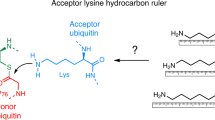
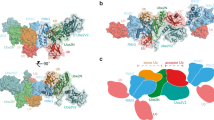
Abstract
E3 ligases carry out the final step in the ubiquitination cascade, catalyzing transfer of ubiquitin from an E2 enzyme to form a covalent bond with a substrate lysine. Three distinct classes of E3 ligases have been identified that stimulate transfer of ubiquitin and ubiquitin-like proteins through either a direct or an indirect mechanism. Only recently have the catalytic mechanisms of E3 ligases begun to be elucidated.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kerscher, O., Felberbaum, R. & Hochstrasser, M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22, 159–180 (2006).
Ulrich, H.D. & Walden, H. Ubiquitin signalling in DNA replication and repair. Nat. Rev. Mol. Cell Biol. 11, 479–489 (2010).
Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012).
Pickart, C.M. & Eddins, M.J. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 1695, 55–72 (2004).
Tokunaga, F. et al. Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nat. Cell Biol. 11, 123–132 (2009).
Scaglione, K.M. et al. The ubiquitin-conjugating enzyme (E2) Ube2w ubiquitinates the N terminus of substrates. J. Biol. Chem. 288, 18784–18788 (2013).
Tatham, M.H., Plechanovová, A., Jaffray, E.G., Salmen, H. & Hay, R.T. Ube2W conjugates ubiquitin to α-amino groups of protein N-termini. Biochem. J. 453, 137–145 (2013).
Deshaies, R.J. & Joazeiro, C.A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 (2009).
Li, W. et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS ONE 3, e1487 (2008).
Metzger, M.B., Hristova, V.A. & Weissman, A.M. HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell Sci. 125, 531–537 (2012).
Budhidarmo, R., Nakatani, Y. & Day, C.L. RINGs hold the key to ubiquitin transfer. Trends Biochem. Sci. 37, 58–65 (2012).
Huibregtse, J.M., Scheffner, M., Beaudenon, S. & Howley, P.M. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. USA 92, 2563–2567 (1995).
Wenzel, D.M. & Klevit, R.E. Following Ariadne's thread: a new perspective on RBR ubiquitin ligases. BMC Biol. 10, 24 (2012).
van Wijk, S.J. & Timmers, H.T. The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB J. 24, 981–993 (2010).
Wenzel, D.M., Stoll, K.E. & Klevit, R.E. E2s: structurally economical and functionally replete. Biochem. J. 433, 31–42 (2011).
Wu, P.Y. et al. A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J. 22, 5241–5250 (2003).
Berndsen, C.E., Wiener, R., Yu, I.W., Ringel, A.E. & Wolberger, C. A conserved asparagine has a structural role in ubiquitin-conjugating enzymes. Nat. Chem. Biol. 9, 154–156 (2013).
Yunus, A.A. & Lima, C.D. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat. Struct. Mol. Biol. 13, 491–499 (2006).
Plechanovová, A., Jaffray, E.G., Tatham, M.H., Naismith, J.H. & Hay, R.T. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 489, 115–120 (2012).
Jin, L., Williamson, A., Banerjee, S., Philipp, I. & Rape, M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 133, 653–665 (2008).
Wickliffe, K.E., Lorenz, S., Wemmer, D.E., Kuriyan, J. & Rape, M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell 144, 769–781 (2011).
Sakata, E. et al. Crystal structure of UbcH5b∼ubiquitin intermediate: insight into the formation of the self-assembled E2∼Ub conjugates. Structure 18, 138–147 (2010).
Eddins, M.J., Carlile, C.M., Gomez, K.M., Pickart, C.M. & Wolberger, C. Mms2–Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat. Struct. Mol. Biol. 13, 915–920 (2006).
McKenna, S. et al. An NMR-based model of the ubiquitin-bound human ubiquitin conjugation complex Mms2.Ubc13: the structural basis for lysine 63 chain catalysis. J. Biol. Chem. 278, 13151–13158 (2003).
Pruneda, J.N., Stoll, K.E., Bolton, L.J., Brzovic, P.S. & Klevit, R.E. Ubiquitin in motion: structural studies of the ubiquitin-conjugating enzyme approximately ubiquitin conjugate. Biochemistry 50, 1624–1633 (2011).
Lorick, K.L. et al. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 96, 11364–11369 (1999).
Bentley, M.L. et al. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. EMBO J. 30, 3285–3297 (2011).
Dou, H., Buetow, L., Sibbet, G.J., Cameron, K. & Huang, D.T. BIRC7–E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat. Struct. Mol. Biol. 19, 876–883 (2012).
Yin, Q. et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nat. Struct. Mol. Biol. 16, 658–666 (2009).
Zheng, N., Wang, P., Jeffrey, P.D. & Pavletich, N.P. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102, 533–539 (2000).
Liew, C.W., Sun, H., Hunter, T. & Day, C.L. RING domain dimerization is essential for RNF4 function. Biochem. J. 431, 23–29 (2010).
Brzovic, P.S., Rajagopal, P., Hoyt, D.W., King, M.C. & Klevit, R.E. Structure of a BRCA1–BARD1 heterodimeric RING–RING complex. Nat. Struct. Biol. 8, 833–837 (2001).
Buchwald, G. et al. Structure and E3-ligase activity of the Ring–Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 25, 2465–2474 (2006).
Campbell, S.J. et al. Molecular insights into the function of RING finger (RNF)-containing proteins hRNF8 and hRNF168 in Ubc13/Mms2-dependent ubiquitylation. J. Biol. Chem. 287, 23900–23910 (2012).
Ohi, M.D., Vander Kooi, C.W., Rosenberg, J.A., Chazin, W.J. & Gould, K.L. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat. Struct. Biol. 10, 250–255 (2003).
Eletr, Z.M., Huang, D.T., Duda, D.M., Schulman, B.A. & Kuhlman, B. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat. Struct. Mol. Biol. 12, 933–934 (2005).
Lydeard, J.R., Schulman, B.A. & Harper, J.W. Building and remodelling Cullin–RING E3 ubiquitin ligases. EMBO Rep. 14, 1050–1061 (2013).
Skaar, J.R., Pagan, J.K. & Pagano, M. Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 14, 369–381 (2013).
Hibbert, R.G., Huang, A., Boelens, R. & Sixma, T.K. E3 ligase Rad18 promotes monoubiquitination rather than ubiquitin chain formation by E2 enzyme Rad6. Proc. Natl. Acad. Sci. USA 108, 5590–5595 (2011).
Mace, P.D. et al. Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. J. Biol. Chem. 283, 31633–31640 (2008).
Ozkan, E., Yu, H. & Deisenhofer, J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc. Natl. Acad. Sci. USA 102, 18890–18895 (2005).
Plechanovová, A. et al. Mechanism of ubiquitylation by dimeric RING ligase RNF4. Nat. Struct. Mol. Biol. 18, 1052–1059 (2011).
Spratt, D.E., Wu, K., Kovacev, J., Pan, Z.Q. & Shaw, G.S. Selective recruitment of an E2∼ubiquitin complex by an E3 ubiquitin ligase. J. Biol. Chem. 287, 17374–17385 (2012).
Dou, H. et al. Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl. Nat. Struct. Mol. Biol. 19, 184–192 (2012).
Reverter, D. & Lima, C.D. Insights into E3 ligase activity revealed by a SUMO–RanGAP1–Ubc9–Nup358 complex. Nature 435, 687–692 (2005).
Pruneda, J.N. et al. Structure of an E3:E2∼Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol. Cell 47, 933–942 (2012).
Bruice, T.C. & Pandit, U.K. Intramolecular models depicting the kinetic importance of “fit” in enzymatic catalysis. Proc. Natl. Acad. Sci. USA 46, 402–404 (1960).
Jencks, W.P. Catalysis in Chemistry and Enzymology (Dover, New York, 1987).
Saha, A., Lewis, S., Kleiger, G., Kuhlman, B. & Deshaies, R.J. Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from cdc34 to substrate. Mol. Cell 42, 75–83 (2011).
Das, R. et al. Allosteric activation of E2-RING finger-mediated ubiquitylation by a structurally defined specific E2-binding region of gp78. Mol. Cell 34, 674–685 (2009).
Brzovic, P.S., Lissounov, A., Christensen, D.E., Hoyt, D.W. & Klevit, R.E.A. UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol. Cell 21, 873–880 (2006).
Das, R. et al. Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine. EMBO J. 32, 2504–2516 (2013).
Notenboom, V. et al. Functional characterization of Rad18 domains for Rad6, ubiquitin, DNA binding and PCNA modification. Nucleic Acids Res. 35, 5819–5830 (2007).
Metzger, M.B. et al. A structurally unique E2-binding domain activates ubiquitination by the ERAD E2, Ubc7p, through multiple mechanisms. Mol. Cell 50, 516–527 (2013).
Huang, L. et al. Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2–E3 enzyme cascade. Science 286, 1321–1326 (1999).
Verdecia, M.A. et al. Conformational flexibility underlies ubiquitin ligation mediated by the WWP1 HECT domain E3 ligase. Mol. Cell 11, 249–259 (2003).
Kamadurai, H.B. et al. Insights into ubiquitin transfer cascades from a structure of a UbcH5B∼ubiquitin-HECT(NEDD4L) complex. Mol. Cell 36, 1095–1102 (2009).
Kamadurai, H.B. et al. Mechanism of ubiquitin ligation and lysine prioritization by a HECT E3. Elife 2, e00828 (2013).
Maspero, E. et al. Structure of a ubiquitin-loaded HECT ligase reveals the molecular basis for catalytic priming. Nat. Struct. Mol. Biol. 20, 696–701 (2013).
Ronchi, V.P., Klein, J.M. & Haas, A.L. E6AP/UBE3A ubiquitin ligase harbors two E2∼ubiquitin binding sites. J. Biol. Chem. 288, 10349–10360 (2013).
Kim, H.C. & Huibregtse, J.M. Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol. Cell. Biol. 29, 3307–3318 (2009).
Aguilera, M., Oliveros, M., Martinez-Padron, M., Barbas, J.A. & Ferrus, A. Ariadne-1: a vital Drosophila gene is required in development and defines a new conserved family of ring-finger proteins. Genetics 155, 1231–1244 (2000).
Dawson, T.M. & Dawson, V.L. The role of parkin in familial and sporadic Parkinson's disease. Mov. Disord. 25, S32–S39 (2010).
Wenzel, D.M., Lissounov, A., Brzovic, P.S. & Klevit, R.E. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 474, 105–108 (2011).
Duda, D.M. et al. Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism. Structure 21, 1030–1041 (2013).
Riley, B.E. et al. Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat. Commun. 4, 1982 (2013).
Trempe, J.F. et al. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science 340, 1451–1455 (2013).
Wauer, T. & Komander, D. Structure of the human Parkin ligase domain in an autoinhibited state. EMBO J. 32, 2099–2112 (2013).
Stieglitz, B. et al. Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP. Nature 503, 422–426 (2013).
Kirisako, T. et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 25, 4877–4887 (2006).
Chaugule, V.K. et al. Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J. 30, 2853–2867 (2011).
Burchell, L., Chaugule, V.K. & Walden, H. Small, N-terminal tags activate Parkin E3 ubiquitin ligase activity by disrupting its autoinhibited conformation. PLoS ONE 7, e34748 (2012).
Cookson, M.R. Parkinsonism due to mutations in PINK1, parkin, and DJ-1 and oxidative stress and mitochondrial pathways. Cold Spring Harb. Perspect. Med. 2, a009415 (2012).
Hofmann, K. & Bucher, P. The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem. Sci. 21, 172–173 (1996).
Walden, H. & Martinez-Torres, R.J. Regulation of Parkin E3 ubiquitin ligase activity. Cell. Mol. Life Sci. 69, 3053–3067 (2012).
Komander, D., Clague, M.J. & Urbe, S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 (2009).
Smit, J.J. et al. The E3 ligase HOIP specifies linear ubiquitin chain assembly through its RING-IBR-RING domain and the unique LDD extension. EMBO J. 31, 3833–3844 (2012).
Stieglitz, B., Morris-Davies, A.C., Koliopoulos, M.G., Christodoulou, E. & Rittinger, K. LUBAC synthesizes linear ubiquitin chains via a thioester intermediate. EMBO Rep. 13, 840–846 (2012).
Smit, J.J. et al. Target specificity of the E3 ligase LUBAC for ubiquitin and NEMO relies on different minimal requirements. J. Biol. Chem. 288, 31728–31737 (2013).
Vijay-Kumar, S., Bugg, C.E. & Cook, W.J. Structure of ubiquitin refined at 1.8 Å resolution. J. Mol. Biol. 194, 531–544 (1987).
Komander, D. et al. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 10, 466–473 (2009).
Datta, A.B., Hura, G.L. & Wolberger, C. The structure and conformation of Lys63-linked tetraubiquitin. J. Mol. Biol. 392, 1117–1124 (2009).
Cook, W.J., Jeffrey, L.C., Carson, M., Chen, Z. & Pickart, C.M. Structure of a diubiquitin conjugate and a model for interaction with ubiquitin conjugating enzyme (E2). J. Biol. Chem. 267, 16467–16471 (1992).
Wu, G. et al. Structure of a β-TrCP1-Skp1-β-catenin complex. Mol. Cell 11, 1445–1456 (2003).
Zheng, N. et al. Structure of the Cul1–Rbx1–Skp1–F boxSkp2 SCF ubiquitin ligase complex. Nature 416, 703–709 (2002).
Duda, D.M. et al. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134, 995–1006 (2008).
Acknowledgements
We thank R. Hay, A. Plechanovová and B. Schulman for providing coordinates of modeled complexes. C.W. acknowledges grant support from the US National Institutes of Health (GM095822) and National Science Foundation (MCB0920082).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Berndsen, C., Wolberger, C. New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol 21, 301–307 (2014). https://doi.org/10.1038/nsmb.2780
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2780
This article is cited by
-
The association of RBX1 and BAMBI gene expression with oocyte maturation in PCOS women
BMC Medical Genomics (2024)
-
Structural snapshots along K48-linked ubiquitin chain formation by the HECT E3 UBR5
Nature Chemical Biology (2024)
-
O-GlcNAcylation of E3 ubiquitin ligase SKP2 promotes hepatocellular carcinoma proliferation
Oncogene (2024)
-
SPHK1 potentiates colorectal cancer progression and metastasis via regulating autophagy mediated by TRAF6-induced ULK1 ubiquitination
Cancer Gene Therapy (2024)
-
Elevated expression of WSB2 degrades p53 and activates the IGFBP3-AKT-mTOR-dependent pathway to drive hepatocellular carcinoma
Experimental & Molecular Medicine (2024)