Abstract
Type XVII collagen (180-kDa bullous pemphigoid antigen) is a structural component of hemidesmosomes. Mutations in the type XVII collagen gene (COL17A1) have been established to be the molecular basis of non-Herlitz junctional epidermolysis bullosa (JEB-nH), an inherited skin blistering disorder. Here we report for the first time truncated type XVII collagen expression, caused by homozygosity for a COL17A1 donor splice-site mutation (4261+1 g → c), which was identified by PCR amplification on genomic DNA. By RT-PCR and sequencing of cDNA derived from mRNA from the patient’s cultured keratinocytes, we provide evidence of cryptic splicing and exon skipping, most abundantly of exon 52. JEB-nH patients with COL17A1 splice-site mutations resulting in an exon skip often have no immunologically detectable type XVII collagen. However, in our patient with the generalized atrophic benign epidermolysis bullosa subtype, a small amount of type XVII collagen was detectable in the skin, and immunoblotting of cultured keratinocytes revealed that the 180-kDa protein was 10 kDa shorter. We hypothesize that the function of this truncated type XVII collagen polypeptide, which is expressed at low levels, is impaired, explaining the JEB-nH phenotype.
Similar content being viewed by others
Introduction
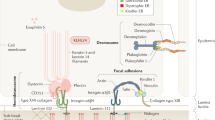
Type XVII collagen (180-kDa bullous pemphigoid antigen; BP180, BPAG2), encoded by the COL17A1 gene, plays a critical role in adhesion of basal keratinocytes to the epidermal basement membrane zone (EBMZ). Its expression is restricted to hemidesmosomes, morphological structures that mediate the adhesion of basal keratinocytes to the underlying EBMZ in stratified and pseudostratified epithelia. Type XVII collagen interacts with other hemidesmosomal components (eg, the α6β4 integrin and 230-kDa bullous pemphigoid antigen [BP230]), and thereby plays a role in the assembly and stabilization of hemidesmosomes (Hopkinson and Jones, 2000; Schaapveld et al, 1998). The molecule is a type II transmembrane protein whose carboxy terminal end is located outside the cell. This extracellular part contains 15 collagenous domains (COL1–15), interspaced by noncollagenous domains (NC1–16) (Giudice et al, 1992), which gives the molecule a certain flexibility. By electron microscopy, type XVII collagen is seen as a rigid central rod (COL15) followed by a movable tail (Hirako et al, 1996). Like other collagens, the molecule is able to trimerize by self assembly in such a way that the extracellular fragment forms a triple helix, α1[XVII]3 (Balding et al, 1997; Hirako et al, 1996; Pas et al, 1999). The length of the rod domain is sufficient to traverse the lamina lucida, and the flexible carboxyl terminus has been seen within the lamina densa (Shimizu, 1998). Its as yet unidentified ligand(s) may reside within the lamina densa or lamina lucida. A candidate ligand is laminin 5, which is mainly localized in the upper lamina densa (Shimizu, 1998). An interaction between the extracellular domain of type XVII collagen and laminin 5 has been reported (Reddy et al, 1998). The globular cytoplasmic domain is sufficient for the localization of type XVII collagen in hemidesmosomes because after removal of the extracellular and transmembrane domains it is still located in hemidesmosomes (Borradori et al, 1997; Hopkinson et al, 1995).
Mutations in the gene for type XVII collagen (COL17A1) lead to non-Herlitz junctional epidermolysis bullosa (JEB-nH), an autosomal recessive nonlethal skin blistering disease with hypoplastic hemidesmosomes and a split level in the lamina lucida (Jonkman et al, 1995; McGrath et al, 1995). Most COL17A1 mutations described to date are nonsense and deletion mutations leading to premature termination of translation and absence of type XVII collagen from the skin (Darling et al, 1997; Floeth et al, 1998; Gatalica et al, 1997; Jonkman et al, 1997; McGrath et al, 1995; 1996a, 1996b; Scheffer et al, 1997; Schumann et al, 1997; Shimizu et al, 1998). To date, three acceptor splice-site mutations (Chavanas et al, 1997; Darling et al, 1998; Pulkkinen et al, 1999), one donor splice-site mutation (Pulkkinen et al, 1999), and four missense mutations (Floeth et al, 1998; McGrath et al, 1996a; Schumann et al, 1997; Tasanen et al, 2000) have been reported. In only one (Schumann et al, 1997) of the patients could type XVII collagen be immunologically detected in skin or cultured keratinocytes. The phenotype of JEB-nH caused by type XVII collagen deficiency may vary from generalized blistering with alopecia, also known as generalized atrophic benign epidermolysis bullosa (GABEB) (Hashimoto et al, 1976; Hintner and Wolff, 1982; Jonkman et al, 1996), to mild localized blistering without loss of hair (Floeth et al, 1998; Mazzanti et al, 1998; Schumann et al, 1997).
In this study, we report for the first time homozygosity for a donor splice-site mutation in COL17A1 leading to multiple aberrant mRNA in a patient with generalized JEB-nH. The most abundant mRNA has an internal deletion of 390 bp, which leads to the expression of a shortened nonfunctional type XVII collagen polypeptide with a 130 amino acid deletion in the carboxy terminal half.
Results
Immunofluorescence Microscopy
There was no reaction with the monoclonal antibody 1D1 (Fig. 1C), which detects the full-length type XVII collagen and the cleaved 120-kDa ectodomain. Reactivity with the monoclonal antibody 123, which is specific for the cleaved 120-kDa ectodomain, was much weaker than in normal intact skin (data not shown). The monoclonal antibody 1A8C, which is specific for the endodomain of the molecule, produced a weak staining along the epidermal basement membrane zone (Fig. 1A), whereas it did not react with the skin of other type XVII-collagen deficient GABEB patients. Uncein recognized by monoclonal antibody 19-DEJ-1 (Fine, 1990), the marker for junctional epidermolysis bullosa, was completely absent. The expression of both BP230 and HD1/plectin was linear but reduced as compared with that in normal controls (not shown). Integrin α6β4, laminin 5, and type VII collagen were normally expressed (not shown). From these results, we conclude that the amount of type XVII collagen was reduced but not altogether absent in this patient.
Immunofluorescence microscopy of the epidermal basement membrane zone using monoclonal antibodies 1A8c (A, B) and 1D1 (C, D), respectively, to the endo- and ectodomains of type XVII collagen on skin specimens of the patient (A, C) and a healthy human control subject (B, D). Expression of epitope 1A8c is severely reduced (A), whereas 1D1 is completely absent in the skin of the patient. Bar is 10 μm.
Type XVII Collagen mRNA Analysis
To find the defect in the COL17A1 gene that underlies the generalized JEB-nH phenotype in this patient, RT-PCR analysis was performed on mRNA extracted from cultured keratinocytes. mRNA from the squamous cell carcinoma cell line UMSSC-22B served as a positive control, and reaction mixtures in which no reverse transcriptase was used served as a negative control. Aberrant amplification was observed for the region spanning bp 3679 to 4596 of the cDNA for type XVII collagen, of which there were fragments of different molecular weight that were all shorter than the expected 917 bp PCR product (Fig. 2, lane 2). At the position of the wild-type 917 bp band only very faint staining was seen, but all attempts to clone and sequence this hypothetical band were unsuccessful. As expected all six controls, UMSSC-22B cells (Fig. 2, lane 4), and five generalized JEB-nH patients only showed a single band (not shown).
Identification of multiple aberrant mRNA fragments in the region spanning bp 3679 to 4596 of type XVII collagen by RT-PCR on poly(A+) RNA isolated from cultured patient keratinocytes (lane 2). The human squamous cell carcinoma cells UMSSC-22B served as a positive control for the wild-type product (917 bp, lane 4). Lane 1 contained a 100-bp calibration marker.
Subcloning and sequencing of the multiple smaller fragments revealed internal deletions of, respectively, 92, 210, 285, 390, or 537 bp (variants A to E, Fig. 3, Table 1). Variant A causes a frameshift at position 1356, which results in a shorter final translation product of which the carboxy-terminal end contains 68 new amino acids that have no resemblance to the normal protein. All other deletions are in-frame: variant B and C remove one, variant D two, and variant E three collagenous domains as well as flanking sequences. Sequencing of the small fragment (F) identified it to be a transcript from the copine I gene of chromosome 20, and it therefore can be considered to be nonspecific.
Top: Schematic representation of the genomic organization of exons 50 to 53 of the type XVII collagen gene. Introns are depicted as dotted lines. Normal splicing is illustrated above the gene. Alternative splicing as a consequence of the gt → ct mutation at nucleotide position 4261+1/2 results in five aberrant transcripts (A to E) depicted under the gene. The activated cryptic splice sites are depicted above its position in exon 52. Bottom: Schematic representation of the COL17A1 polypeptide variants (A to E) found. As a reference, the wild-type polypeptide (wt) is illustrated above the altered splicing variants; the first nucleotide of each exon is depicted above the bar. The collagenous domains are cross-hatched. Variant A contains a unique 68-amino acid tail which is depicted by a solid line.
Identification of Homozygosity for a Donor Splice-Site Mutation
In variant E, exon 51 and 52 were deleted, in variant D the deletion exactly spanned exon 52, and in the other variants there are partial deletions of exon 52. All deletions had the same downstream join; that is, the acceptor site of exon 53. Therefore, we presumed that the mutation underlying the cryptic splicing and exon skipping most probably was located in the exon/intron boundaries of exon 52. We determined the DNA sequence of these exon/intron boundaries by PCR amplification of exon 52 and its flanking sequences from genomic DNA of the patient. Direct sequencing of the PCR product and sequencing of four independent clones gave a g-to-c substitution at nucleotide 4261+1 in the donor splice site of intron 52. No mutation was found in the 3′ acceptor splice site of intron 51 or in exon 52 itself.
Analysis of Type XVII Collagen Protein Expression
The expression of type XVII collagen in the keratinocytes of the patient was investigated by immunoblotting. Whereas in lysates of normal keratinocytes a band occurs at 180 kDa (Fig. 4B, lane 4*), in those of keratinocytes from the patient there was a band at approximately 170 kDa (Fig. 4B, lane 2*). The intensity of this band was weaker than that representing the normal keratinocyte protein (Fig. 4A, lanes 2 and 4), although the extract had been concentrated before analysis. Thus, as in skin, the expression level of type XVII collagen was severely decreased.
Patient cells synthesize a shortened type XVII collagen polypeptide. Immunoblot analysis of cellular protein extracts of cultured patient keratinocytes (lanes 1 and 2) and normal keratinocytes (lanes 3 and 4). A, Lanes 1 and 3, 230-kDa bullous pemphigoid antigen (BP230); lanes 2 and 4, type XVII collagen. Staining for BP230 was performed with BP230-specific bullous pemphigoid patient serum and for type XVII collagen with monoclonal antibody 1A8C. Intensity scans for the mutant type XVII collagen band from this blot and other blots (not shown) gave a maximal expression level of 15% mutant protein compared with normal keratinocyte type XVII collagen expression. B, Redevelopment of the immunoblot in A for mixed-in biotinylated calibration proteins. This shows the drop in molecular weight of the patient type XVII collagen band (lane 2*) compared with wild-type protein (lane 4*). The molecular weight of the mutant protein was thus calculated as approximately 170 kDa. (* indicates redevelopment of the corresponding lanes shown in A).
Discussion
Here we describe the effects of homozygosity for a donor splice-site mutation of exon 52 in the COL17A1 genes of a patient with a generalized variant of JEB-nH. Analysis of splice-site mutations of this region of the COL17A1 genes demonstrated homozygosity for a g-to-c [4261+1 g → c] transition in the donor splice site (Fig. 3). This mutation was revealed by inbreeding and must have occurred not later than 1794 in the rural municipality 0527 of East Norway (Gedde-Dahl, 1971). Surprisingly, no similar patient has so far been recorded in Norway.
By RT-PCR, we were able to show different aberrant mRNA with internal deletions in the region spanning exon 51 and 52. One of these deletions (ie, 92 bp [variant A]) gives a frameshift leading to a shortened polypeptide with an aberrant tail. The other deletions, of 210, 285, 390, and 537 bp (variants B to E), are in-frame and lead to removal of 70 (1317 to 1386), 95 (1292 to 1386), 130 (1257 to 1386), and 179 (1208 to 1386) amino acid fragments from the corresponding proteins. These deletions include the COL3 domain in variant B and C, both COL3 and COL4 in variant D, and COL3, COL4, and COL5 in variant E. The deduced joins of the shortened polypeptides, however, are all located in noncollagenous domains, NC4-NC3, NC5-NC3, and NC6-NC3, respectively. In case of variant, D and E, the amino acid at the site of the join is changed.
The most abundant transcript, D, contains the 390-bp deletion that exactly spans exon 52. Therefore, we presume that the splice-site mutation leads to exon skipping and cryptic splicing (Fig. 3). Splicing of higher eukaryotic pre-mRNA is thought to be influenced by three main elements: the 5′- and 3′-splice sites and the branch point sequence. Consensus sequence analysis has shown that, in contrast to the branch point sequence, the 5′- and 3′-splice sites must be absolutely conserved gt and ag dinucleotides, respectively. Mutations at these sites abolish and/or greatly reduce normal splicing. The integrity of the overall 5′ donor splice site is required for correct processing of nuclear pre-mRNA (Ohshima and Gotoh, 1987).
Remarkably, the 5′ donor splice site between exon 52 and intron 52 is weak because it does not follow the consensus; that is, exon 52 ends with AGCgt, whereas the consensus is AGgt. The 5′ donor splice site was analyzed by the splice-site score as described by Shapiro and Senapathy (1987). In this procedure, splice sites are scored relative to the frequency with which the same bases have been found in 542 normal splice sites of primate genes. The splice-site score for the wild-type 5′ donor splice site of intron 52 is 78.7% and for the mutated form of the 5′ donor splice site of intron 52 is 61.2% (a sequence identical to the consensus gives a 100% score). It is unknown if such nonconforming splice sites have a specific role and/or need specific components of the splicing machinery for correct splicing. An overview of the DNA sequence used as (cryptic) splice site in the five variants and the splice-site scores for these sequences are given in Table 2). Note that the most abundant transcript, D, has the highest splice-site score (77.8%) among the alternative transcripts.
The protein product(s) in the keratinocytes from the patient appeared in immunoblotting as a band approximately 10 kDa shorter than normal, at about 170 kDa. This agrees with the RT-PCR, which gives the 390 bp deleted transcript as the major mRNA. The splicing out of exon 52 would result in a loss of 130 amino acids corresponding with of 12.4 kDa molecular weight. There was no staining of the skin with monoclonal 1D1. This antibody is believed to bind to type XVII collagen at an epitope located in the most-carboxy-terminal 20 kDa portion of the molecule (Hirako et al, 1996). Because this includes the COL3 domain and flanking regions, the deletion of this region would explain the nonreactivity of the skin of our patient with 1D1 antibody. Possibly, some full-length type XVII collagen may be expressed at a subthreshold level because the RT-PCR showed a very faint band at the position of the expected wild-type amplimer. However, neither immunofluorescence microscopy nor immunoblotting revealed expression of the wild-type protein.
Thus, the final 170-kD protein product is a mutant form of type XVII collagen. The relative decrease in molecular weight of the different proteins encoded by the transcripts varies from 7.2 to 12.5 kDa (excluding possible post-translational modification). This, combined with a possible broadening or shift of bands caused by phosphorylated intermediates (Kitajima et al, 1995), makes it hard to identify the character of the protein. However, based on the relative amounts of the different transcripts it is most likely that the transcript in which exon 52 is deleted (D) produced the smaller 170 kDa polypeptide. The polypeptide product of the out-of-frame 92 bp deletion (C) is unlikely to be stable; the shortened product of this transcript contains 68 new, nonsense, amino acids at the carboxy-terminus, which probably affects protein stability. All other transcripts contain joins outside of the helical domains and therefore have correct carboxy-termini. Normally, type XVII collagen is assembled as a triple-helical homotrimer. Because the helical winding of type XVII collagen is proposed to be N- to C-terminal (Areida et al, 2001), and mRNA truncations were found at the C-terminal side, the initiation of the helical winding of the mutant polypeptides is likely to be normal. Dominant negative interference, such as that described for a glycine substitution mutation in COL17A1 (McGrath, 1996a), is not likely to occur with an in-frame truncation mutation because helical winding may not be completed when heterologous type XVII collagen polypeptides attempt to associate into heterotrimer complexes. In contrast, homologous type XVII collagen polypeptides, although mutant, may assemble into homotrimeric complexes because the homologous domains may position in the correct intermolecular position. Because the exon 52 deletion transcript is the most abundant (Fig. 2, D), and also because its translation product may exert a dominant negative effect on the other mutant polypeptides, this polypeptide most probably is the one present in the skin of the patient.
This is the first patient with a COL17A1 splice-site mutation in which a mutant protein is expressed in the skin. Donor splice-site mutations of genes other than COL17A1 have been observed in epidermolysis bullosa patients. In a number of cases this resulted in a similar effect on transcription, namely activation of cryptic splice sites and exon skipping (Gardella et al, 1996; Posteraro et al, 1998; Terracina et al, 1998; Weil et al, 1988). In some patients, this was accompanied by a milder phenotype. In our patient, the preferentially expressed deletion of the mRNA does not interfere with the coding frame and results in a shortened polypeptide lacking two collagen domains. Because the clinical phenotype of the JEB-nH patient was GABEB, the resulting mutant type XVII collagen is either not functional or is expressed in too small amounts to prevent a normal clinical phenotype. Recently, another splice-site mutation in the same region was published (Pulkkinen et al, 1999). In this case the patient was heterozygous and had two splice-site mutations. One of them was located at the donor splice site of exon 50 and led to three transcriptional variants, two out-of-frame and one in-frame skip of exon 51. In contrast with our patient, this patient had no protein expression, presumably because of the amount of stable transcripts; that is, mRNA with the in-frame exon skip was too low to be detected.
In summary, we here describe a case of homozygosity for a donor splice-site mutation in the COL17A1 gene. This results in cryptic splicing and exon skipping and the expression of a truncated 170-kDa type XVII collagen polypeptide.
Materials and Methods
Patient
The patient (EB6-1), a female aged 69 years at the end of the present study, was first investigated at age 33 years and preliminarily diagnosed as having dystrophic epidermolysis bullosa by Dr. T. Gedde-Dahl (Gedde-Dahl, 1971; patient VI 1; pages 64, 64, 155). She was the only child of second cousins and had herself a single child who was clinically normal. She was reexamined at age 61 and classified as having epidermolysis bullosa atrophicans, mitis type. At 33 years of age, the retrospective history revealed onset of blistering before she was 1 year old. The blisters then occurred only on the extremities and until adult age, no oral blisters developed. The toenails were dystrophic and the finger nails largely normal, despite repeated loss of individual nails. The dentition was poor and had been completely extracted. When she was 30, thinning of the hair was noted, particularly on the sides of the head above the ears. In her 60s she presented with pronounced diffuse alopecia; few eyelashes and sparse hair in the eyebrows, axillae, and pubic regions. Blistering was generalized and blisters often occurred in groups and most frequently on the limbs (but not particularly over bends). The lesions healed without scarring or milia but with atrophic skin to variable degrees, and some pigmented maculae closely resembling those in the phenotype known as GABEB (Hashimoto et al, 1976; Hintner and Wolff, 1982).
At age 61 biopsies of affected skin of the lower back and forearm were examined by electron microscopy (I. Anton-Lamprecht, Heidelberg, Germany), which revealed junctional split. A biopsy from unaffected skin of the forearm revealed areas with a reduced number of immature hemidesmosomes, and between them areas with normal-appearing hemidesmosomes. The expression of type XVII collagen was reduced in the skin (Jonkman et al, 1996, patient 6). By microsatellite typing she had been found to be heterozygous at all three loci of the laminin 5 subunits (Gedde-Dahl et al, 1994, but homozygous at intragenic markers at the type XVII collagen locus COL17A1 (Scheffer et al, 1997, patient D).
Skin Specimens
A knife-biopsy specimen was obtained from the skin over the scapula of the patient. One part of the skin specimen was used for keratinocyte culturing and the other part for immunofluorescence microscopy.
Keratinocyte cultures were started from the biopsy according to the method of Mitra and Nickoloff (1994) and grown in serum-free medium (BioWhittaker Europe SPRL, Belgium).
Antibodies
Monoclonal antibodies 1D1 and 1A8C (kind gift of Dr. Owaribe, Nagoya, Japan) are directed against the ectodomain and the endodomain of type XVII collagen, respectively (Nishizawa et al, 1993). Monoclonal antibody 123 (kind gift of Dr. Marinkovich, Stanford, California) is directed against the 120-kDa cleaved ectodomain of type XVII collagen (LAD-1); R815 (the kind gift of Dr. Owaribe) is directed against the 230-kDa bullous pemphigoid antigen (BP230). Monoclonal antibody GB3 (the kind gift of Dr. Meneguzzi, Nice, France) is directed against laminin 5; HD121 (the kind gift of Dr. Owaribe) is directed against 500-kDa HD1/plectin, 450-11A (the kind gift of Dr. Kennel, Oak Ridge National Laboratory, Oak Ridge, Tennessee) is directed against the β4 integrin subunit; and 19-DEJ-1 (a generous gift of Dr. Fine, University of North Carolina School of Medicine, Chapel Hill, North Carolina) is directed against uncein, a component of the lamina lucida. Monoclonal antibody LH7:2 (the kind gift of Dr. Leigh, London Hospital, London, United Kingdom) is directed against type VII collagen. All monoclonal antibodies were mouse IgG1.
Immunofluorescence Studies
Cryostat sections (4 μm) of skin specimens were processed for immunofluorescence as described previously (Jonkman et al, 1992). In combination with the primary mouse monoclonal antibodies, biotinylated goat antimouse IgG1, in a dilution 1:100 (SBA, Birmingham, Alabama), and DTAF (dichlorotriazinyl-amino-fluorescein)-conjugated streptavidin, in a dilution 1:200 (Jackson Immuno Research, West Grove, Pennsylvania), were used in secondary and tertiary steps. Digital video microscopic images of tissue sections were obtained using an imaging system with long exposure times to detect very low levels of fluorescence (Bruins et al, 1994).
cDNA Synthesis and PCR
Cells were grown to subconfluence and then harvested by extraction with 1 ml/15 cm2 TRIZOL reagent (Life Technologies BV, Breda, Holland). The RNA was isolated according to the manufacturer’s protocol and poly(A+) mRNA was purified from total RNA using an mRNA Isolation Kit (Roche Molecular Biochemicals, Mannheim, Germany).
First strand type XVII collagen cDNA was prepared by reverse transcription of 5 μg of mRNA using the M-MLV reverse transcriptase cDNA synthesis system (Life Technologies BV) with 0.5 μg oligo dT primer in a total volume of 20 μl. The PCR was performed in a total volume of 50 μl containing 20 mm Tris (pH 8.4), 50 mm NaCl, 1.5 mm MgSO4, 0.2 mm of each dNTP, 100 ng of each primer, and 1 U of Taq DNA polymerase (Life Technologies BV). All reactions described had a pre-PCR step of 2 minutes at 94° C followed by an extension step of 60 minutes at 72° C, and a post-PCR terminal extension step of 8 minutes at 72° C. A 971-bp fragment (bp 3679–4596) in the carboxy-terminal end of type XVII collagen was amplified by PCR using the following primers: downstream 5′-GTGTGGTCCAGCATCAGCGT-3′ (bp 3679–3698) and upstream 5′-CGGCTTGACAGCAATACTTC-3′ (bp 4577–4596) on 1 μl of the cDNA. The PCR conditions were 32 cycles of 45 seconds at 94° C, 45 seconds at 60° C, and 90 seconds at 72° C. The resulting DNA fragments were isolated from a 2% agarose gel and cloned into the pCRII-Topo vector (Topo II TA cloning Kit; Invitrogen, San Diego, California). Sequencing of the subclones was performed in an automated system and/or by using the dideoxy chain termination method (T7 Sequencing Kit; Pharmacia LKB Biotechnology, Uppsala, Sweden).
Determination of the Genomic Sequence
The fragment containing the skipped type XVII collagen exon and flanking sequences was amplified by PCR. The following two primers were used: 5′-AGGAGCCCTGGCAACCTATG-3′ (downstream primer, 3801–3820) and upstream primer 5′-CCACAAACAAGAAAGCCAGT-3′ (intronic). The PCR was performed in a total volume of 25 μl containing 20 mm Tris-HCl (pH 8.4), 50 mm NaCl, 1.5 mm MgCl2, 0.2 mm of each dNTP, 150 ng of each primer, and 2 U of Taq DNA polymerase (Life Technologies BV). The PCR condition was: a denaturation step of 5 minutes at 94° C, followed by 35 cycles of 60 seconds at 94° C, 60 seconds at 54° C, 60 seconds at 72° C, and a terminal extension step of 8 minutes at 72° C. The PCR products were cloned into the pGEM-T vector (Promega, Madison, Wisconsin) and processed for sequencing. In addition, an independent PCR-product was directly sequenced.
Immunoblotting
Keratinocytes were grown in serum-free medium until subconfluence, lysed with cold water containing 20 U/ml benzonase (Roche Molecular Biochemicals), and then extracted with SDS-PAGE sample buffer. Extracts from the patient cells were concentrated by ultrafiltration before electrophoresis. Extracts were separated on 5% acrylamide slabgels, electrophoretically transferred to nitrocellulose filters, and stained with type XVII collagen-specific antibody 1A8C. Molecular weights were calibrated by mixing in biotinylated marker proteins as previously described (Pas et al, 1997).
References
Areida SK, Reinhardt DP, Müller PK, Fietzek PP, Köwitz J, Marinkovich MP, and Notbohm H (2001). Properties of the collagen type XVII ectodomain: Evidence for N- to C-terminal triple helix folding. J Biol Chem 276: 1594–1601.
Balding S, Diaz LA, and Giudice GJ (1997). A recombinant form of the human BP180 ectodomain forms collagen-like homotrimeric complex. Biochemistry 36: 8821–8830.
Borradori L, Koch PJ, Niessen CM, Erkeland S, van Leusden MR, and Sonnenberg A (1997). The localization of bullous pemphigoid antigen 180 (BP180) in hemidesmosomes is mediated by its cytoplasmic domain and seems to be regulated by the beta4 integrin subunit. J Cell Biol 136: 1333–1347.
Bruins S, de Jong MJCM, Heeres K, Wilkinson MHF, Jonkman MF, and van der Meer JB (1994). Fluorescence overlay antigen mapping of the epidermal basement membrane zone: I. Geometric errors. J Histochem Cytochem 42: 555–560.
Chavanas S, Gache Y, Tadini G, Pulkkinen L, Uitto J, Ortonne JP, and Meneguzzi G (1997). A homozygous in-frame deletion in the collagenous domain of bullous pephoid antigen BP180 (type XVII collagen) causes generalized atrophic benign epidermolysis bullosa. J Invest Dermatol 109: 74–78.
Darling TN, McGrath JA, Yee C, Gatalica B, Hametner R, Bauer JW, Pohla-Gubo G, Christiano AM, Uitto J, Hintner H, and Yancey KB (1997). Premature termination codons are present on both alleles of the bullous pemphigoid antigen 2/type XVII collagen gene in five Austrian families with generalized atrophic benign epidermolysis bullosa. J Invest Dermatol 108: 463–468.
Darling TN, Yee C, Koh B, McGrath JA, Bauer JW, Uitto J, Hintner H, and Yancey KB (1998). Cycloheximide facilitates the identification of aberrant transcripts resulting from a novel splice-site mutation in COL17A1 in a patient with generalized atrophic benign epidermolysis bullosa. J Invest Dermatol 110: 165–169.
Fine JD (1990). 19-DEJ-1, a monoclonal antibody to the hemidesmosome-anchoring filament complex, is the only reliable immunohistochemical probe for all major forms of junctional epidermolysis bullosa. Arch Dermatol 126: 1187–1190.
Floeth M, Fiedowowicz J, Schäcke H, Hammami-Hauasli N, Owaribe K, Trüeb RM, and Bruckner-Tuderman L (1998). Novel homozygous and compound heterozygous COL17A1 mutations associated with junctional epidermolysis bullosa. J Invest Dermatol 111: 528–533.
Gardella R, Belletti L, Zoppi N, Marini D, Barlati S, and Colombi M (1996). Identification of two splicing mutations in the collagen type VII gene (COL7A1) of a patient affected by the localisata variant of recessive dystrophic epidermolysis bullosa. Am J Hum Genet 59: 292–300.
Gatalica B, Pulkkinen L, Li K, Kuokkanen K, Ryyananen M, McGrath JA, and Uitto J (1997). Cloning of the human type XVII collagen gene (COL17a1), and detection of novel mutations in generalized atrophic benign epidermolysis bullosa. Am J Hum Genet 60: 352–365.
Gedde-Dahl T Jr (1971). Epidermolysis bullosa: A clinical, genetic and epidemiological study. The Johns Hopkins Press, Baltimore: 180.
Gedde-Dahl T Jr, Dupuy BM, Jonassen R, Winberg JO, Anton-Lamprecht I, and Olaisen B (1994). Junctional epidermolysis bullosa inversa (locus EBR2A) assigned to 1q31 by linkage and association to LAMC1. Hum Mol Genet 3: 1387–1391.
Giudice GJ, Emery DJ, and Diaz LA (1992). Cloning and primary structural analysis of the bullous pemphigoid autoantigen BP180. J Invest Dermatol 99: 243–250.
Hashimoto I, Schnyder UW, and Anton-Lamprecht I (1976). Epidermolysis bullosa hereditaria with junctional blistering in an adult. Dermatologica 152: 72–86.
Hintner H and Wolff K (1982). Generalized atrophic benign epidermolysis bullosa. Arch Dermatol 118: 375–384.
Hirako Y, Usukura J, Nishizawa Y, and Owaribe K (1996). Demonstration of the molecular shape of BP180, a 180-kDa pemphigoid antigen and its potential for trimer formation. J Biol Chem 271: 13739–13745.
Hopkinson SB, Baker SE, and Jones JCR (1995). Molecular genetic studies of a human epidermal autoantigen (the 180 kD bullous pemphigoid antigen/BP180): Identification of functionally important sequences within the BP180 molecule and evidence for an interaction between BP180 and α6 integrin. J Cell Biol 130: 117–125.
Hopkinson SB and Jones JC (2000). The N terminus of the transmembrane protein BP180 interacts with the N-terminal domain of BP230, thereby mediating keratin cytoskeleton anchorage to the cell surface at the site of the hemidesmosome. Mol Biol Cell 11: 277–286.
Jonkman MF, De Jong MCJM, Heeres K, Pas HH, Van der Meer JB, Martinez de Velasco AM, Niessen CM, and Sonnenberg A (1995). 180 kD bullous pemphigoid antigen (BP180) is deficient in generalized atrophic benign epidermolysis bullosa. J Clin Invest 95: 1345–1352.
Jonkman MF, De Jong MCJM, Heeres K, and Sonnenberg A (1992). Expression of integrin α6β4 in junctional epidermolysis bullosa. J Invest Dermatol 99: 489–496.
Jonkman MF, De Jong MCJM, Heeres K, Steijlen PM, Owaribe K, Kuster W, Meurer M, Gedde-Dahl T Jr, Sonnenberg A, and Bruckner-Tuderman L (1996). Generalized atrophic benign epidermolysis bullosa: Either 180-kd bullous pemphoid antigen or laminin 5 deficiency. Arch Dermatol 132: 145–150.
Jonkman MF, Scheffer H, Stulp R, Pas HH, Nijenhuis M, Heeres K, Owaribe K, Pulkkinen L, and Uitto J (1997). Revertant mosaicism in epidermolysis bullosa caused by mitotic gene conversion. Cell 88: 543–551.
Kitajima Y, Owada MK, Fujisawa Y, Seishima M, Yaoita H, Hirako Y, and Owaribe K (1995). A hemidesmosomal transmembrane collagenous molecule, the 180-kDa bullous pemphigoid antigen (BPA II), is phosphorylated with 12-O- tetradecanoylphorbol-13-acetate in a human squamous cell carcinoma cell line (DJM-1). Epithelial Cell Biol 4: 70–75.
Mazzanti C, Gobello T, Posteraro P, Paradisi M, Meneguzzi G, Chinni L, and Zambruno G (1998). 180-kDa bullous pemphigoid antigen defective generalized atrophic benign epidermolysis bullosa: Report of four cases with an unusually mild phenotype. Br J Dermatol 138: 859–866.
McGrath JA, Darling T, Gatalica B, Pohla-Gubo G, Hintner H, Christiano AM, Yancey K, and Uitto J (1996b). A homozygous deletion mutation in the gene encoding the 180-kDa bullous pemphigoid antigen (BPAG2) in a family with generalized atrophic benign epidermolysis bullosa. J Invest Dermatol 106: 771–774.
McGrath JA, Gatalica B, Christiano AM, Li K, Owaribe K, McMillan JR, Eady RAJ, and Uitto J (1995). Mutations in the 180-kD bullous pemphigoid antigen (BPAG2), a hemidesmosomal transmembrane collagen (COL17A1), in generalized atrophic benign epidermolysis bullosa. Nat Genet 11: 83–86.
McGrath JA, Gatalica B, Li K, Dunnill GS, McMillan JR, Christiano AM, Eady RA, and Uitto J (1996a). Compound heterozygosity for a dominant glycine substitution and a recessive internal duplication mutation in the type XVII collagen gene results in junctional epidermolysis bullosa and abnormal dentition. Am J Pathol 148: 1787–1796.
Mitra R and Nickoloff BJ (1994). Cultivation of human epidermal keratinocytes in serum-free growth medium. In: Leigh IM, Watt FM, editors. Keratinocyte methods. Cambridge, UK: Cambridge University Press, 17–19.
Nishizawa Y, Uematsu J, and Owaribe K (1993). HD4, a 180 kDa bullous pemphigoid antigen, is a major transmembrane glycoprotein of the hemidesmosome. J Biochem (Tokyo) 113: 493–501.
Ohshima Y and Gotoh Y (1987). Signals for the selection of a splice site in pre-mRNA: Computer analysis of splice junction sequences and like sequences. J Mol Biol 195: 247–259.
Pas HH, Kloosterhuis GJ, Heeres K, van der Meer JB, and Jonkman MF (1997). Bullous pemphigoid and linear IgA dermatosis sera recognize a similar 120-kDa keratinocyte collagenous glycoprotein with antigenic cross-reactivity to BP180. J Invest Dermatol 108: 423–429.
Pas HH, Kloosterhuis GJ, Nijenhuis M, de Jong MCJM, van der Meer JB, and Jonkman MF (1999). Type XVII collagen (BP180) and LAD-1 are present as separate trimeric complexes. J Invest Dermatol 112: 58–61.
Posteraro P, Sorvillo S, Gagnoux-Palacios L, Angelo C, Paradisi M, Meneguzzi G, Castiglia D, and Zambruno G (1998). Compound heterozygosity for an out-of-frame deletion and a splice site mutation in the LAMB3 gene causes nonlethal junctional epidermolysis bullosa. Biochem Biophys Res Commun 243: 758–764.
Pulkkinen L, Marinkovich MP, Hoang TT, Lin Lin G, Herron S, and Uitto J (1999). Compound heterozygosity for novel splice site mutations in the BPAG2/COL17A1 gene underlies generalized atrophic benign epidermolysis bullosa. J Invest Dermatol 113: 1114–1118.
Reddy D, Muller P, Tran H, Nguyn N, Schaecke H, Bruckner-Tudermann L, Guidice G, and Marinkovich M (1998). The extracellular domain of BP180 binds laminin-5 (Abstract 723). J Invest Dermatol 110: 593.
Schaapveld RQ, Borradori L, Geerts D, van Leusden MR, Kuikman I, Nievers MG, Niessen CM, Steenbergen RD, Snijders PJ, and Sonnenberg A (1998). Hemidesmosome formation is initiated by the β4 integrin subunit, requires complex formation of β4 and HD1/plectin, and involves a direct interaction between β4 and the bullous pemphigoid antigen 180. J Cell Biol 142: 271–284.
Scheffer H, Stulp RP, van der Meulen M, Bruckner-Tuderman L, Gedde-Dahl T Jr, te Meerman GJ, Sonnenberg A, Buys CHCM, and Jonkman MF (1997). Implications of intragenic marker homozygosity and haplotype sharing in a rare autosomal recessive disorder: The example of the collagen type XVII (COL17A1) locus in generalised atrophic benign epidermolysis bullosa. Hum Genet 100: 230–235.
Schumann H, Hammami-Hauasli N, Pulkkinen L, Mauviel A, Kuester W, Luethi U, Owaribe K, Uitto J, and Bruckner-Tuderman L (1997). Three novel homozygous point mutations and a new polymorphism in the COL17A1 gene: Relation to biological and clinical phenotypes of junctional epidermolysis bullosa. Am J Hum Genet 60: 1344–1353.
Shapiro MB and Senapathy P (1987). RNA splice junctions of different classes of eukaryotes: Sequence statistics and functional implications in gene expression. Nucleic Acids Res 15: 7155–7174.
Shimizu H (1998). New insights into the immunoultrastructural organization of cutaneous basement membrane zone molecules. Exp Dermatol 7: 303–313.
Shimizu H, Takizawa Y, Pulkkinen L, Zone JJ, Matsumoto K, Uitto J, and Nishikawa T (1998). The 97 kDa linear IgA bullous dermatosis antigen is not expressed in a patient with generalized atrophic benign epidermolysis bullosa with a novel homozygous G258X mutation in COL17A1. J Invest Dermatol 111: 887–892.
Tasanen K, Floeth M, Schumann H, and Bruckner-Tuderman L (2000). Hemizygosity for a glycine substitution in collagen XVII: Unfolding and degradation of the ectodomain. J Invest Dermatol 115: 207–212.
Terracina M, Posteraro P, Schubert M, Sonego G, Atzori F, Zambruno G, Bruckner-Tuderman L, and Castiglia D (1998). Compound heterozygosity for a recessive glycine substitution and a splice site mutation in the COL7A1 gene causes an unusually mild form of localized recessive dystrophic epidermolysis bullosa. J Invest Dermatol 111: 744–750.
Weil D, Bernard M, Combates N, Wirtz MK, Hollister DW, Steinmann B, and Ramirez F (1988). Identification of a mutation that causes exon skipping during collagen pre-mRNA splicing in an Ehlers-Danlos syndrome variant. J Biol Chem 263: 8561–8564.
Acknowledgements
This work was supported by a grant from the Netherlands Organization for Scientific Research (NWO 902–11–060). The follow-up of the epidermolysis bullosa patients of Gedde-Dahl (1971) was supported by Nasenfondet og de dermed forbundne fonds, Oslo. We acknowledge the previous electron microscopical examinations by Professor I. Anton-Lamprecht, Institute for Ultrastructure Research on the Skin, University of Heidelberg, Germany. The authors thank Mr. Klaas Heeres and Ms. Miranda Nijenhuis for technical assistance. We thank Dr. Gerrit van der Steege and Dr. Hans Scheffer for their critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
van Leusden, M., Pas, H., Gedde-Dahl, T. et al. Truncated Type XVII Collagen Expression in a Patient with Non-Herlitz Junctional Epidermolysis Bullosa Caused by a Homozygous Splice-Site Mutation. Lab Invest 81, 887–894 (2001). https://doi.org/10.1038/labinvest.3780297
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.3780297