Article
|
Open Access
Featured
-
-
Article
| Open Access Simultaneous spatiotemporal super-resolution and multi-parametric fluorescence microscopy
Simultaneous spatiotemporal super-resolution and multi-parametric fluorescence microscopy
Super-resolution microscopy and single molecule fluorescence spectroscopy require optimisation of the temporal or spatial resolution, which are usually mutually exclusive. Here the authors report a GPU-supported, camera-based strategy to achieve high spatial and temporal resolution from the same dataset.
- Jagadish Sankaran
- , Harikrushnan Balasubramanian
- & Thorsten Wohland
-
Article
| Open Access Joint analysis of expression levels and histological images identifies genes associated with tissue morphology
Joint analysis of expression levels and histological images identifies genes associated with tissue morphology
Image features from histological slides can be used as informative endophenotypes in association studies for tissue-localized pathologies. Here, the authors develop ImageCCA, a framework for joint analysis of paired gene expression and histology data derived from automatically extracted image features.
- Jordan T. Ash
- , Gregory Darnell
- & Barbara E. Engelhardt
-
Article
| Open Access Whole-brain tissue mapping toolkit using large-scale highly multiplexed immunofluorescence imaging and deep neural networks
Whole-brain tissue mapping toolkit using large-scale highly multiplexed immunofluorescence imaging and deep neural networks
It is challenging to map complex processes in brain tissue. Here the authors report a toolkit enabling large-scale multiplexed IHC and automated cell classification whereby they use a conventional epifluorescence microscope and deep neural networks to phenotype all major cell classes of the brain.
- Dragan Maric
- , Jahandar Jahanipour
- & Badrinath Roysam
-
Article
| Open Access An annotation-free whole-slide training approach to pathological classification of lung cancer types using deep learning
An annotation-free whole-slide training approach to pathological classification of lung cancer types using deep learning
Deep learning for digital pathology is hindered by the extremely high spatial resolution of whole slide images (WSIs), which requires researchers to adopt patch-based methods and laborious free-hand contouring. Here, the authors develop a whole-slide training method to classify types of lung cancers using slide-level diagnoses with deep learning.
- Chi-Long Chen
- , Chi-Chung Chen
- & Cheng-Yu Chen
-
Article
| Open Access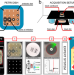 AI-based mobile application to fight antibiotic resistance
AI-based mobile application to fight antibiotic resistance
Antimicrobial resistance is a major global health threat and its development is promoted by antibiotic misuse. Here, the authors present an offline smartphone application for automated and standardized antibiotic susceptibility testing, to be deployed in resource-limited settings.
- Marco Pascucci
- , Guilhem Royer
- & Mohammed-Amin Madoui
-
Article
| Open Access Direct supercritical angle localization microscopy for nanometer 3D superresolution
Direct supercritical angle localization microscopy for nanometer 3D superresolution
Supercritical angle localisation microscopy (SALM) allows the z-positions of single fluorophores to be extracted from the intensity of supercritical angle fluorescence. Here the authors improve the z-resolution of SALM, and report nanometre isotropic localisation precision on DNA origami structures.
- Anindita Dasgupta
- , Joran Deschamps
- & Jonas Ries
-
Article
| Open Access Automatic deep learning-driven label-free image-guided patch clamp system
Automatic deep learning-driven label-free image-guided patch clamp system
Patch clamp recording of neurons is slow and labor-intensive. Here the authors present a method for automated deep learning driven label-free image guided patch clamp physiology to perform measurements on hundreds of human and rodent neurons.
- Krisztian Koos
- , Gáspár Oláh
- & Peter Horvath
-
Article
| Open Access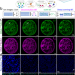 Nanoscopic subcellular imaging enabled by ion beam tomography
Nanoscopic subcellular imaging enabled by ion beam tomography
Secondary ion beam mass spectrometry (SIMS) is a method to obtain a chemical snapshot of biological tissue, but the spatial resolution is low. Here, the authors develop a computational and technology pipeline to localise a chemical signal in SIMS in 3D and sub-25 nm accuracy, called Ion Beam Tomography
- Ahmet F. Coskun
- , Guojun Han
- & Garry P. Nolan
-
Article
| Open Access A network-based framework for shape analysis enables accurate characterization of leaf epidermal cells
A network-based framework for shape analysis enables accurate characterization of leaf epidermal cells
While cell shape is crucial for function and development of organisms, versatile frameworks for cell shape quantification, comparison, and classification remain underdeveloped. Here, the authors use a network-based framework for Arabidopsis leaf epidermal cell shape characterization and classification.
- Jacqueline Nowak
- , Ryan Christopher Eng
- & Zoran Nikoloski
-
Article
| Open Access Establishment of a morphological atlas of the Caenorhabditis elegans embryo using deep-learning-based 4D segmentation
Establishment of a morphological atlas of the Caenorhabditis elegans embryo using deep-learning-based 4D segmentation
The systematic characterization of C. elegans morphology during development has yet to be performed. Here, the authors produce a 3D atlas of C. elegans morphology from 17 embryos and 54 developmental stages, using an automated pipeline, CShaper (combining segmentation of fluorescently labeled membranes with automated cell lineage tracing).
- Jianfeng Cao
- , Guoye Guan
- & Hong Yan
-
Article
| Open Access A convolutional neural network segments yeast microscopy images with high accuracy
A convolutional neural network segments yeast microscopy images with high accuracy
Current cell segmentation methods for Saccharomyces cerevisiae face challenges under a variety of standard experimental and imaging conditions. Here the authors develop a convolutional neural network for accurate, label-free cell segmentation.
- Nicola Dietler
- , Matthias Minder
- & Sahand Jamal Rahi
-
Article
| Open Access TranSPHIRE: automated and feedback-optimized on-the-fly processing for cryo-EM
TranSPHIRE: automated and feedback-optimized on-the-fly processing for cryo-EM
High-throughput single particle cryo-EM, for instance in drug research, requires the automation of the single particle analysis workflow. Here, the authors present TranSPHIRE, a software package that allows the fully-automated, feedback-driven processing of cryo-EM datasets during data acquisition.
- Markus Stabrin
- , Fabian Schoenfeld
- & Stefan Raunser
-
Article
| Open Access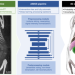 Deep learning-enabled multi-organ segmentation in whole-body mouse scans
Deep learning-enabled multi-organ segmentation in whole-body mouse scans
Organ segmentation of whole-body mouse images is essential for quantitative analysis, but is tedious and error-prone. Here the authors develop a deep learning pipeline to segment major organs and the skeleton in volumetric whole-body scans in less than a second, and present probability maps and uncertainty estimates.
- Oliver Schoppe
- , Chenchen Pan
- & Bjoern H. Menze
-
Article
| Open Access Rapid vessel segmentation and reconstruction of head and neck angiograms using 3D convolutional neural network
Rapid vessel segmentation and reconstruction of head and neck angiograms using 3D convolutional neural network
Manual postprocessing of computed tomography angiography (CTA) images is extremely labor intensive and error prone. Here, the authors propose an artificial intelligence reconstruction system that can automatically achieve CTA reconstruction in healthcare services.
- Fan Fu
- , Jianyong Wei
- & Jie Lu
-
Article
| Open Access Motion corrected MRI differentiates male and female human brain growth trajectories from mid-gestation
Motion corrected MRI differentiates male and female human brain growth trajectories from mid-gestation
The human fetal brain may exhibit early sex differences. Here, the authors present MRI based analysis female and male brain growth. Cortical development follows very similar trajectories at larger scales, while differing in focal regions. White matter growth differs at both large and small scales.
- Colin Studholme
- , Christopher D. Kroenke
- & Manjiri Dighe
-
Article
| Open Access Transfer functions linking neural calcium to single voxel functional ultrasound signal
Transfer functions linking neural calcium to single voxel functional ultrasound signal
Neurovascular coupling refers to changes in cerebral blood flow in response to neuronal stimulation, but to what extent this change can report neuronal activation is not known. Here the authors develop transfer functions between neural calcium signals and functional ultrasound changes in blood volume in co-registered single voxel brain volumes.
- Ali-Kemal Aydin
- , William D. Haselden
- & Davide Boido
-
Article
| Open Access Sexual signaling pattern correlates with habitat pattern in visually ornamented fishes
Sexual signaling pattern correlates with habitat pattern in visually ornamented fishes
Sensory drive theory posits that selection on sexual signals should depend on the environmental background. Here, Hulse et al. analyze the spatial statistics of body patterning in 10 darter fish species and find a correlation with habitat spatial statistics only for males, consistent with sexual selection via sensory drive.
- Samuel V. Hulse
- , Julien P. Renoult
- & Tamra C. Mendelson
-
Article
| Open Access Computational analysis of pathological images enables a better diagnosis of TFE3 Xp11.2 translocation renal cell carcinoma
Computational analysis of pathological images enables a better diagnosis of TFE3 Xp11.2 translocation renal cell carcinoma
Translocation renal cell carcinoma is an aggressive form of renal cancer that is often misdiagnosed to other subtypes. Here the authors demonstrated that by using machine learning and H&E stained whole-slide images, an accurate diagnose of this particular type of renal cancer can be achieved.
- Jun Cheng
- , Zhi Han
- & Jie Zhang
-
Article
| Open Access Dictionary learning in Fourier-transform scanning tunneling spectroscopy
Dictionary learning in Fourier-transform scanning tunneling spectroscopy
Aperiodic structure imaging suffers limitations when utilizing Fourier analysis. The authors report an algorithm that quantitatively overcomes these limitations based on nonconvex optimization, demonstrated by studying aperiodic structures via the phase sensitive interference in STM images.
- Sky C. Cheung
- , John Y. Shin
- & Abhay N. Pasupathy
-
Article
| Open Access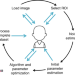 An interactive ImageJ plugin for semi-automated image denoising in electron microscopy
An interactive ImageJ plugin for semi-automated image denoising in electron microscopy
Large 3D electron microscopy data sets frequently contain noisy data due to accelerated imaging, and denoising techniques require specialised skill sets. Here the authors introduce DenoisEM, an ImageJ plugin that democratises denoising EM data sets, enabling fast parameter tuning and processing through parallel computing.
- Joris Roels
- , Frank Vernaillen
- & Yvan Saeys
-
Article
| Open Access Democratized image analytics by visual programming through integration of deep models and small-scale machine learning
Democratized image analytics by visual programming through integration of deep models and small-scale machine learning
Deep learning approaches for image preprocessing and analysis offer important advantages, but these are rarely incorporated into user-friendly software. Here the authors present an easy-to-use visual programming toolbox integrating deep-learning and interactive data visualization for image analysis.
- Primož Godec
- , Matjaž Pančur
- & Blaž Zupan
-
Article
| Open Access Label-free neuroimaging in vivo using synchronous angular scanning microscopy with single-scattering accumulation algorithm
Label-free neuroimaging in vivo using synchronous angular scanning microscopy with single-scattering accumulation algorithm
A major challenge of in vivo imaging is imaging deeper, including in turbid tissue. The authors report an adaptive optics based microscope that uses coherent single scattering signal to reduce sample-induced aberrations and enable fast deep-tissue imaging of in vivo larval zebrafish brain.
- Moonseok Kim
- , Yonghyeon Jo
- & Wonshik Choi
-
Article
| Open Access Fourier ring correlation simplifies image restoration in fluorescence microscopy
Fourier ring correlation simplifies image restoration in fluorescence microscopy
Fourier ring correlation (FRC) analysis is commonly used in fluorescence microscopy to measure effective image resolution. Here, the authors demonstrate that FRC can also be leveraged in blind image restoration methods, such as image deconvolution.
- Sami Koho
- , Giorgio Tortarolo
- & Giuseppe Vicidomini
-
Article
| Open Access Coherent diffractive imaging of microtubules using an X-ray laser
Coherent diffractive imaging of microtubules using an X-ray laser
XFEL radiation is providing new opportunities for probing biological systems. Here the authors perform nanoscale x-ray imaging of microtubules with helical symmetry, by using imaging sorting and reconstruction techniques.
- Gisela Brändén
- , Greger Hammarin
- & Richard Neutze
-
Article
| Open Access Facial recognition from DNA using face-to-DNA classifiers
Facial recognition from DNA using face-to-DNA classifiers
Prediction of face from DNA followed by matching to facial images has been proposed for forensic applications. Here, Sero et al. present a different approach that can establish facial identity from DNA without directly predicting the face but is based on classifying given faces by individual DNA-encoded traits.
- Dzemila Sero
- , Arslan Zaidi
- & Peter Claes
-
Article
| Open Access Interpretable classification of Alzheimer’s disease pathologies with a convolutional neural network pipeline
Interpretable classification of Alzheimer’s disease pathologies with a convolutional neural network pipeline
Convolutional neural networks have been applied to various areas of medical imaging and histology. Here the authors develop an automated approach using interpretable neural networks to determine Alzheimer’s disease plaque and cerebral amyloid angiopathy burden in post-mortem human brain tissue.
- Ziqi Tang
- , Kangway V. Chuang
- & Brittany N. Dugger
-
Article
| Open Access Information-rich localization microscopy through machine learning
Information-rich localization microscopy through machine learning
Single-molecule methods often rely on point spread functions that are tailored to interpret specific information. Here the authors use a neural network to extract complex PSF information from experimental images, and demonstrate this by classifying color and axial positions of emitters.
- Taehwan Kim
- , Seonah Moon
- & Ke Xu
-
Article
| Open Access Resolution limit of image analysis algorithms
Resolution limit of image analysis algorithms
The resolution limitations when using the ubiquitous algorithms that process images obtained using modern techniques are not straightforward to define. Here, the authors examine the performance of localization algorithms and use spatial statistics to provide a metric for assessing the resolution limit of such algorithms.
- Edward A. K. Cohen
- , Anish V. Abraham
- & Raimund J. Ober
-
Article
| Open Access Ultrafast data mining of molecular assemblies in multiplexed high-density super-resolution images
Ultrafast data mining of molecular assemblies in multiplexed high-density super-resolution images
Analyzing the organization of molecular complexes in multi-color single-molecule localization microscopy data requires heavy computation resources that are impractical for laboratory computers. Here the authors develop a coordinate-based Triple-Correlation algorithm with improved speed and reduced computational cost.
- Yandong Yin
- , Wei Ting Chelsea Lee
- & Eli Rothenberg
-
Article
| Open Access Adaptive particle representation of fluorescence microscopy images
Adaptive particle representation of fluorescence microscopy images
Modern microscopes can generate high volumes of 3D images, driving difficulties in data handling and processing. Here, the authors present a content-adaptive image representation as an alternative to standard pixels that goes beyond data compression to overcome storage, memory, and processing bottlenecks.
- Bevan L. Cheeseman
- , Ulrik Günther
- & Ivo F. Sbalzarini
-
Article
| Open Access Automated quantification of bioluminescence images
Automated quantification of bioluminescence images
Analysis of bioluminescence images of bacterial distributions in living animals is mostly manual and semiquantitative. Here, the authors present an analysis platform featuring an animal mold, a probabilistic organ atlas, and a mirror gantry to perform automatic in vivo bioluminescence quantification.
- Alexander D. Klose
- & Neal Paragas
-
Article
| Open Access Quantitative spatial analysis of haematopoiesis-regulating stromal cells in the bone marrow microenvironment by 3D microscopy
Quantitative spatial analysis of haematopoiesis-regulating stromal cells in the bone marrow microenvironment by 3D microscopy
The bone marrow microenvironment modulates haematopoiesis, stem cell maintenance and differentiation. Here, the authors use 3D microscopy to map the topography of haematopoietic stem cell niche stromal components.
- Alvaro Gomariz
- , Patrick M. Helbling
- & César Nombela-Arrieta
-
Article
| Open Access ZOLA-3D allows flexible 3D localization microscopy over an adjustable axial range
ZOLA-3D allows flexible 3D localization microscopy over an adjustable axial range
3D single-molecule localization is limited in depth and often requires using a wide range of point spread functions (PSFs). Here the authors present an optical solution featuring a deformable mirror to generate different PSFs and easy-to-use software for super-resolution imaging up to 5 µm deep.
- Andrey Aristov
- , Benoit Lelandais
- & Christophe Zimmer
-
Article
| Open Access Deconvolution of subcellular protrusion heterogeneity and the underlying actin regulator dynamics from live cell imaging
Deconvolution of subcellular protrusion heterogeneity and the underlying actin regulator dynamics from live cell imaging
Cell protrusion dynamics are heterogeneous at the subcellular level, but current analyses operate at the cellular or ensemble level. Here the authors develop a computational framework to quantify subcellular protrusion phenotypes and reveal the underlying actin regulator dynamics at the leading edge.
- Chuangqi Wang
- , Hee June Choi
- & Kwonmoo Lee
-
Article
| Open Access Mapping molecular assemblies with fluorescence microscopy and object-based spatial statistics
Mapping molecular assemblies with fluorescence microscopy and object-based spatial statistics
Elucidating molecular organisation requires precise localisation and analysis. Here the authors develop SODA software for automatic and quantitative mapping of statistically coupled molecules, and use it to unravel spatial organisation of thousands of synaptic proteins in SIM and 3DSTORM microscopy.
- Thibault Lagache
- , Alexandre Grassart
- & Jean-Christophe Olivo-Marin
-
Article
| Open Access Monitoring single-cell gene regulation under dynamically controllable conditions with integrated microfluidics and software
Monitoring single-cell gene regulation under dynamically controllable conditions with integrated microfluidics and software
How gene regulatory pathways control cell fate decisions in single cells is not fully understood. Here the authors present an integrated dual-input microfluidic chip and a linked analysis software, enabling tracking of gene regulatory responses of single bacterial cells to changing conditions.
- Matthias Kaiser
- , Florian Jug
- & Erik van Nimwegen
-
Article
| Open Access Intelligent image-based in situ single-cell isolation
Intelligent image-based in situ single-cell isolation
The isolation of single cells while retaining context is important for quantifying cellular heterogeneity but technically challenging. Here, the authors develop a high-throughput, scalable workflow for microscopy-based single cell isolation using machine-learning, high-throughput microscopy and laser capture microdissection.
- Csilla Brasko
- , Kevin Smith
- & Peter Horvath
-
Article
| Open Access Reconstructing cell cycle and disease progression using deep learning
Reconstructing cell cycle and disease progression using deep learning
The interpretation of information-rich, high-throughput single-cell data is a challenge requiring sophisticated computational tools. Here the authors demonstrate a deep convolutional neural network that can classify cell cycle status on-the-fly.
- Philipp Eulenberg
- , Niklas Köhler
- & F. Alexander Wolf
-
Article
| Open Access A BaSiC tool for background and shading correction of optical microscopy images
A BaSiC tool for background and shading correction of optical microscopy images
Accurate quantification of bioimaging data is often confounded by uneven illumination (shading) in space and background variation in time. Here the authors present BaSiC, a Fiji plugin solving both issues. It requires fewer input images and is more robust to artefacts than existing shading correction tools.
- Tingying Peng
- , Kurt Thorn
- & Nassir Navab
-
Article
| Open Access Smooth 2D manifold extraction from 3D image stack
Smooth 2D manifold extraction from 3D image stack
Maximum Intensity Projection is a common tool to represent 3D biological imaging data in a 2D space, but it creates artefacts. Here the authors develop Smooth Manifold Extraction, an ImageJ/Fiji plugin, to preserve local spatial relationships when extracting the content of a 3D volume to a 2D space.
- Asm Shihavuddin
- , Sreetama Basu
- & Auguste Genovesio
-
Article
| Open Access Reconstructing 3D deformation dynamics for curved epithelial sheet morphogenesis from positional data of sparsely-labeled cells
Reconstructing 3D deformation dynamics for curved epithelial sheet morphogenesis from positional data of sparsely-labeled cells
Quantifying deformation patterns of curved epithelial sheets is challenging owing to imaging difficulties. Here the authors develop a method to obtain a quantitative description of 3D tissue deformation dynamics from a small set of cell positional data and applied it to chick forebrain morphogenesis.
- Yoshihiro Morishita
- , Ken-ichi Hironaka
- & Daisuke Ohtsuka
-
Article
| Open Access Asynchronous fate decisions by single cells collectively ensure consistent lineage composition in the mouse blastocyst
Asynchronous fate decisions by single cells collectively ensure consistent lineage composition in the mouse blastocyst
Early embryonic cell fate and lineage specification is tightly regulated in the preimplantation mammalian embryo. Here, the authors quantitatively examine the ratio of epiblast to primitive endoderm lineages in the blastocyst and show composition of the inner cell mass is conserved, independent of its size.
- Néstor Saiz
- , Kiah M. Williams
- & Anna-Katerina Hadjantonakis
-
Article
| Open Access Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features
Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features
Diagnosis of lung cancer through manual histopathology evaluation is insufficient to predict patient survival. Here, the authors use computerized image processing to identify diagnostically relevant image features and use these features to distinguish lung cancer patients with different prognoses.
- Kun-Hsing Yu
- , Ce Zhang
- & Michael Snyder
-
Article
| Open Access High-resolution imaging and computational analysis of haematopoietic cell dynamics in vivo
High-resolution imaging and computational analysis of haematopoietic cell dynamics in vivo
It is difficult to image haematopoietic stem cells (HSC) in their niche. Here, the authors present a new high-throughput computational approach to visualise HSCs in vivoat a high spatial and temporal resolution and also use a Msi2-reporter to label endogenous HSCs and progenitors, enabling cell tracking
- Claire S. Koechlein
- , Jeffrey R. Harris
- & Tannishtha Reya
-
Article
| Open Access A workflow to process 3D+time microscopy images of developing organisms and reconstruct their cell lineage
A workflow to process 3D+time microscopy images of developing organisms and reconstruct their cell lineage
Quantitative analysis of embryonic cell dynamics from large data sets remains a major challenge in the field of developmental biology. Here the authors develop software and a workflow to reconstruct cell lineage trees from 3D time lapse imaging data sets from several developing organisms including zebrafish, tunicates and sea urchins.
- Emmanuel Faure
- , Thierry Savy
- & Paul Bourgine

 Markerless tracking of an entire honey bee colony
Markerless tracking of an entire honey bee colony