Abstract
Mutations of the p53 tumor suppressor gene are reported in various kinds of malignancies including lymphomas. However, p53 gene mutations in nasal NK/T-cell lymphoma have not been reported because most parts of tumors are necrotic and a small amount of living tumor tissues is available for the molecular study. Expression and mutations of the p53 gene were examined in the paraffin-embedded specimens of the nasal lesions from 42 Chinese (Beijing and Chengdu) and Japanese (Okinawa and Osaka) patients with nasal NK/T-cell lymphoma by the immunohistochemistry and single strand conformation polymorphism (SSCP) analysis of polymerase chain reaction (PCR) amplified products followed by direct sequencing. Thirty single-nucleotide substitution mutations were observed in 20 of 42 cases (47.6%). Among the 30 mutations, 18 were missense (mainly G:C to A:T transitions), 9 were silent, and 1 was a nonsense mutation. The remaining 2 mutations involved intron 5 and exon 5 terminal points. Abnormal expression of the p53 protein was also observed in 19 of 42 (45.2%) cases. The incidence was significantly (4-fold) higher in the cases of Osaka than those in other areas, although the incidence of p53 mutations in the cases of Osaka was one-half to one-third of those in the other three areas. The results may suggest some racial, environmental, or lifestyle differences in the cause of nasal tumorigenesis.
Similar content being viewed by others

Introduction
Nasal lymphoma frequently shows a polymorphous pattern of proliferation, consisting of large atypical cells, small lymphocytes, plasma cells, benign looking macrophages occasionally showing with phagocytic figures, neutrophils, and eosinophils much less frequently. Thus the term polymorphic reticulosis (PR) was proposed for this condition (Eichel et al, 1966). PR constitutes lethal midline granuloma (LMG), which is characterized by necrotic and granulomatous lesions mainly affecting the nasal cavity. Malignant lymphoma with monomorphous proliferation and Wegener's granulomatosis also show clinical features of LMG (Kassel et al, 1969). Majority of nasal lymphoma, especially PR, was recently categorized as NK/T cell lymphoma (Jaffe et al, 1996). Through immunophenotypic and genotypic studies, we have shown that PR is an NK cell lymphoma (Ohsawa et al, 1999). Epidemiologic studies of PR revealed the clustering of patients in the east Asian countries (Aozasa et al, 1989) and Epstein-Barr virus (EBV) association (Harabuchi et al, 1990; Tomita et al, 1995), however little is known about mechanisms for development of PR.
p53 is a well-known tumor suppressor gene that causes cells with damaged DNA to arrest at the G1 phase of cell cycle or stimulating expression of the bax gene, the protein that promotes apoptosis (Levine et al, 1991). In a wide variety of human cancers, p53 gene mutations have been detected mainly in exons 5 though 8 (Hollstein et al, 1991). High incidence of malignant lymphoma in the p53 knockout mice has been reported (Donehower et al, 1992), suggesting an important role of p53 gene mutations in lymphomagenesis. The mutated p53 gene encodes mutant p53 protein, which has a much longer half-life time than that of wild type p53 protein, thus accumulating in the cytoplasm in an amount sufficient for immunohistochemical detection. Previous immunohistochemical study showed that p53 is overexpressed in a high percentage of cases with nasal NK/T-cell lymphoma (Quintanilla-Martinez et al, 1998). These findings suggest that the p53 gene mutations might be frequent in nasal NK/T-cell lymphoma. In this study, we analyzed the mutations of the p53 gene in a series of 42 cases of nasal lymphomas of PR type collected from China and Japan by polymerase chain reaction-single strand conformation polymorphism (PCR-SSCP) and direct sequencing.
Results
Histological and Immunohistochemical Findings
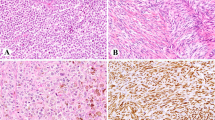
Histologically, varying degrees of necrosis were found in the upper respiratory lesions. Diffuse proliferation of large atypical mono- or multinucleated cells was observed with various numbers of lymphocytes, plasma cells, and macrophages, giving a polymorphous appearance. An angiocentric pattern of proliferation was occasionally observed. Histologic grade based on the criteria described by Lipford et al (1988) was Grade II in 17 cases and Grade III in 25 cases. Percentage of large atypical cells among infiltrating cells was more than 30% and 80% in Grade II and III lesions, respectively. Immunohistochemically, tumor cells were CD20−, TIA-1+, CD56+, CD3+, or CD43+ (Fig. 1).
Expression and Mutation of the p53 Gene
Positive immuno-reactivity for DO-7, suggestive of abnormal expression or stabilization of p53 protein, was found in 19 of 42 (45.2%) cases (Table 1). The incidence of abnormal p53 expression was 4-fold higher in the cases in Osaka than those in Okinawa and 2-fold higher than those in China (Table 2).
By the direct sequencing of SSCP products, 30 single-nucleotide substitution mutations were detected in 20 of 42 cases: 13 cases had a single mutation, 5 had two mutations, 1 had three mutations, and 1 had four mutations (Fig. 2) (Table 1). With the exception of 2 mutations involving the intron 5 and exon 5 terminal point (codon 187), 18 were missense mutations leading to amino acid substitutions, 1 was a nonsense mutation, and 9 were silent mutations resulting in no amino acid changes. No specific mutations were observed, although G:C to A:T transition was predominant (19 lesions), followed by A:T to G:C (3 lesions) transition. For mutation frequency by histologic grade, 10 of 17 cases (58.8%) in Grade II and 10 of 25 cases (40%) in Grade III showed p53 mutations.
Nonradioactive single strand conformation polymorphism (cold SSCP) analysis of p53 exon 5 from representative cases with nasal lymphoma. Aberrant migration patterns compared with normal wild type (WT) bands are seen in cases 12, 15, and 19 and with point mutation at codons 142, 151, and 162, respectively.
There were no significant differences in the incidence of p53 mutation among the cases in the four areas. However, the incidence in the cases in Osaka was one-half to one-third of those in other areas, although the incidence of abnormal p53 expression was very high in the cases in Osaka (Table 2). The results suggest the difference in the cause of nasal lymphomagenesis between the cases in Osaka and the three other areas, Beijing, Chengdu, and Okinawa. Histologic grade of the cases that stained positive for p53, but were negative by SSCP, was Grade II in 3 cases and Grade III in 8.
EBV
The proliferating cells in 8 of 10 cases showed positive signals in the nucleus. Percentage of positive cells per 1000 cells counted under high power fields ranged from 11% to 64% (mean, 35%).
Discussion
Pathological characteristics of 42 nasal lymphomas, ie, varying degrees of necrotic change in the upper respiratory tract, and the polymorphous pattern of proliferation were identical with those in LMG of PR type (Eichel et al, 1966; Kassel et al, 1969), which is now classified as an NK/T- or NK-cell lymphoma (Jaffe et al, 1996). Indeed immunohistochemical findings of the present 42 cases were in agreement with those of NK/T- or NK-cell lymphoma, ie, TIA-1+, CD56+, CD3+, or CD43+.
In the non-nasal and ordinary lymphomas, p53 mutations were rare; none of 43 cases in the United States (Gaidano et al, 1991) and 8 of 48 (17%) in Japan (Ichikawa et al, 1992) had mutations. Meanwhile it was reported that aggressive high-grade B-cell non–Hodgkin's lymphoma (NHL) had approximately 30% incidence of p53 mutations, whereas its indolent counterpart rarely had incidence (Lo Coco et al, 1993). Among T-cell lymphomas, frequency of p53 gene mutations was reported to be less than 10%. Conversely frequency of p53 mutations was high in the specific category of lymphomas; greater than 50% in Burkitt's lymphoma (Villuendas et al, 1993) and approximately 70% in cases with pyothorax-associated lymphoma (Hongyo et al, 1998). Both types of lymphoma are known to be EBV-associated. The present study on NK/T-cell lymphoma revealed a high frequency (47.6%) of p53 gene mutations. NK/T-cell lymphoma is also EBV-associated (Harabuchi et al, 1990; Tomita et al, 1995). There have been no reports on p53 gene mutations in NK/T-cell lymphoma by using the PCR-SSCP method followed by direct sequencing. This must be due to availability of a small amount of samples with necrotic change from PR lesions.
The ability to find a mutation is partially dependent upon the percentage of tumor cells. Based on the criteria proposed by Lipford et al (1988), our cases had Grade II or III histology, composing more than 30% of large atypical cells. Nasal NK/T-cell lymphoma is one of the EBV-associated lymphomas, and the percentage of EBV genome-containing cells was more than 10% (mean, 35%) in our cases. We previously reported that the PCR-SSCP method used in this study could detect the alterations of the p53 gene in 3% cells of population (Hongyo et al, 1993). Therefore p53 mutations could be detected in all of our cases if they occurred.
Transitions at the CpG dinucleotides site were the predominant pattern of substitutions in NHL cases, and G:C to T:A transversion is uncommon (Hollstein et al, 1991). In our series of NK/T-cell lymphoma, more than 70% of substitutions were transitions, however the CpG site was involved in only three lesions. The predominance of transition mutations (G:C to A:T) in our series of NK/T-cell lymphoma suggests that some “endogenous” mutagens act in lymphomagenesis. The transition pattern of the p53 mutation is known to be more susceptible to spontaneous genetic instability than transversion. Meanwhile 75% of the cases in the current series had at least one mutation that changed an amino acid, which might have provided the selection pressure for expansion.
Mutational analysis of the p53 gene has been confined principally to exons 5 through 8, because 90% of the mutations in human tumors occurred in this region (Hollstein et al, 1991). In NHL, the predominant site for mutations did not present in previous reports (Adamson et al, 1995; Ichikawa et al, 1992). In our cases with NK/T-cell lymphoma, exon 5 is the most common site for mutations, ie, 13 of 30 (43%) mutations occurred in exon 5. Some carcinogens might cause mutation in specific codons, as was observed in the mutation of codon 249 in lung and liver cancers induced by irradiation (Anderson et al, 1995; Taylor et al, 1994). Relatively restricted distribution of mutational spots were found in our cases; codon 273, one of the so-called mutational “hot spots” (Hollstein et al, 1991), was involved in 2 cases, and codons 151, 193, and 251 were involved in 2 or 3 cases. Distribution of hot spots in p53 mutations might reflect the function of specific mutant alleles being selected for promoting cell proliferation or the nature and activity of mutagens in different tissue fashion.
Alteration of the p53 gene results in accumulation of the p53 mutant protein due to its longer half-life time than that of wild type. Thus immunohistochemical detection of the p53 protein could be performed in various kinds of malignancies under supposition that p53 gene mutations might occur frequently in these tumors. On the nasal lymphomas, Quintanilla-Martines et al (1998) reported that the p53 protein was overexpressed in 22 of 29 cases (76%) examined. In our cases with nasal NK/T-cell lymphomas, 19 of 42 cases (45%) overexpressed the p53 protein. Our study showed a discordance between p53 protein overexpression and the presence of mutations: 11 of 19 cases (58%) overexpressing the p53 protein had no mutations. This kind of discordance had been reported in many types of tumors including lymphomas (Adamson et al, 1995; Martinez et al, 1997), and reasons for this were variously speculated. Recently a link between accumulation of a wild type p53 protein and EBV has been postulated in the nasopharyngeal carcinoma (Effert et al, 1992). Nasal NK/T-cell lymphoma is also an EBV-associated disease (Harabuchi et al, 1990; Tomita et al, 1995). Accumulation of a wild type p53 protein was also reported in the acquired immunodeficiency syndrome-related lymphomas (Nakamura et al, 1993). EBV was studied only in 10 of the current cases, therefore we could not evaluate the relationship between abnormal p53 expression and EBV.
Functional inactivation of p53 through interaction of the wild type gene product with various viral products such as SV 40 large T antigen, adenovirus E1B, human papilloma virus (HPV) E6 protein, or cellular protein MDM2 was reported (Capoulade et al, 1998). In HPV-positive tumors, binding of the E6 protein obviates the need for p53 mutations in the genesis of such tumors, but this association was not noted in EBV infection and Hodgkin's disease (Niedobitek et al, 1993). However, EBV immediate-early protein, BZLF1, which is highly expressed in immunodeficiency syndrome-related lymphoma, can interact with p53 and inhibit its function (Zhang et al, 1994). EBNA-5, another EBV encoded protein necessary for transformation of infected B cells, can also form complexes with both wild type and mutant p53 protein (Szekely et al, 1993). Taken together, it is possible that EBV gene products could indirectly suppress p53 function, thus resulting in overproduction and accumulation of wild type p53 as a compensating function. The lowest incidence of p53 gene mutations and the highest incidence of p53 protein expression in the Osaka cases in our series suggest the different causes for nasal lymphomagenesis including racial, environmental, or lifestyle causes.
In conclusion, mutations of the p53 gene are frequent in nasal NK/T-cell lymphoma with rather restricted sites for mutations. These findings give an insight on the lymphomagenesis of nasal NK/T-cell lymphoma and also give some suggestions for its treatment.
Materials and Methods
Forty-two cases with PR, 19 from China and 23 from Japan, were selected for the current study: they were admitted to hospitals during the period of 1986 to 1997 (Table 1). All patients presented with the necrotic and granulomatous lesions in the upper respiratory tract, which were biopsied for histologic diagnosis before treatment. Age of patients ranged from 18 to 79 years (median, 49.5) with a male to female ratio (M/F) of 6:1. There were differences in the distribution of age and sex between Chinese and Japanese patients: in China, a median age of 42 years and all men, and in Japan, a median age of 53 years and M/F ratio of 2.8:1. The nasal cavity and paranasal sinuses were the most common sites (37 cases) for involvement followed by naso-/oropharynx (3 cases), tonsil (1 case), and trachea (1 case). Histologic specimens were fixed in 10% formalin and routinely processed for paraffin-embedding. All of the paraffin blocks were gathered from Osaka University, and 3 μm sections were cut and stored at 4° C before staining with hematoxylin-eosin and immunohistochemical procedures at the same time. Histologic slides were reviewed by two of the authors (LT, KA) for diagnosis. To show the number of the proliferating cells in the lesions, histologic grade was determined based on the criteria described by Lipford et al (1988).
Immunohistochemistry
Immunohistochemical study of the paraffin sections was carried out using the ABC method. The primary antibodies used in the study, their suppliers, and dilutions were as follows: CD3 (1:100; Dakopatts, Glostrup, Denmark), MT-1(CD43) (1:50; Bioscience, Emmenbrucke, Switzerland), Mx-PanB (CD20) (1:200; Kyowa Medex, Tokyo, Japan), 123C3 (CD56) (1:40; Zymed, South San Francisco, California), ZH7 (CD16) (1:200; Novocastra, Newcastle, United Kingdom), and TIA-1(1:500; Coulter, Hialeah, Florida). The alkaline phosphatase-anti-alkaline-phosphatase (APAAP) method was used in p53 protein detection with use of monoclonal anti-human p53 protein (DO-7) (Dakopatts) diluted at 1:10 as the primary antibody. When CD3, CD56, and DO-7 were used as primary antibody, sections were treated in a microwave oven for 15 minutes in 10 mm citrate buffer (10 mm citrate monohydrate in distilled water, pH 6.0) for antigen retrieval. Cases with more than 10% of tumor cells positive for DO-7 were regarded as positive.
DNA Extraction and PCR for p53 Gene
DNA for PCR amplification was extracted using chelation resin. Three 10-μm thick paraffin sections were cut, transferred into sterile distilled water containing 20% chelating resin iminodiacetic acid (Sigma, St. Louis, Missouri), and boiled for 30 minutes. After centrifugation, the supernatant was transferred to a sterile 500 μl tube and stored at −20° C. The PCR primer pairs for the amplification of the p53 gene exons 5 through 8 were: (a) 5′ -GTACTCCCCTGCCCTCAACA-3′ and 5′ -CTCACC-ATCGCTATCTGAGCA-3′ for exon 5; (b) 5′ -TTGCT-CTTAGGTCTGGCCCC-3′ and 5′ -CAGACCTCAGGC-GGCTCATA-3′ for exon 6; (c) 5′ -TAGGTTGGCTCTG-ACTGTACC-3′ and 5′ -TGACCTGGAGTCTTCCAGT-GT-3′ for exon 7; and (d) 5′ -AGTGGTAATCTACTG-GGACGG-3′ and 5′ -ACCTCGCTTAGTGCTCCCTG-3′ for exon 8.
Hot start PCR was performed as follows: 45 cycles of denaturation at 95° C for 30 seconds; annealing at 58° C, 62° C, 60° C, and 60° C for 30 seconds for exon 5, 6, 7, and 8, respectively; extension at 72° C for 1 minute, and final extension at 72° C for 7 minutes. Paraffin blocks containing no sample were cut and used as negative controls throughout the procedures. The amplified products were subjected to electrophoresis in 1.5% agarose gel containing 2 μg/ml ethidium bromide in TBE buffer. After electrophoresis, the gels were examined under an ultraviolet light transilluminator.
SSCP
Nonradioactive SSCP was performed as previously reported (Hongyo et al, 1995). Twenty microliters of reaction mixtures containing 5 μl of PCR product (20–200 ng of DNA), 0.2 μl of 1M methylmercury hydroxide, 3 μl of loading buffer (15% Ficoll, 0.25% bromphenol blue, 0.25% xylene cyanol), and TBE buffer were heated to 90° C for 4 minutes and put on ice and then electrophoresed in 18% polyacrylamide TBE gel at 300 volts, while maintaining the temperature at 35° C for exon 5, 5° C for exon 6, and 25° C for exons 7 and 8. The gels were stained with 0.5 μg/ml ethidium bromide in TBE buffer for 20 minutes at room temperature. The bands migrated apart from that of wild type were determined as SSCP positive. The bands possibly mutated by SSCP were extracted from the gels and amplified by 25 cycles of PCR to enrich the mutated alleles.
Direct Sequencing
Sequencing was carried out on PCR products of SSCP positive cases. To purify single or double stranded PCR products with a range of 100 bp, the PCR products were processed using the QIAquick PCR purification kit (QIAGEN Inc., Valencia, California) according to the manufacturer's protocol. Sequencing was performed by the dideoxy chain termination method using the Big Dye Terminator cycle-sequencing kit (Perkin-Elmer Corporation, Foster City, California). The same primers were used as for PCR. Cycle sequencing was performed following the protocol, ie, 30 cycles of denaturation (95° C, 20 seconds), annealing (54° C, 30 seconds), and extension (72° C, 3 minutes). After ethanol precipitation, the samples were analyzed by the Genetic Analyzer (ABI Prism 310, Perkin-Elmer Corporation). The PCR-SSCP analysis and sequencing of the possible positive cases were repeated three times to rule out the contamination and artifacts.
In Situ Hybridization for EBV
EBV RNA in situ hybridization (ISH) was performed in 10 cases as previously described (Weiss et al, 1991). As a positive control, the Raji cell line was used. Hodgkin's disease of mixed cellularity type with the EBV genome was also included as positive control. As negative controls, the hybridizing mixture was used with (a) sense probe and (b) antisense probe after RNase (Sigma) treatment.
References
Adamson DJA, Thompson WD, Dawson AA, Bennett B, and Haites NE (1995). p53 mutation and expression in lymphoma. Br J Cancer 72: 150–154.
Anderson M, Jonsson M, Nielsen LL, Vyberg M, Visfeldt J, Storm HH, and Wallin H (1995). Mutations in the tumor suppressor gene p53 in human liver cancer induced by alpha-particles. Cancer Epidemiol Biomarkers Prev 4: 765–770.
Aozasa K, Ohsawa M, Tajima K, Sasaki R, Maede H, Matsunaga T, and Friedmann I (1989). Nation-wide study of lethal midline granuloma in Japan: frequencies of Wegener's granulomatosis, polymorphic reticulosis, malignant lymphoma and other related conditions. Int J Cancer 44: 63–66.
Capoulade C, Bressac PB, Lefrere I, Ronsin M, Feunteun J Tursz T, and Wiels J (1998). Overexpression of MDM2, due to enhanced translation, results in inactivation of wild-type p53 in Burkitt's lymphoma cells. Oncogene 16: 1603–1610.
Donehower LA, Harrey M, Slagle BL, McArthur MJ, Montgomery CA Jr, Butel J, and Bradley A (1992). Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumors. Nature 356: 215–221.
Effert P, McCoy R, Abdel-Hamid M, Flynn K, Zhang Q, Busson P, Tursz T, Liu E, and Raab-Traub N (1992). Alterations of the p53 gene in nasopharyngeal carcinoma. J Virol 66: 3768–3775.
Eichel BS, Harrison EGJ, Devine KD, Scanlon PW, and Brown HA (1966). Primary lymphomas of the nose including relationship to lethal midline granuloma. Am J Surg 112: 597–605.
Gaidano G, Ballerini P, Gong JZ, Inghirami G, Neri A, Newcomb EW, Magrath IT, Knowles DM, and Dalla-Favera R (1991). p53 mutations in human lymphoid malignancies: association with Burkitt lymphoma and chronic lymphocytic leukemia. Proc Natl Acad Sci USA 88: 5413–5417.
Harabuchi Y, Yamanaka N, Kataura A, Imai S, Kinoshita T, Mizuno A, and Osato T (1990). Epstein-Barr virus in nasal T-cell lymphomas in patients with lethal midline granuloma. Lancet 335: 128–130.
Hollstein M, Sidransky D, Vogelstein B, and Harris SR (1991). p53 mutations in human cancers. Science 253: 49–53.
Hongyo T, Buzard GS, Calvert RJ, and Weghorst CM (1993). ‘Cold SSCP’: a simple, rapid and non-radioactive method for optimized single strand conformation polymorphism analyses. Nucl Acid Res 21: 3637–3642.
Hongyo T, Buzard GS, Palli D, Weghorst CM, Amorosi A, Galli M, Caporaso NE, Fraumeni JF, and Rice JM (1995). Mutations of the k-ras and p53 gene in gastric adenocarcinoma from a high incidence region around Florence, Italy. Cancer Res 55: 2665–2672.
Hongyo T, Kurooka M, Taniguchi E, Iuchi K, Nakajima Y, Aozasa K, and Nomura T (1998). Frequent p53 mutations at dipyrimidine sites in patients with pyothorax-associated lymphoma. Cancer Res 58: 1105–1107.
Ichikawa A, Hotta T, Takagi N, Tsushita K, Kinoshita T, Nagai H, Murakami Y, Hayashi K, and Saito H (1992). Mutations of p53 gene and their relation to disease progression in B-cell lymphoma. Blood 79: 2701–2707.
Jaffe ES, Chan JKC, Su I-J, Frizzera G, Mori S, Feller AC, and Ho FCS (1996). Report of the workshop on nasal and related extranodal angiocentric T/natural killer cell lymphomas. Am J Surg Pathol 20: 103–111.
Kassel S, Echevaria RA, and Guzzo FP (1969). Midline malignant reticulosis (so-called lethal midline granuloma). Cancer 23: 920–935.
Levine AJ, Momand J, and Finley CA (1991). The p53 tumor suppressor gene. Nature 351: 453–456.
Lipford EH Jr, Margolick JB, Longo DL, Fauci AS, Jaffe ES (1988). Angiocentric lymphoproliferative lesions: A clinicopathologic spectrum of post-thymic T-cell proliferations. Blood 72: 1674–1681.
Lo Coco F, Gaidano G, Louie DC, Offit K, Chaganti RSK, and Dalla-Favera R (1993). p53 mutations are associated with histologic transformation of follicular lymphoma. Blood 82: 2289–2295.
Martinez DB, Robledo M, Arranz E, Infantes F, Echezarreta G, Marcos B, Sanz C, Rivas C, and Benitez J (1997). Correlation between mutations in p53 gene and protein expression in human lymphomas. Am J Hematol 55: 1–8.
Nakamura H, Said JW, Miller CW, and Koeffer HP (1993). Mutation and expression of p53 in acquired immunodeficiency syndrome-related lymphoma. Blood 82: 920–926.
Niedobitek G, Rowlands DC, Yong LS, Herbst H, Williams A, Hall P, Padfield J, Rooney N, and Jones EL (1993). Overexpression of p53 in Hodgkin's disease: lack of correlation with Epstein-Barr virus infection. J Pathol 169: 207–212.
Ohsawa M, Nakatsuka S, Kanno H, Miwa H, Kojya S, Harabachi Y, Yang WI, and Aozasa K (1999). Immunophenotypic and genotypic characterization of nasal lymphoma with polymorphic reticulosis morphology. Int J Cancer 81: 865–870.
Quintanilla-Martinez L, Guerrero I, Franklin JL, Naresh KN, Krenacs L, Rama-Rao C, Bhatia K, Magrath IT, and Raffeld M (1998). Nasal NK/T-cell lymphoma from Peru. High prevalence of p53 overexpression. Mod Pathol 11: 139A.
Szekely L, Selivanova G, Magnusson KP, Klein G, and Wiman KG (1993). EBNA-5, an Epstein-Barr virus-encoded nuclear antigen, binds to the retinoblastoma and p53 proteins. Proc Natl Acad Sci USA 90: 5455–5459.
Taylor JA, Watson MA, Devereux TR, Michels RY, Saccomanno G, and Anderson M (1994). p53 mutation hotspot in radon-associated lung cancer. Lancet 8: 86–87.
Tomita Y, Ohsawa M, Mishiro Y, Kubo T, Maeshiro N, Kojya S, Noda Y, and Aozasa K (1995). The presence and sub-type of Epstein-Barr virus in B- and T-cell lymphomas of the sino-nasal region from the Osaka and Okinawa districts of Japan. Lab Invest 73: 190–196.
Villuendas R, Piris MA, Algara P, Sanchez-Beato M, Sanchez-Verde L, Martinez JC, Orradre JL, Garcia P, Lopez C, and Martinez P (1993). The expression of p53 protein in non-Hodgkin's lymphoma is not always dependent on p53 gene mutations. Blood 82: 3151–3156.
Weiss LM, Chen YY, Liu XF, and Shibata D (1991). Epstein-Barr virus Hodgkin's disease: a correlative in situ hybridization and polymerase-chain-reaction study. Am J Pathol 139: 1259–1265.
Zhang Q, Gutsch D, and Kenney S (1994). Functional and physical interaction between p53 and BZLF1: implications for Epstein-Barr virus latency. Mol Cell Biol 14: 1929–1938.
Acknowledgements
The authors thank Dr. Wei-ping Liu (Department of Pathology, West-China University of Medical Science, Chengdu, China) for providing histologic materials.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported by a grant from the Vehicle Racing Commemorative Foundation, grants from the Ministry of Education, Science and Culture, Japan (09670184, 09770148, 10042005, 11470353, 11670212, 11680546), and Research for the Future.
Rights and permissions
About this article
Cite this article
Li, T., Hongyo, T., Syaifudin, M. et al. Mutations of the p53 Gene in Nasal NK/T-Cell Lymphoma. Lab Invest 80, 493–499 (2000). https://doi.org/10.1038/labinvest.3780055
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.3780055
This article is cited by
-
Nasal natural killer (NK)/T-cell lymphoma: clinical, histological, virological, and genetic features
International Journal of Clinical Oncology (2009)
-
Nasal NK/T-cell lymphoma: epidemiology and pathogenesis
International Journal of Hematology (2008)
-
Analysis of p53 and Bak gene mutations in lymphoproliferative disorders developing in rheumatoid arthritis
Journal of Cancer Research and Clinical Oncology (2006)
-
Leukemia and lymphoma of natural killer lineage cells
International Journal of Hematology (2003)
-
Diagnosis and treatment of epstein-barr virus-associated natural killer cell lymphoproliferative disease
International Journal of Hematology (2003)