Abstract
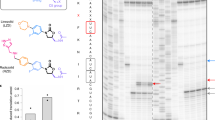
Lysine riboswitches are bacterial RNA structures that sense the concentration of lysine and regulate the expression of lysine biosynthesis and transport genes. Members of this riboswitch class are found in the 5′ untranslated region of messenger RNAs, where they form highly selective receptors for lysine. Lysine binding to the receptor stabilizes an mRNA tertiary structure that, in most cases, causes transcription termination before the adjacent open reading frame can be expressed. A lysine riboswitch conceivably could be targeted for antibacterial therapy by designing new compounds that bind the riboswitch and suppress lysine biosynthesis and transport genes. As a test of this strategy, we have identified several lysine analogs that bind to riboswitches in vitro and inhibit Bacillus subtilis growth, probably through a mechanism of riboswitch-mediated repression of lysine biosynthesis. These results indicate that riboswitches could serve as new classes of antibacterial drug targets.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Wolfson, W. Holding back the tide of antibiotic resistance. Chem. Biol. 13, 1–3 (2006).
D'Costa, V.M., McGrann, K.M., Hughes, D.W. & Wright, G.D. Sampling the antibiotic resistome. Science 311, 374–377 (2006).
Winkler, W.C. & Breaker, R.R. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 59, 487–517 (2005).
Sudarsan, N., Wickiser, J.K., Nakamura, S., Ebert, M.S. & Breaker, R.R. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 17, 2688–2697 (2003).
Grundy, F., Lehman, S. & Henkin, T. The L box regulon: lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc. Natl. Acad. Sci. USA 100, 12057–12062 (2003).
Rodionov, D.A., Vitreschak, A.G., Mironov, A.A. & Gelfand, M.S. Regulation of lysine biosynthesis and transport genes in bacteria: yet another RNA riboswitch? Nucleic Acids Res. 31, 6748–6757 (2003).
Wimberly, B., Varani, G. & Tinoco, I.J. The conformation of loop E of eukaryotic 5S ribosomal RNA. Biochemistry 32, 1078–1087 (1993).
Klein, D.J., Schmeing, T.M., Moore, P.B. & Steitz, T.A. The kink-turn: a new RNA secondary structure motif. EMBO J. 20, 4214–4221 (2001).
Hutton, C.A., Southwood, T.J. & Turner, J.J. Inhibitors of lysine biosynthesis as antibacterial agents. Mini Rev. Med. Chem. 3, 115–127 (2003).
Bugg, T.D. & Brandish, P.E. From peptidoglycan to glycoproteins: common features of lipid-linked oligosaccharide biosynthesis. FEMS Microbiol. Lett. 119, 255–262 (1994).
Sudarsan, N., Cohen-Chalamish, S., Nakamura, S., Emilsson, G.M. & Breaker, R.R. Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chem. Biol. 12, 1325–1335 (2005).
Woolley, D.W. & White, A.G.C. Selective reversible inhibition of microbial growth with pyrithiamine. J. Exp. Med. 78, 489–497 (1943).
Shiota, T., Folk, J.E. & Tietze, F. Inhibition of lysine utilization in bacteria by S-(beta-aminoethyl) cysteine and its reversal by lysine peptides. Arch. Biochem. Biophys. 77, 372–377 (1958).
Lu, Y., Chen, N.Y. & Paulus, H. Identification of aecA mutations in Bacillus subtilis as nucleotide substitutions in the untranslated leader region of the aspartokinase II operon. J. Gen. Microbiol. 137, 1135–1143 (1991).
Patte, J.C., Akrim, M. & Mejean, V. The leader sequence of the Escherichia coli lysC gene is involved in the regulation of LysC synthesis. FEMS Microbiol. Lett. 169, 165–170 (1998).
Soukup, G.A. & Breaker, R.R. Relationship between internucleotide linkage geometry and the stability of RNA. RNA 5, 1308–1325 (1999).
Matsumoto, N. Isolation and identification of S-2-aminoethyl-L-cysteine from Rozites caperta and 2-amino-3-butenoic acid from Rhodophyllus crassipes and their antibacterial activity. J. Med. Soc. Toho, Japan 31, 249–264 (1984).
Wickiser, J.K., Winkler, W.C., Breaker, R.R. & Crothers, D.M. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell 18, 49–60 (2005).
Wickiser, J.K., Cheah, M.T., Breaker, R.R. & Crothers, D.M. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry 44, 13404–13414 (2005).
Zhang, J.J., Hu, F.M., Chen, N.Y. & Paulus, H. Comparison of the three aspartokinase isozymes in Bacillus subtilis Marburg and 168. J. Bacteriol. 172, 701–708 (1990).
Higgins, C.F. & Gibson, M.M. Peptide transport in bacteria. Methods Enzymol. 125, 365–377 (1986).
Zhang, R., Ou, H.-Y. & Zhang, C.-T. DEG, a database of essential genes. Nucleic Acids Res. 32, D271–D272 (2004).
Blount, K.F. & Breaker, R.R. Riboswitches as antibacterial drug targets. Nat. Biotechnol. (in the press).
Anagnostopoulos, C. & Spizizen, J. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81, 741–746 (1961).
Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically: Approved Standard: M7–A7 (National Committee for Clinical Laboratory Standards, Wayne, Pennsylvania, USA, 2006).
Lazazzera, B.A., Solomon, J.M. & Grossman, A.D. An exported peptide functions intracellularly to contribute to cell density signalling in B. subtilis. Cell 89, 917–925 (1997).
Kawasaki, T. & Nose, Y. Thiamine regulatory mutants in Escherichia coli. J. Biochem. 65, 417–425 (1969).
Landick, R., Wang, D. & Chan, C.L. Quantitative analysis of transcriptional pausing by Escherichi coli RNA polymerase: His leader pause site as a paradigm. Methods Enzymol. 274, 334–353 (1996).
Acknowledgements
The authors wish to thank J. Barrick for determining the consensus sequence and secondary structure for the lysine riboswitch. This work was supported by the US National Institutes of Health and the US Defense Advanced Research Projects Agency (DARPA). Riboswitch discovery science is also supported by the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
K.F.B. and R.R.B. coordinated project plans, execution and preparation of the manuscript. J.L. conducted chemical design and synthesis work. J.X.W. carried out in vitro biochemical analyses of the compounds. J.X.W. and J.L. conducted in vivo expression reporter assays. K.F.B., J.X.W. and N.S. conducted experiments to assess the antibiotic action of the compounds.
Corresponding author
Ethics declarations
Competing interests
K.F.B. and R.R.B. are cofounders of BioRelix, a company that is pursuing intellectual property related to the use of riboswitches as drug targets.
Supplementary information
Supplementary Fig. 1
The consensus and secondary structure for lysine riboswitches. (PDF 413 kb)
Supplementary Fig. 2
The pathways for lysine biosynthesis and import in bacteria. (PDF 772 kb)
Supplementary Fig. 3
Growth of Bacillus subtilis lysine auxotroph strain 1A40 with supplementation of minimal media with various lysine analogs. (PDF 251 kb)
Supplementary Table 1
The distribution of lysine riboswitches among bacterial species. (PDF 554 kb)
Supplementary Table 2
Sporulation efficiency of B. subtilis when grown in the presence of various lysine analogs. (PDF 329 kb)
Rights and permissions
About this article
Cite this article
Blount, K., Wang, J., Lim, J. et al. Antibacterial lysine analogs that target lysine riboswitches. Nat Chem Biol 3, 44–49 (2007). https://doi.org/10.1038/nchembio842
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio842