Abstract
Microscopically detectable deletions and X;autosome translocations have previously facilitated the construction of a high-resolution interval map of the Xq21 region. Here, we have generated three yeast artificial chromosome contigs spanning approximately 7 megabases of the Xq13.3-q21.31 region. In addition, a novel deletion associated with choroideremia and mental retardation was identified and mapped in detail. The proximal deletion endpoint was positioned between the loci DXS995 and DXS232, which enabled us to confirm the critical region for a locus involved in mental retardation. The distal deletion endpoint is situated in the Xq21.33 band, which allowed us to refine the order of several markers in this region.
Similar content being viewed by others
Introduction
In the Giemsa-dark-staining Xq21 band, numerous large male-viable deletions have been described which encompass up to 15 megabases (Mb) of DNA, or 8–10% of the estimated size of the human X chromosome [1, 2]. These deletions often give rise to contiguous gene syndromes including choroideremia (CHM), X-linked deafness (DFN3) and nonspecific mental retardation (MR) [3–7]. Sizeable deletions have also been found in patients with nonsyndromic CHM or DFN3 [4–6, 8, 9].
Employing positional cloning strategies, the genes underlying CHM and DFN3 have been isolated. Analysis of patients with Xq21 deletions and syndromic and nonsyndromic forms of MR has enabled us to map the gene for X-linked MR to a region between CHM and DFN3 [4, 7, 10–13].
As a prerequisite for the isolation of this and other genes in Xq21, we have set out to generate a ladder of overlapping yeast artificial chromosomes (YACs) spanning the entire Xq21 region. To this end, we first employed numerous X;autosome translocations and deletions to subdivide the Xq21 band into 24 different intervals [8, 14; Philippe, unpubl. results]. Moreover, we have previously described a 850-kb YAC and cosmid contig encompassing the DFN3 locus and a 350-kb YAC encompassing the CHM gene [9,12,13]. Here, we report on three YAC contigs spanning approximately 7 Mb of the Xq13.3-q21.31 region and on the localization of a novel deletion associated with CHM and MR.
Materials and Methods
Patients
All patients except patient AP have been described elsewhere and are listed in table 1.
YAC Clones
YAC clones were initially identified by PCR amplification of YAC DNA pools or by hybridization of probes to filters containing gridded YAC DNA. YACs were isolated from previously described CEPH [15] and ICRF YAC libraries [16]. All relevant markers used in this study are listed in table 2.
Positive YAC clones were plated on NZYM agar [17], and three yeast clones each were picked and cultured as described elsewhere [18]. Yeast cells were embedded in low-melting agarose, incubated for 2 h in the presence of zymolyase (Seikagu) and incubated overnight with pronase (Boehringer Mannheim). Yeast chromosomes were separated using pulsed-field gel electrophoresis (PFGE) and blotted to a GeneScreen Plus membrane (NEN Dupont). YAC blots were hybridized with 32P-labelled total human DNA to estimate the size of the human insert. Subsequently, the identity of the YACs was checked with the marker originally used for their isolation. YACs were further characterized by screening of adjacent markers and through the generation of left- and right-end fragments by ligation-mediated PCR (LM-PCR; see below).
In addition, we screened the CEPH YAC data base with new DNA markers from this region and for YAC clones which overlap with previously mapped YACs.
Southern Blot Analysis
Hybridization of 32P-labelled DNA fragments to nylon filters containing DNA from patients or from YAC clones was essentially done as described elsewhere [9].
LM-PCR and Sequence Analysis
The LM-PCR method was employed essentially as described by Kere et al. [19]. Yeast DNA was digested with AluI, EcoRV, RsaI or PvuII prior to ligation of adaptors. LM-PCR end fragment products of the YAC clones were separated on an agarose gel and purified with the Qiaquick gel extraction kit (Quiagen). Purified DNA was sequenced using fluorescent dideoxynucleotides on an Applied Biosystems 373A DNA sequencer. Sequencing reactions were done with a Taq DyeDeoxy Terminator Cycle Sequencing Kit (Applied Biosystems) according to the manufacturer’s instructions. The oligonucleotides of end fragment 753e10EV/L (ATTTATTCTTCCAGGCCCATA and CGGAATTCTGAAAAAGAGATC) were designed (Isogen Bioscience, The Netherlands) to generate a product of 94 bp. For end fragment 5045H5, the oligonucleotides TGGATAAAGTAAAAAGCACACAAG and TGGTAGTTGTCTCATAGCTCTTG amplify a product of 260 bp. Finally, a 152-bp PCR product (A0822EV/L) can be amplified from the left end of YAC A0822 with the oligonucleotides CCTAATTTGGGAAGAACATATC and AATTTAAGAGAGAAATTATTCATATT.
Results
Construction of a YAC Contig
Several markers from the Xq21 region were employed to screen the ICRF and CEPH YAC libraries [15, 16]. All YAC clones were extensively screened for the presence of X-linked markers and for the absence of autosomal sequences. In this way, a YAC map was generated consisting of three nonoverlapping contigs encompassing the Xq13.3-q21.31 region. From these contigs, YAC 4893 and YAC 5045 have been published elsewhere [9]. To establish novel sequence-tagged sites (STSs) and to check the integrity of the YACs, we employed the LM-PCR technique [19]. LM-PCR products were hybridized to adjacent YACs and to DNAs of a large number of patients with deletions in Xq21 to localize and orient the YACs (fig. 1). End fragments mapping to the Xq21 region were sequenced, and oligonucleotides were designed for PCR amplification to generate new STSs.
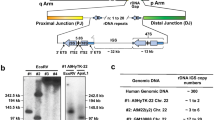
Physical map of the Xq13.3-q21.3 region. On the upper horizontal bar, the previously and newly mapped DNA loci are indicated. The YACs are represented by solid bars for X chromosomal sequences and wavy bars for autosomal sequences. Vertical bars indicate the presence of the markers on the YACs and in the DNA of patients with deletions. Open circles are right-end fragments while solid circles indicate left-end fragments of the YACs. Solid and open diamonds represent the outermost X-chromosomal cosmid clones derived from their respective YACs used for direct-visual-hybridization analysis. The identification numbers of the YACs are depicted adjacent to the YACs, while their estimated sizes are depicted underneath in parentheses. The gaps between the three YAC contigs are indicated with double arrowheads. The deletions found in several patients used in this study are depicted at the bottom of the figure. The solid bar represents the chromosomal segments present in the patients. The molecular characterization of all patients has been reported elsewhere except for patient AP (indicated with an arrow), who suffers from MR and CHM. The mapping results of the YAC end fragments are only indicated for overlapping YACs. For purposes of clarity, the region between POU3F4 and DXS233 is not true to scale but expanded twofold. At the bottom, the intervals from this region as defined by Philippe et al. [14; unpubl. data] are depicted.
The middle and most distal YAC contigs span approximately 3 and 2.5 Mb, respectively. Although the size of the most proximal contig has not been determined precisely, we estimate that together, the resulting YAC contigs encompass a chromosomal segment of approximately 7 Mb, extending from PGK1 proximally to DXS1002 distally with a gap between DXS986 and DXS738 and between end fragment 5045H5 and the DXS232 locus. The most proximal contig is linked to a previously published YAC contig spanning the PGK1 locus [20, 21]. The most distal contig overlaps a previously reported YAC contig in the Xq21.3 region [22].
Identification and Fine Mapping of a Deletion Associated with CHM and MR
Patient AP is a 40-year-old mentally retarded man with CHM. Ophthalmologic findings suggest that two of his sisters (II-1 and II-4) and his mother (I-2) are carriers of CHM. Apart from a microdeletion in the Xq21 band (see below) patient AP carries a balanced translocation 46,XY,t(10;15) (q26.3;q24.3), which is also present in his mentally normal sister (II-4) and father, and apparently is not associated with a clinical phenotype (fig. 2). II-1 and II-2 have not been investigated for the presence of the translocation.
Pedigree of kindred with CHM and MR. Patient AP (II-3) was ascertained via his sister (II-4) who was prenatally counselled. Two of his sisters (II-1 and II-4) and his mother (I-2) are carriers of the deletion. AP, his father (I-1) and one of his sisters (II-4) also carry a balanced translocation t(10;15)(q26.3;q24.3). II-1 and II-2 have not been tested for chromosomal abnormalities.
Patient AP was investigated for the presence of a deletion by PCR amplification of several exons from the CHM gene. All exons tested were absent, which prompted us to investigate the extent of the deletion in greater detail. On the proximal side, the markers DXS26 and DXS995 were found to be present in patient AP, while DXS232 and DXS121 were absent. Also, the POU3F4 gene, previously shown to be involved in DFN3, appeared to be present in this patient [13]. These findings located the proximal breakpoint of patient AP between the previously defined intervals 7 and 8 of the Xq21 band (fig. 1) [14; Philippe, unpubl. data]. Subsequently, several cosmids from a previously published 850-kb cosmid contig [9] were hybridized to precisely map the proximal end-point of this deletion. The breakpoint is located in cosmid 5045H3, which detects a novel restriction fragment carrying the deletion junction (fig. 3). A cosmid located proximal to this breakpoint (cosmid 5045A6) is entirely-present in patient AP, while the first cosmid (5045E7) distal to cosmid 5045H3 is completely absent. These results confirm the initial localization of the breakpoint in cosmid 5045H3 (fig. 3). It is noteworthy that the distal breakpoint of another deletion, detected previously in a patient with DFN3 (TD) [9], is also located in cosmid 5045H3, approximately 10 kb from the proximal deletion breakpoint in patient AP (data not shown).
Mapping of the proximal deletion breakpoint in patient AP. Hybridization of the cosmids 5045E7 (a), 5045H3 (b) and 5045A6 (c) to a Southern blot of EcoRI-digested DNA of patients with deletions and a female control. In patient AP, cosmid 5045H3 detects a novel restriction fragment indicative of the proximal breakpoint of the deletion.
Several markers from the Xq21.2-q21.31 region, i.e. DXS1002, DXS95, DXS262, DXS110 and DXS472, appeared to be absent in patient AP, thus the distal breakpoint of the deletion in patient AP could be assigned to the Xq21.32-q21.33 segment. To localize this breakpoint more precisely, several markers from a previously defined interval (interval 20) [14; Philippe, unpubl. data] were employed. PCR analysis revealed that DXS3 is absent, while DXS1170 and DXS990 are present in the DNA of AP. Conventional Southern blot analysis showed that the DXS112 locus is also located proximal to this deletion breakpoint (data not shown). Therefore, the distal breakpoint of the AP deletion subdivides this interval [Philippe, unpubl. data] in two distinct intervals: 20A and 20B. The loci DXS3 and DXS112 are located in interval 20A, whereas interval 20B harbors DXS1170and DXS990.
Mapping of New Markers with Respect to Known Markers
These data and the previously established deletion interval map [14; Philippe et al., unpubl. data] have enabled us to define the physical order for several markers. Four markers have been mapped to interval 2 based on their presence in patient XL62 and in C56N, a human-rodent cell hybrid containing the Xq21.1-qter segment [Philippe et al., unpubl. data]. DXS1225 is present on YAC 910h1 and absent from YAC 752e10. Therefore, this marker is located proximal to DXSH97 and DXS986 which are both present on YAC 753e10. Since DXS1197 is present on both above-mentioned YACs, the order must be Xcen-DXS1225-DXS1197-DXS986-Xqter. DXS346 also maps to interval 2 and is absent in both YACs. Since the left-end fragment of YAC 753e10 (753e10EV/L) is also located in this interval (data not shown), DXS346 can be placed distal to this end fragment.
DXS738 is located in interval 3, based on its presence in patient 1/10 and absence from patient XL62 (fig. 1) [Philippe et al., unpubl. data]. DXS738 is absent from both YAC 753e10 and 321f8, which positions it proximal to DXS72. Since the end fragment A0822EV/L of YAC A0822 is also present in YAC 321f8 while DXS72 is absent from YAC A0822, the end fragment is positioned proximal from DXS72 in interval 3. By combining these results with previous mapping data [14; Philippe et al., unpubl. data] we propose the following order: Xcen-PGK1-DXS566-DXS1225-DXS1197-DXS896-753e10EV/L-DXS346 — DXS738 — A0822EV/L — DXS72 — DXS169-Xqter.
A third relevant end fragment, 5045H5, maps distal to POU3F4 and proximal to DXS232. This end fragment is located ouside the deletions found in patients TD and LGL2905, which assigns this marker to interval 8. 753e10EV/L, A0822EV/Land 5045H5 were sequenced and oligonucleotides were designed for PCR amplification. In this way, three new STSs could be generated (see Materials and Methods).
More distally, DXS1167 bould be placed between DXS326 and DXS165 because of its localization on YAC 949e11 and its absence from YAC 4887. This marker has been mapped previously to interval 10 [14; Philippe et al., unpubl. data] (fig. 1). DXS1209 and ZNF6, a gene containing a zinc finger motif, which has been mapped to Xq21 [23] could be localized in the same way. DXS95, DXS349, DXS364 and DXS1168 could be placed distal to DXS1002 on the basis of their absence from YAC 949e11. A comprehensive overview of the Xq21 region, including the YAC contigs, all above-mentioned markers and the relevant deletions is given in figure 1.
Discussion
In this study we have established three YAC contigs encompassing the Xq13.3-q21.31 region except for a gap between DXS986 and DXS738, and a gap between POU3F4 and DXS232. To estimate the size of the latter gap, cosmids from the proximal segment of YAC 4886 and the most distal cosmid of YAC 5045 were employed as probes for direct-visual-hybridization analyses [24] (fig. 1). These experiments revealed that the size of the gap does not exceed 100 kb [Merkx and van der Maarel, unpubl. data]. We are currently screening for cosmids from the ICRF Cosmid Reference Library [25] and Chromosome X Cosmid Library LL0XNC01 ‘U’ to fill this gap. Despite extensive screening of the ICRF and CEPH YAC libraries [15, 16], we have not yet been able to fill the remaining two gaps. The generation of the end clone 753e10EV/L will hopefully lead to the identification of new YACs between DXS986 and DXS738.
We confirmed the localization of 19 DNA markers and 4 genes from the Xq13.3-Xq21.31 region and were able to map them in the correct order. In addition, three STS clones were generated and positioned in the contigs.
We assume that the DXS566 locus maps to an internal deletion in YAC 910h1, because in this YAC, the respective primer pair has failed to yield a PCR product. We cannot rule out the possibility that DXS346 also maps to this deletion interval. If so, DXS346 should be located between DXS566 and DXS1225.
Many of the microscopically detectable deletions in the Xq21 region are associated with DFN3, MR and CHM [2, 6, 7, 26, 27]. The identification of submicroscopic deletions associated with either CHM [28] or DFN3 [6,9] has enabled the subsequent cloning of the relevant genes [10–13]. Characterization of these deletions has revealed that the physical order of these disease genes is: Xcen-DFN3-MR-CHM-qter. Some patients with deletions in Xq21 show clinical features which are not observed in others and which do not seem to fit the contiguous gene syndrome model. The most striking additional features are obesity [29], hypogonadism [6] and cleft lip and palate [30]. The presence of a gene in Xq21 involved in palate closure is also suggested by linkage studies in three families with X-linked cleft palate and/or ankyloglossia (CPX). The critical region for CPX was demarcated proximally by DXS1002 and distally by DXYS1 [Stanier, pers. commun.; 31–35]. In view of the large number of deletions known in Xq21, it is striking that thus far no small deletions have been found with nonspecific mental retardation or with a combination of MR with either DFN3 or CHM.
Here, we are the first to report on a deletion associated with MR and CHM. The deletion encompasses the CHM gene and extends into the chromosomal region between the DFN3 and CHM genes confirming the initial localization of a MR locus in Xq21.1. Since the deletion in AP encompasses a large segment of Xq21, one could speculate that the deletion disrupts a MR gene in Xq21.33, distal to the CHM gene. There are two arguments against this hypothesis. First, the MR gene would be located distal to several deletions associated with a complex phenotype including MR, e.g. XL45, SD, and DM. Second, two patients with nonsyndromic CHM have been described [8], in whom deletions extend into or beyond interval 20 (patient LUN3 and LUN1). Together, these data strongly suggest that a locus for XLMR is situated in the chromosomal region defined by intervals 8, 9 and 10. At the proximal side, this region is demarcated by the deletions found in DFN3 patients II/7, 1/10, and TD [9]. The proximal deletion breakpoint in AP does not significantly narrow down the localization of the MR locus, since it is located only 10 kb from the distal deletion breakpoint in TD. At the distal side, deletions associated with CHM in patients C759, 3.5, 25.6, MS, and 7.6 [8] commence between ZNF6 and DXS165 and extend telomerically. The deletion in patient LGL2905 who has CHM but no MR, restricts the critical region for MR to interval 8.
It is remarkable that apart from DFN3, patient TD shows hypogonadism and mild MR. Five affected family members are not mentally retarded but show marked antisocial and immature behavior and possibly mild learning deficits [36]. Since the deletion in patient TD is close to the MR locus [7], it could affect the proper transcription of the MR gene. This might explain the complex clinical findings in TD and his family.
So far, we found no evidence for microdeletions in the critical region for MR (interval 8) in 40 patients with XLMR [van der Maarel, unpubl. results], whereas in patients with CHM or DFN3, deletions were detected in approximately 25% of cases [8, 9; de Kok and Cremers, unpubl. results]. It is very likely that the main reason for this discrepancy is the genetic heterogeneity of XLMR [37]. Indeed, the Xq21.1 locus may only be involved in a small proportion of cases with XLMR. Thus far, several syndromic forms of MR, like Allan-Herndon-Dudley syndrome, Juberg-Marsidi, and α-thalassemia/MR (ATR-X) have been mapped to Xq13-q21 [38–43], but recent linkage studies of our group in families with nonspecific XLMR point to a clustering in the Xp11 region [44]. Recently, mutations in a putative global transcriptional regulator (XH2) have been shown to be causative for ATR-X [45].
Definite proof for the existence of a gene for MR in Xq21.1 can only be obtained by cloning of candidate genes from the relevant region and identification of mutations in syndromic or nonsyndromic cases of XLMR. These studies will be greatly facilitated by the Xq13.3-q21.31 YAC contigs reported here.
References
Cremers FPM, van de Pol TJR, Wieringa B, Hofker MH, Pearson PL, Pfeiffer RA, Mikkelsen M, Tabor A, Ropers HH: Molecular analysis of male-viable deletions and duplications allows ordering of 52 DNA probes on proximal Xq. Am J Hum Genet 1988;43:452–461.
Merry DE, Lesko JG, Sosnoski DM, Lewis RA, Lubinsky M, Trask B, van den Engh G, Collins FS, Nussbaum RL: Choroideremia and deafness with stapes fixation: A contiguous gene deletion syndrome in Xq21. Am J Hum Genet 1989;45: 530–540.
Nussbaum RL, Lesko JG, Lewis RA, Ledbetter SA, Ledbetter DH: Isolation of anonymous DNA sequences from within a submicroscopic X chromosomal deletion in a patient with choroideremia, deafness, and mental retardation. Proc Natl Acad Sci USA 1987;84:6521–6525.
Cremers FPM, Sankila EM, Brunsmann F, Jay M, Jay B, Wright A, Pinckers AJLG, Schwartz M, van de Pol TJR, Wieringa B, de la Chapelle A, Pawlowitzki IH, Ropers HH: Deletions in patients with classical choroideremia vary in size from 45 kb to several megabases. Am J Hum Genet 1990;47:622–628.
Sankila EM, Bruns GAP, Schwartz M, Nikoskelainen E, Niebuhr E, Hodgson SV, Wright AF, de la Chapelle A: DXS26 (HU16) is located in Xq21.1. Hum Genet 1990;85:117–120.
Bach I, Brunner HG, Beighton P, Ruvalcaba RHA, Reardon W, Pembrey ME, van der Velde-Visser SD, Bruns GAP, Cremers CWRJ, Cremers FPM, Ropers HH: Microdeletions in patients with gusher-associated, X-linked mixed deafness (DFN3). Am J Hum Genet 1992;50: 38–44.
Bach I, Robinson D, Thomas N, Ropers HH, Cremers FPM: Physical fine mapping of genes underlying X-linked deafness and non fra(X)-X-hnked mental retardation at Xq21. Hum Genet 1992;89:620–624.
van Bokhoven H, Schwartz M, Andréasson S, van den Hurk JAJM, Bogerd L, Jay M, Rüther K, Jay B, Pawlowitzki IH, Sankila EM, Wright A, Ropers HH, Rosenberg T, Cremers FPM: Mutation spectrum in the CHM gene of Danish and Swedish choroideremia patients. Hum Mol Genet 1994;3:1047–1051.
Huber I, Bitner-Glindzicz M, de Kok YJM, van der Maarel SM, Ishikawa-Brush Y, Monaco AP, Robinson D, Malcolm S, Pembrey ME, Brunner HG, Cremers FPM, Ropers HH: X-linked mixed deafness (DFN3): Cloning and characterization of the critical region allows the identification of novel microdeletions. Hum Mol Genet 1994;3: 1151–1154.
Cremers FPM, van de Pol TJR, van Kerkhoff EPM, Wieringa B, Ropers HH: Cloning of a gene that is rearranged in patients with choroideraemia. Nature 1990;347:674–677.
Merry DE, Jänne PA, Landers JE, Lewis RA, Nussbaum RL: Isolation of a candidate gene for choroideremia. Proc Natl Acad Sei USA 1992; 89:2135–2139.
van Bokhoven H, van den Hurk JAJM, Bogerd L, Philippe C, Gilgenkrantz S, de Jong P, Ropers HH, Cremers FPM: Cloning and characterization of the human choroideremia gene. Hum Mol Genet 1994;3: 1041–1046.
de Kok YJM, van der Maarel SM, Bitner-Glindzicz M, Huber I, Monaco AP, Malcolm S, Pembrey ME, Ropers HH, Cremers FPM: Association between X-linked mixed deafness and mutations in the POU domain gene POU3F4. Science 1995; 267:685–688.
Philippe C, Cremers FPM, Chery M, Bach I, Abbadi N, Ropers HH, Gilgenkrantz S: Physical mapping of DNA markers in the q13-q22 region of the human X chromosome. Genomics 1993;17:147–152.
Albertsen HM, Abderrahim H, Cann HM, Dausset J, Le Paslier D, Cohen D: Construction and characterization of a yeast artificial chromosome library containing seven haploid human genome equivalents. Proc Natl Acad Sci USA 1990;87: 4256–4260.
Larin Z, Monaco AP, Lehrach H: Yeast artificial chromosome libraries containing large inserts from mouse and human DNA. Proc Natl Acad Sci USA 1991;88:4123–4127.
Maniatis T, Fritsch EF, Sambrook J: Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, Cold Spring Harbor Laboratory, 1982.
Green ED, Olson MV: Systematic screening of yeast artificial-chromosome libraries by use of the polymerase chain reaction. Proc Natl Acad Sci USA 1990;87:1213–1217.
Kere J, Nagaraja R, Mumm S, Ciccodicola A, D’Urso M, Schlessinger D: Mapping human chromosomes by walking with sequence-tagged sites from end fragments of yeast artificial chromosome inserts. Genomics 1992;14:241–248.
Gecz J, Villard L, Lossi AM, Millasseau P, Djabali M, Fontes M: Physical and transcriptional mapping of DXS56-PGK1 1 Mb region: Identification of three new transcripts. Hum Mol Genet 1993;2:1389–1396.
Villard L, Gecz J, Colleaux L, Lossi AM, Chelly J, Ishikawa-Brush Y, Monaco AP, Fontes M: Construction of a YAC contig spanning the Xq13.3 subband. Genomics 1995; 26:115–122.
Stanier P, Forbes S, Richardson M, Brennan L, Cole CG, Bentley DR, Moore GE: Physical mapping of the X-linked cleft palate locus. Cytogenet Cell Genet 1994;67:351.
Lloyd SL, Sargent CA, Chalmers J, Lim E, Habeebu SSM, Affara NA: An X-linked zinc finger gene mapping to Xq21.1-q21.3 closely related to ZFX and ZFY: Possible origins from a common ancestral gene. Nucleic Acids Res 1991;19:4835–4841.
Parra I, Windle B: High resolution visual mapping of stretched DNA by fluorescent hybridization. Nat Genet 1993;5:17–21.
Zehetner G, Lehrach H: The reference library system-sharing biological material and experimental data. Nature 1994;367:489–491.
Cremers FPM, van de Pol TJR, Wieringa B, Collins FS, Sankila EM, Siu VM, Flintoff WF, Brunsmann F, Blonden LAJ, Ropers HH: Chromosomal jumping from the DXS165 locus allows molecular characterization of four microdeletions and a de novo chromosome X/13 translocation associated with choroideremia. Prox Natl Acad Sci USA 1989;86: 7510–7514.
Cremers FPM, van de Pol TJR, Diergaarde PJ, Wieringa B, Nussbaum RL, Schwartz M, Ropers HH: Physical fine mapping of the choroideremia locus using Xq21 deletions associated with complex syndromes. Genomics 1989;4:41–46.
Cremers FPM, Brunsmann F, van de Pol TJR, Pawlowitzki IH, Paulsen K, Wieringa B, Ropers HH: Deletion of the DXS165 locus in patients with classical choroideremia. Clin Genet 1987;32:421–423.
Ayazi S: Choroideremia, obesity and congenital deafness. Am J Ophthalmol 1981;92:63–69.
Rosenberg T, Schwartz M, Niebuhr E, Yang HM, Sardemann H, Andersen O, Lundsteen C: Choroideremia in interstitial deletion of the X chromosome. Ophthalmic Paediatr Genet 1986;7:205–210.
Moore GE, Ivens A, Chambers J, Farrall M, Williamson R, Page DC, Bjornsson A, Arnason A, Jensson O: Linkage of an X-chromosome cleft palate gene. Nature 1987;326:91–92.
Ivens A, Moore GE, Chambers J, Arnason A, Jensson O, Bjornsson A, Williamson R: X-linked cleft palate: The gene is localized between polymorphic DNA markers DXYS12 and DXS17. Hum Genet 1988;78: 356–358.
Gorski SM, Adams KJ, Birch PH, Friedman JM, Goodfellow PJ: The gene responsible for X-linked cleft palate (CPX) in a British Columbia native kindred is localized between PGK1 and DXYS1. Am J Hum Genet 1992;50:1129–1136.
Stanier P, Forbes SA, Arnason A, Bjornsson A, Sveinbjornsdottir E, Williamson R, Moore G: The localization of a gene causing X-linked cleft palate and ankyloglossia (CPX) in an Icelandic kindred is between DXS326 and DXYS1X. Genomics 1993;17:549–555.
Gorski SM, Adams KJ, Birch PH, Chodirker BN, Greenberg CR, Goodfellow PJ: Linkage analysis of X-linked cleft palate and ankyloglossia in Manitoba Mennonite and British Columbia native kindreds. Hum Genet 1994;94:141–148.
Myhre SA, Ruvalcaba RHA, Kelley VC: Congenital deafness and hypogonadism: A new X-linked recessive disorder. Clin Genet 1982;22:299–307.
Neri G, Chiurazzi P, Arena JF, Lubs A: XLMR genes: Update 1994. Am J Med Genet 1994;51:542–549.
Schwartz CE, Ulmer J, Brown A, Pancoast I, Goodman HO, Stevenson RE: Allan-Herndon syndrome. II: Linkage to DNA markers in Xq21. Am J Hum Genet 1990;47: 454–458.
Schwartz CE, Martin J, Ouzts L, Arena JF, Lubs HA, Stevenson RE: Allan-Herndon-Dudley syndrome: Linkage analysis in a third family and refinement of the localization in Xq21. Cytogenet Cell Genet 1994; 67:351.
Saugier-Veber P, Abadie V, Moncla A, Mathieu M, Piussan C, Turleau C, Mattei JF, Munnich A, Lyonnet S: The Juberg-Marsidi syndrome maps to the proximal long arm of the X chromosome (Xq12-q21). Am J Hum Genet 1993;52:1040–1045.
Gibbons RJ, Suthers GK, Wilkie AOM, Buckle VJ, Higgs DR: X-linked α-thalassemia/mental retardation (ATR-X) syndrome: Localization to Xq12-q21.31 by X inactivation and linkage analysis. Am J Hum Genet 1992;51:1136–1149.
Houdayer C, Toutain A, Ronce N, Lefort G, Sarda P, Taib J, Briault S, Lambert JC, Moraine C: X-linked α-thalassemia/mental retardation syndrome: Linkage analysis in a new family further supports localization in proximal Xq. Ann Genet 1993; 36:194–199.
Lefort G, Taib J, Toutain A, Houdayer C, Moraine C, Humeau C, Sarda P: X-linked o-thalassemia/mental retardation (ATR-X) syndrome. Ann Genet 1993;36:200–205.
Ropers HH, van der Maarel S, Knoers N, Kremer H, Smits A, Hamel B, Cremers F, Gilgenkrantz S, Philippe C, Monaco T, Ishikawa-Brush Y, Smeets D, Mariman E: ‘Unspecific’ X-linked mental retardation: Clinical, genetic and molecular studies. Am J Hum Genet 1994; 55:A49.
Gibbons RJ, Picketts DJ, Villard L, Higgs DR: Mutations in a putative global transcriptional regulator cause X-linked mental retardation with α-thalassemia (ATR-X syndrome). Cell 1995;80:837–845.
Hammerstein W, Böhm T: Fluoreszenzangiographische Verlaufsbeobachtungen bei Chorioideremie. Stuttgart, Enke, 1985, pp 300–303.
Hodgson SV, Robertson ME, Fear CN, Goodship J, Malcolm S, Jay B, Bobrow M, Pembrey ME: Prenatal diagnosis of X-linked choroideremia with mental retardation, associated with a cytologically detectable X-chromosome deletion. Hum Genet 1987;75:286–290.
Wells S, Mould S, Robins D, Robinson D, Jacobs P: Molecular and cytogenetic analysis of a familial microdeletion of Xq. J Med Genet 1991;28:163–166.
Wallis C, Ballo R, Wallis G, Beighton P, Goldblatt J: X-linked mixed deafness with stapes fixation in a Mauritian kindred: Linkage to Xq probe pDP34. Genomics 1988,3: 299–301.
Tabor A, Andersen O, Lundsteen C, Niebuhr E, Sardemann H: Interstitial deletion in the ‘critical region’ of the long arm of the X chromosome in a mentally retarded boy and his normal mother. Hum Genet 1983; 64:196–199.
Willard HF, Goss SJ, Holmes MT, Munroe DL: Regional localization of the phosphoglycerate kinase gene and pseudogene on the human X chromosome and assignment of a related DNA sequence to chromosome 19. Hum Genet 1985;71:138–143.
Porteous MEM, Curtis A, Lindsay S, Williams O, Goudie D, Kamakari S, Bhattacharya SS: The gene for Aarskog syndrome is located between DSX255 and DXS566 (Xp11.2-Xq13). Genomics 1992; 14: 298–301.
Weissenbach J, Gyapay G, Dib C, Vignal A, Morissette J, Millasseau P, Vaysseix G, Lathrop M: A second-generation linkage map of the human genome. Nature 1992,359: 794–801.
Gyapay G, Morissette J, Vignal A, Dib C, Fizames C, Millasseau P, Marc S, Bernardi G, Lathrop M, Weissenbach J: The 1993–94 Genethon human genetic linkage map. Nat Genet 1994;7:246–339.
Barker DF, Fain PR: Definition and mapping of STSs at STR and RFLP loci in Xp11-Xq22. Genomics 1993; 18:712–716.
Hudson TJ, Engelstein M, Lee MK, Ho EC, Rubenfield MJ, Adams CP, Housman DE, Dracopoli NC: Isolation and chromosomal assignment of 100 highly informative human simple sequence repeat polymorphisms. Genomics 1992,13:622–629.
Riddell DC, Wang HS, Beckett J, Chan A, Holden JJA, Mulligan LM, Phillips MA, Simpson NE, Wrogemann K, Hamerton JL, White BN: Regional localization of 18 human X-linked DNA sequences. Cytogenet Cell Genet 1986;42:123–128.
Hofker MH, Bergen AAB, Skraastad MI, Carpenter NJ, Veenema H, Connor JM, Bakker E, van Ommen GJB, Pearson PL: Efficient isolation of X chromosome-specific single-copy probes from a cosmid library of a human X/hamster hybrid-cell line: Mapping of new probes close to the locus for X-linked mental retardation. Am J Hum Genet 1987;40: 312–328.
Goodfellow PN, Davies KE, Ropers HH: Report of the committee on the genetic constitution of the X and Y chromosomes. Human Gene Mapping 8: Eighth International Workshop on Human Gene Mapping. Cytogenet Cell Genet 1985;40:296–352.
Paulsen K, Forrest S, Scherer G, Ropers HH, Davies K: Regional localisation of X chromosome short arm probes. Hum Genet 1986;74: 155–159.
van de Pol TJR, Cremers FPM, Brohet RM, Wieringa B, Ropers HH: Derivation of clones from the choroideremia locus by preparative field inversion gel electrophoresis. Nucleic Acids Res 1990; 18:725–731.
Davatelis G, Siniscalco M, Szabo P: Toward a more complete linkage map of the human X-chromosome. Cytogenet Cell Genet 1985;40:611.
Dietz-Band JN, Turco AE, Willard HF, Vincent A, Skolnick MH, Barker DF: Isolation, characterization, and physical localization of 33 human X-chromosome RFLP markers. Cytogenet Cell Genet 1990;54: 137–141.
Aldridge J, Kunkel L, Bruns G, Tantravahi U, Lalande M, Brewster T, Moreau E, Wilson M, Bromley W, Roderick T, Latt S: A strategy to reveal high-frequency RFLPs along the human X chromosome. Am J Hum Genet 1984;36:546–564.
Weber JL, Kwitek AE, May PE, Polymeropoulos MH, Ledbetter S: Dinucleotide repeat polymorphisms at the DXS453, DXS454 and DXS458 loci. Nucleic Acids Res 1990; 18:4037.
Acknowledgements
The authors thank Mrs. S.D. van der Velde-Visser and E. Boender-van Rossum for expert technical assistance in cell culturing and W.K.C. van Loon and Y. Ishikawa-Brush for screening the YAC libraries in the context of the YAC Screening Center grant GENECT-0093-0088 of the European Community. The chromosome-specific gene library LL0XNC01 used in this work was constructed at the Biology and Biotechnology Research Program, Lawrence Livermore National Laboratory, Livermore, CA 94550, USA, under the auspices of the National Gene Library Project sponsored by the U.S. Department of Energy. We thank Drs. M. Ross, G. Zehetner and H. Lehrach for access to the ICRF cosmid library filters. The research of FPMC has been made possible by a fellowship of the Royal Netherlands Academy of Arts and Sciences. This study was supported by the Netherlands Organization for Scientific Research (NWO grants 901-04-125 and 900-716-801), the Dutch Praeventiefonds (grant 28-2447) and the European Community (grant GENE-CT93-0022).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
van der Maarel, S.M., Scholten, I.H.J.M., Maat-Kievit, J.A. et al. Yeast Artificial Chromosome Cloning of the Xq13.3-q21.31 Region and Fine Mapping of a Deletion Associated with Choroideremia and Nonspecific Mental Retardation. Eur J Hum Genet 3, 207–218 (1995). https://doi.org/10.1159/000472301
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1159/000472301