFR and ErbB-2 seem to be associated with rhabdomyogenesis and appear to play a role in rhabdomyosarcoma development in the mouse. These data, coupled with the fact that EGFR and ErbB-2 are expressed by rhabdomyosarcoma tumors, suggest that targeting these two molecules for therapeutic purposes in preclinical trials might be warranted.
Rhabdomyosarcoma is a common soft tissue neoplasm that includes two primary histologic subtypes—the alveolar and embryonal subtypes—with divergent morphologic, molecular, and clinical features. It is well-established that embryonal rhabdomyosarcoma tumors generally behave less aggressively than alveolar rhabdomyosarcoma tumors.9, 10 However, despite advances in the treatment of patients with rhabdomyosarcoma, the overall 5-year failure-free survival does not exceed 80% even among patients with embryonal rhabdomyosarcoma or low stage disease,27, 36 highlighting the need for novel approaches in stratifying rhabdomyosarcoma patients for therapeutic and prognostic purposes. The correlation of EGFR expression with prognosis has been identified in several human cancers. In cancers of the head and neck, ovary, cervix, and bladder, EGFR expression inversely correlates with patient survival.37, 38 Whether EGFR expression connotes a favorable outcome—since it correlates with the more favorable embryonal subtype—or delineates a subset of embryonal rhabdomyosarcoma with poor outcome remains to be determined. The identification of a subset of rhabdomyosarcoma tumors with coexpression of EGFR and ErbB-2 might also point to a distinct subset of rhabdomyosarcoma tumors that could be particularly susceptible to targeted therapies. Analysis of this subset in our study group reveals that all six embryonal rhabdomyosarcoma tumors with EGFR and ErbB-2 coexpression have advanced stage disease (stage III or IV). Interestingly, the only two alveolar rhabdomyosarcoma cases with coexpression of EGFR and ErbB-2 were low stage tumors (stage I and II).
The expression of EGFR and ErbB-2 does not appear to be as prevalent among other small round cell neoplasms that commonly present in children and adolescents as it is in rhabdomyosarcoma. In a group of Ewing sarcoma family tumors with confirmed EWS translocations, we have identified only 2/47 (4.2%) EGFR-positive tumors and 11/49 (22.4%) ErbB-2-positive tumors.39, 40 Likewise, neuroblastoma tumors predominantly lack EGFR expression (J Khoury, unpublished data), and in a published survey of a large group of neuroblastoma tumors only a small subset expressed ErbB-2.41 These data seem to suggest that the prevalence of EGFR and ErbB-2 expression is distinctly higher among rhabdomyosarcoma tumors compared to the Ewing sarcoma family of tumors and neuroblastoma.
Although immunohistochemistry is useful in identifying EGFR expression, its utility as a screening tool for identifying subjects who may respond to current anti-EGFR therapies has not been established. Among patients with colon cancer, for example, cetuximab—a chimeric monoclonal antibody that targets the extracellular domain of EGFR—appears to show activity in patients whose tumors are negative for EGFR by immunohistochemistry.42 Accordingly, whether anti-EGFR therapies might benefit more patients with rhabdomyosarcoma than is suggested by our immunohistochemistry results remains to be determined in controlled prospective studies. The latter would also overcome another limitation of this study, which involved retrospective analysis of convenience samples from several multi-institutional clinical trials.
Aberrations of the EGFR gene carry significant implications in a variety of human tumors.43 For example, EGFR amplification has been found to be an independent predictor of prolonged survival in patients with glioblastoma multiforme who were older than 60 years of age.44 Using FISH, we demonstrate the absence of ErbB-2 and EGFR amplification in all rhabdomyosarcoma tumors in our study group. In patients with non-small cell lung carcinoma, mutations in the EGFR tyrosine kinase domain have been associated with responsiveness to EGFR inhibitors.17, 18, 19 In our hands, there was no evidence of mutations in the EGFR tyrosine kinase domain in 11 rhabdomyosarcoma tumor samples, as evidenced by the lack of detectable mutations in EGFR exons 18 through 21. Similarly, in a recent study by Baird et al, mutation analysis of all 28 exons of EGFR in 275 soft tissue sarcomas that included six rhabdomyosarcoma tumors revealed no evidence of mutations.45 Despite the biological insight gained from these data, they remain limited by small sample numbers, and by themselves do not rule out a possible role for EGFR inhibitors with anti-tyrosine kinase activity in treating rhabdomyosarcoma. Among patients with recurrent malignant glioma, response to EGFR inhibitors was observed in a subset of patients despite the lack of mutations in the EGFR kinase domain, and such a response was correlated with loss of PTEN and expression of EGFR class III variant (EGFRvIII), a mutant form of EGFR that lacks 267 amino acids from its extracellular domain.46, 47
In conclusion, expression of EGFR and/or ErbB-2 can be detected in a sizeable subset of rhabdomyosarcoma tumors, with no evidence of amplification at 7p11.2 and 17q12 or mutations in the EGFR tyrosine kinase domain. Notably, EGFR expression correlates with the embryonal subtype independent of other variables such as stage, age, and gender, and it is also more likely to coexpress EGFR and ErbB-2.
References
Ries LAG, Eisner MP, Kosary CL, et al (eds). SEER Cancer Statistics Review, 1975–2002. National Cancer Institute: Bethesda, MD, http://seer.cancer.gov/csr/1975_2002/, based on November 2004 SEER data submission, posted to the SEER Web site 2005.
Hawkins WG, Hoos A, Antonescu CR, et al. Clinicopathologic analysis of patients with adult rhabdomyosarcoma. Cancer 2001;91:794–803.
Esnaola NF, Rubin BP, Baldini EH, et al. Response to chemotherapy and predictors of survival in adult rhabdomyosarcoma. Ann Surg 2001;234:215–223.
Qualman SJ, Coffin CM, Newton WA, et al. Intergroup Rhabdomyosarcoma Study: update for pathologists. Pediatr Dev Pathol 1998;1:550–561.
Galili N, Davis RJ, Fredericks WJ, et al. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat Genet 1993;5:230–235.
Shapiro DN, Sublett JE, Li B, et al. Fusion of PAX3 to a member of the forkhead family of transcription factors in human alveolar rhabdomyosarcoma. Cancer Res 1993;53:5108–5112.
Davis RJ, D'Cruz CM, Lovell MA, et al. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res 1994;54:2869–2872.
Parham DM, Barr FG, Kapadia SB, et al. Skeletal muscle tumors. In: Fletcher CDM, Unni KK, Mertens F (eds). World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of Soft Tissue and Bone. IARC Press: Lyon, 2002, pp 146–152.
Punyko JA, Mertens AC, Baker KS, et al. Long-term survival probabilities for childhood rhabdomyosarcoma. A population-based evaluation. Cancer 2005;103:1475–1483.
Koscielniak E, Harms D, Henze G, et al. Results of treatment for soft tissue sarcoma in childhood and adolescence: a final report of the German Cooperative Soft Tissue Sarcoma Study CWS-86. J Clin Oncol 1999;17:3706–3719.
Yarden Y, Sliwkowski MX . Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2001;2:127–137.
Hynes NE, Lane HA . ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer 2005;5:341–354.
Holbro T, Hynes NE . ErbB receptors: directing key signaling networks throughout life. Annu Rev Pharmacol Toxicol 2004;44:195–217.
Slamon DJ, Clark GM, Wong SG, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987;235:177–182.
Ohgaki H, Dessen P, Jourde B, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res 2004;64:6892–6899.
Sunpaweravong P, Sunpaweravong S, Puttawibul P, et al. Epidermal growth factor receptor and cyclin D1 are independently amplified and overexpressed in esophageal squamous cell carcinoma. J Cancer Res Clin Oncol 2005;131:111–119.
Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497–1500.
Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129–2139.
Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from ‘never smokers’ and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA 2004;101:13306–13311.
Roskoski Jr R . The ErbB/HER receptor protein-tyrosine kinases and cancer. Biochem Biophys Res Commun 2004;319:1–11.
Daw NC, Furman WL, Stewart CF, et al. Phase I and pharmacokinetic study of gefitinib in children with refractory solid tumors: a Children's Oncology Group Study. J Clin Oncol 2005;23:6172–6180.
Ricci C, Landuzzi L, Rossi I, et al. Expression of HER/erbB family of receptor tyrosine kinases and induction of differentiation by glial growth factor 2 in human rhabdomyosarcoma cells. Int J Cancer 2000;87:29–36.
De Giovanni C, Landuzzi L, Frabetti F, et al. Antisense epidermal growth factor receptor transfection impairs the proliferative ability of human rhabdomyosarcoma cells. Cancer Res 1996;56:3898–3901.
Ricci C, Polito L, Nanni P, et al. HER/erbB receptors as therapeutic targets of immunotoxins in human rhabdomyosarcoma cells. J Immunother 2002;25:314–323.
Breneman JC, Lyden E, Pappo AS, et al. Prognostic factors and clinical outcomes in children and adolescents with metastatic rhabdomyosarcoma—a report from the Intergroup Rhabdomyosarcoma Study IV. J Clin Oncol 2003;21:78–84.
Donaldson SS, Meza J, Breneman JC, et al. Results from the IRS-IV randomized trial of hyperfractionated radiotherapy in children with rhabdomyosarcoma—a report from the IRSG. Int J Radiat Oncol Biol Phys 2001;51:718–728.
Crist WM, Anderson JR, Meza JL, et al. Intergroup rhabdomyosarcoma study-IV: results for patients with nonmetastatic disease. J Clin Oncol 2001;19:3091–3102.
Qualman SJ, Bowen J, Parham DM, et al. Protocol for the examination of specimens from patients (children and young adults) with rhabdomyosarcoma. Arch Pathol Lab Med 2003;127:1290–1297.
Ginestier C, Charafe-Jauffret E, Bertucci F, et al. Distinct and complementary information provided by use of tissue and DNA microarrays in the study of breast tumor markers. Am J Pathol 2002;161:1223–1233.
Fuller CE, Wang H, Zhang W, et al. High-throughput molecular profiling of high-grade astrocytomas: the utility of fluorescence in situ hybridization on tissue microarrays (TMA-FISH). J Neuropathol Exp Neurol 2002;61:1078–1084.
Wakeling EL, Abu-Amero SN, Stanier P, et al. Human EGFR, a candidate gene for the Silver–Russell syndrome, is biallelically expressed in a wide range of fetal tissues. Eur J Hum Genet 1998;6:158–164.
Nanney LB, Stoscheck CM, King Jr LE, et al. Immunolocalization of epidermal growth factor receptors in normal developing human skin. J Invest Dermatol 1990;94:742–748.
Andrechek ER, Hardy WR, Girgis-Gabardo AA, et al. ErbB2 is required for muscle spindle and myoblast cell survival. Mol Cell Biol 2002;22:4714–4722.
Nanni P, Nicoletti G, De Giovanni C, et al. Development of rhabdomyosarcoma in HER-2/neu transgenic p53 mutant mice. Cancer Res 2003;63:2728–2732.
Taylor AC, Shu L, Danks MK, et al. P53 mutation and MDM2 amplification frequency in pediatric rhabdomyosarcoma tumors and cell lines. Med Pediatr Oncol 2000;35:96–103.
Wharam MD, Meza J, Anderson J, et al. Failure pattern and factors predictive of local failure in rhabdomyosarcoma: a report of group III patients on the third Intergroup Rhabdomyosarcoma Study. J Clin Oncol 2004;22:1902–1908.
Nicholson RI, Gee JM, Harper ME . EGFR and cancer prognosis. Eur J Cancer 2001;37 (Suppl 4):S9–S15.
Xia W, Lau YK, Zhang HZ, et al. Combination of EGFR, HER-2/neu, and HER-3 is a stronger predictor for the outcome of oral squamous cell carcinoma than any individual family members. Clin Cancer Res 1999;5:4164–4174.
Khoury JD, Navid F, Rodriguez-Galindo C, et al. EGFR and ERBB2 protein expression and gene amplification status in Ewing sarcoma family of tumors [Abstract]. Mod Pathol 2005;18:304A.
Lai R, Navid F, Rodriguez-Galindo C, et al. STAT3 is activated in a subset of the Ewing sarcoma family of tumors. J Pathol 2006;208:624–632.
Gambini C, Sementa AR, Boni L, et al. Expression of HER2/neu is uncommon in human neuroblastic tumors and is unrelated to tumor progression. Cancer Immunol Immunother 2003;52:116–120.
Chung KY, Shia J, Kemeny NE, et al. Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. J Clin Oncol 2005;23:1803–1810.
Dancey JE, Freidlin B . Targeting epidermal growth factor receptor—are we missing the mark? Lancet 2003;362:62–64.
Smith JS, Tachibana I, Passe SM, et al. PTEN mutation, EGFR amplification, and outcome in patients with anaplastic astrocytoma and glioblastoma multiforme. J Natl Cancer Inst 2001;93:1246–1256.
Baird K, Davis S, Antonescu CR, et al. Gene expression profiling of human sarcomas: insights into sarcoma biology. Cancer Res 2005;65:9226–9235.
Mellinghoff IK, Wang MY, Vivanco I, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med 2005;353:2012–2024.
Kuan CT, Wikstrand CJ, Bigner DD . EGF mutant receptor vIII as a molecular target in cancer therapy. Endocr Relat Cancer 2001;8:83–96.
Acknowledgements
We acknowledge the outstanding services of St Jude Children's Research Hospital's Tissue Resources Facility and the Hartwell Center for Bioinformatics and Biotechnology. This work was supported in part by NIH grant CA-023099 (JDK) and by the American Lebanese Syrian Associated Charities (ALSAC).
Author information
Authors and Affiliations
Corresponding author
Additional information
Presented in part as a proffered paper at the 95th Annual United States and Canadian Academy of Pathology Meeting in Atlanta, Georgia, February 2006.
Rights and permissions
About this article
Cite this article
Ganti, R., Skapek, S., Zhang, J. et al. Expression and genomic status of EGFR and ErbB-2 in alveolar and embryonal rhabdomyosarcoma. Mod Pathol 19, 1213–1220 (2006). https://doi.org/10.1038/modpathol.3800636
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800636
Keywords
This article is cited by
-
CD276-CAR T cells and Dual-CAR T cells targeting CD276/FGFR4 promote rhabdomyosarcoma clearance in orthotopic mouse models
Journal of Experimental & Clinical Cancer Research (2023)
-
Rhabdomyosarcoma
Nature Reviews Disease Primers (2019)
-
Redirecting T cells to treat solid pediatric cancers
Cancer and Metastasis Reviews (2019)
-
Clinicopathological characteristics and treatment outcomes of Chinese patients with genitourinary embryonal rhabdomyosarcoma
World Journal of Surgical Oncology (2015)
-
Novel EGFR-specific immunotoxins based on panitumumab and cetuximab show in vitro and ex vivo activity against different tumor entities
Journal of Cancer Research and Clinical Oncology (2015)