Abstract
The epidermal growth factor receptor (EGFR) is amplified in approximately 40% of glioblastomas making it a compelling molecular target for therapy. Before starting a therapy targeting the EGFR pathway, accurate determining of EGFR status is a prerequisite. We evaluated the reliability of the novel automated silver enhanced in situ hybridization for the detection of EGFR gene amplification in human glioblastomas. EGFR-amplification status was assessed in 93 cases of glioblastoma by silver enhanced in situ hybridization and compared with results of fluorescence in situ hybridization and immunohistochemistry. In a second cohort, silver enhanced in situ hybridization status was correlated with EGFR gene expression data. The EGFR gene was amplified in 25/90 tumours (28%) by silver enhanced in situ hybridization, and in 28/93 tumours (30%) by fluorescence in situ hybridization. The concordance rate for silver enhanced in situ hybridization and fluorescence in situ hybridization was 98%. Two glioblastomas were scored as being amplified by fluorescence in situ hybridization but not by silver enhanced in situ hybridization. Polymerase chain reaction-based EGFR-amplification data were highly correlated with EGFR silver enhanced in situ hybridization. Altogether, 81 of 91 cases (89%) showed positivity for EGFR expression by immunohistochemistry. Although EGFR protein over expression was associated with gene amplification (r=0.40, P<0.001), there were 29 of 91 cases that showed a high EGFR protein level and no EGFR amplification by fluorescence in situ hybridization. The high concordance rate of silver enhanced in situ hybridization and fluorescence in situ hybridization for the detection of EGFR amplification in paraffin-embedded glioblastomas samples demonstrates that silver enhanced in situ hybridization is a valid and attractive alternative to fluorescence in situ hybridization. Silver enhanced in situ hybridization combines the advantages of bright field microscopy with fully automated analysis in a cost-effective way thereby emphasizing its use for routine application in surgical pathology.
Similar content being viewed by others
Introduction
Glioblastoma is the most aggressive form of adult primary astroglial tumour with median survival of <12 months.1 One reason for the dismal prognosis is the lack of therapeutic agents to eliminate remaining diffusely infiltrating tumour cells after surgery. Molecular profiling of genetic alterations in glioblastoma enables classifying tumours into categories amenable to novel targeted treatment modalities. One of these therapeutic targets is the epidermal growth factor receptor (EGFR). EGFR is a transmembrane glycoprotein that belongs to the erb tyrosine kinase receptor family including EGFR (HER1 or erb-B1), erbB-2 (HER2), erbB-3 (HER3) and erbB-4 (HER4). EGFR overexpression has been found in approximately 40% of patients with glioblastoma.2 It has been reported that EGFR amplification is a negative prognostic factor.3 In contrast, there are publications opposing a prognostic relevance.4, 5 Currently, the possibility of EGFR-targeted therapies has initiated the development of a variety of agents directed either towards the extracellular ligand-binding domain or the intracellular tyrosine kinase domain including the monoclonal antibody Erbitux (IMC-C225, ImClone/Bristol Myers Squibb), and the EGFR tyrosine kinase inhibitors Gefitinib (ZD1839, Iressa, AtraZeneca) and Erlotinib (OSI-774, OSI/Genentech/Roche).6 Although these EGFR-targeted therapies are already in phase III clinical trials, it still remains to be determined, which subgroup of patients will most likely benefit.7 Several methods are available for testing EGFR status, including Southern blot analysis, polymerase chain reaction, fluorescence in situ hybridization and chromogenic in situ hybridization. Disadvantages of the first two methods are the lack of consideration of cell morphology. The advantage of both chromogenic in situ hybridization and fluorescence in situ hybridization is the ability to illustrate gene alterations on a tumour tissue section, which enables the comparison of tissue morphology with gene abnormality. Fluorescence in situ hybridization is globally accepted and used as a standard method to determine HER2 status in breast cancer.8, 9 Disadvantages of fluorescence in situ hybridization testing are an expensive fluorescent microscope, extensive user training and rapid loss of gene signals at room temperature, thereby hindering convenient slide storage. Currently, fluorescence in situ hybridization is the gold standard. However, it has been recently reported that chromogenic in situ hybridization is an attractive alternative to fluorescence in situ hybridization.10 Chromogenic in situ hybridization is based on the same principle as fluorescence in situ hybridization but uses a chromogen to detect gene signals. Even though chromogenic in situ hybridization can deal with some of the mentioned problems it has not yet found wide acceptance in routine laboratories. Chromogenic in situ hybridization requires manual processing, overnight hybridization, and two hybridizations, one for the gene of interest and one for detection of chromosomal copy number. Furthermore, due to signal weakness, it can be difficult to discriminate gene signals. An alternative may be a novel fully automated silver enhanced in situ hybridization protocol. This rapid method yields a stable and discrete chromogenic reaction product, which is analysed by standard light microscopy. Moreover, the manual steps that have to be performed for silver enhanced in situ hybridization are reduced to minimum. However, technical and clinical validation is required before silver enhanced in situ hybridization might be accepted as a standard practice. This study compares the different methods and validates silver enhanced in situ hybridization as a method for assessing EGFR gene amplification in glioblastoma tumour samples.
Materials and methods
Tumour Materials
A series of 95 randomly selected glioblastomas, which had been diagnosed between 2006 and 2008, was collected from the tumour bank archive of the Department of Neuropathology, University Hospital Heidelberg. The study was approved by the local ethics committee. Before paraffin embedding, tumour tissues were fixed in 4% neutral buffered formalin for 5–36 h. The slides were cut to 3-μm thick sections and then processed in parallel for fluorescence in situ hybridization (LSI EGFR SpectrumOrange/CEP 7 SpectrumGreen Probe, Vysis/Abbott), silver enhanced in situ hybridization (INFORM EGFR Probe and INFORM chromosome 7 probe, Ventana Medical Systems) and immunohistochemistry (CONFIRM anti-EGFR (3C6), Ventana). The mean patient age was 57 years and 60% (57/95) of the patients were male. From the 95 glioblastomas, 92 samples had been derived from gross total resections, two samples had been biopsied stereotactically, and one was frozen section tissue. All cases were diagnosed according to the current classification criteria of the World Health Organization.11 In a second step, a group of patients (n=26), with known EGFR-amplification status determined by a semiquantitative polymerase chain reaction-based assay was screened for EGFR amplification by silver enhanced in situ hybridization. These samples were kindly provided by the Brain Tumour Reference Centre located at the Institute of Neuropathology, University of Bonn (Director Prof Dr Pietsch).
Immunohistochemistry
The sections were immunostained with CONFIRM anti-EGFR primary antibody (clone 3C6, Ventana Medical Systems, Tucson, AZ) on the BENCHMARK XT automated stainer (Ventana). Immunohistochemistry was performed following the manufacturer's recommendations. Immunohistochemical semiquantitative assessment of EGFR protein expression was based on the fraction of stained cells and the intensity of the membranous staining. Regardless of a possible cytoplasmic staining, the membranous staining for EGFR was specified on a scale of 0 to 3 as follows: 0=negative (no staining of glioblastoma cells); 1=weak staining; 2=moderate staining; 3=strong staining. EGFR-positive staining was defined as any immunohistochemical staining of tumour cell membranes above background level regardless of complete or incomplete circumferential staining. Cytoplasmic staining without membranous staining was scored as negative. Same slides of all tissue samples were scored by two primary independent observers according to the Dako EGFR pharmDx Interpretation Manual. Additionally, the scoring algorithm according to Hirsch et al12 was used. This algorithm is based on multiplication of the percent fraction of stained cells with the staining intensity of tumour cells, thereby resulting in an immunohistochemistry score. Scores of 0–149 were considered negative/no expression of EGFR, and scores of 150–300 were considered positive/high expression of EGFR. Endothelial cells within the glioblastoma tumour samples served as an internal negative control for the EGFR staining procedures. Additional negative controls were obtained by the omission of the primary antibody. In each immunohistochemistry run one glioblastoma sample with known strong staining reaction was included as a positive control.
Silver Enhanced In Situ Hybridization
Automated silver enhanced in situ hybridization was performed according to the manufacturer's protocol for the INFORM EGFR probe and chromosome 7 probe. Both probes were labelled with dinitrophenol and optimally formulated for use with ultraVIEW SISH Detection Kit and accessory reagents on BENCHMARK series of automated slide stainers (Ventana). The EGFR DNA probe was denatured at 95°C for 12 min; hybridization was performed at 52°C for 3 h for EGFR and 52°C for 2 h for centromere 7 probe. After washing the EGFR and chromosome 7 dinitrophenol-labelled probes were visualized using the rabbit anti-dinitrophenol antibody and the ultraVIEW silver SISH Detection Kit. For details see Dietel et al.13 The silver precipitation was deposited in the nuclei and a single copy of the EGFR gene or the centromere 7 was visualized as a black dot. After counterstaining 60 tumour cells, nuclei were evaluated in three different areas (20 nuclei each).
Fluorescence In Situ Hybridization
For fluorescence in situ hybridization, consecutive sections from the same blocks used for silver enhanced in situ hybridization and immunohistochemistry were cut and mounted on SuperFrost +/+ slides. Deparaffinising, pre-treatment and protease digestion procedures were performed according to the DAKO Histology FISH Accessory Kit. Probe mixes were hybridized at 37°C between 14 and 24 h, washed and counterstained with DAPI. A minimum of 60 tumour cell nuclei in three different areas (20 nuclei in each area) were scored using LEICA epi-fluorescence microscopes equipped with × 100 oil immersion objective and DAPI/Spectrum Green/Orange single filters.
Fluorescence In Situ Hybridization and Silver Enhanced In Situ Hybridization Scoring Criteria and Algorithms
The scoring procedure for EGFR fluorescence in situ hybridization and silver enhanced in situ hybridization was performed similar to the INFORM Her2 DNA probe (for details see http://www.her2sish.com). The slide was screened for a suitable tumour target area. For fluorescence in situ hybridization: EGFR gene and centromere 7 signals were counted in 60 cells in three different areas (20 nuclei in each area) and the sum was built for both probes. Then the ratio between EGFR gene signals and centromere 7 signals was calculated. For silver enhanced in situ hybridization: First, the EGFR gene signals were counted in 20 cells. Then the same area was located in the centromere 7 stained slide and again 20 cells were counted. This was repeated two times until 60 cells were examined. Finally the ratio between EGFR gene signals and centromere 7 signals was calculated in analogy to fluorescence in situ hybridization. To determine the cut-off value for EGFR amplification, the EGFR gene status of normal human brain tissue samples was examined. The normalized ratio (EGFR Gene number/centromere 7 number) varied between 0.7 and 1.8 (data not shown). As previously reported cut-off level of ≥2 has been proven to be useful for EGFR gene status assessment.3, 14 We decided to perform the analyses in analogy to the Her2 guidelines of the American Society of Clinical Oncology/College of American Pathologists. This scoring system includes an equivocal/borderline group. EGFR gene amplification status was classified applying the following criteria:
A EGFR/centromere 7 ratio less than 1.8 was defined as negative for EGFR gene amplification. A EGFR/centromere 7 ratio between 1.8 and 2.2 was defined as ambiguous (not determinable) for EGFR gene amplification. A EGFR/centromere 7 ratio greater than 2.2 was defined as positive for EGFR gene amplification.
Semiquantitative Polymerase Chain Reaction-Based Assay
Semiquantitative polymerase chain reaction-based assay was performed to determine the ratio between EGFR and a control gene, Interferon-γ (IFN-γ), as described earlier.15 Amplification was defined as a ratio of EGFR and IFN-γ higher than 4.16. The ratio of 2.09–4.16 was defined to be ambiguous for EGFR and a EGFR/IFN-γ ratio of <2.09 was defined as normal/negative for EGFR. For comparison with silver enhanced in situ hybridization, 10 semiquantitative polymerase chain reaction-based assay amplified, 4 ambiguous and 12 nonamplified samples were screened.
Statistics
Statistical analysis was done with SAS version 9.1WIN. Concordance between silver enhanced in situ hybridization and fluorescence in situ hybridization methods and between silver enhanced in situ hybridization and semiquantitative polymerase chain reaction-based assay were determined using the Pearson's correlation coefficient. The kappa statistics were calculated with Cohen's kappa.
Results
Failure Rate
The failure rate for both methods was very low. Fluorescence in situ hybridization failed in 2 of 95 cases (2%), silver enhanced in situ hybridization showed no signals in 3 of 95 cases (3%).
EGFR Gene Amplification Pattern
In total, 86 of 90 dual evaluable (for fluorescence in situ hybridization and silver enhanced in situ hybridization informative) cases showed a homogeneous EGFR-amplification pattern with rather constant gene signals in all tumour cells. Four cases illustrated a heterogeneous expression pattern with amplified gene signals in some interspersed tumour cells. This observation was independent from the applied method (Figure 1a and b). In EGFR amplified cases, the gene ratio was rather high (mean value=13.7). The EGFR gene signals in amplified cases reached numbers up to 60 in each cell.
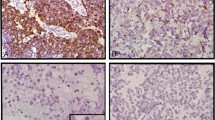
Amplified single cells by fluorescence in situ hybridization and silver enhanced in situ hybridization. (a) Fluorescence in situ hybridization: in this example, the cell in the centre has two green signals (centromere 7 probe) with clusters of red signals (EGFR gene locus), while all other cells have two green and two red signals. (b) Silver enhanced in situ hybridization: in this example, the cell in the centre has clusters of black signals (EGFR gene locus), while the surrounding tumour cells show only 1–3 signals. Magnification: (a) 1000 × oil objective; (b) 630 × air objective.
EGFR-Amplification Status by Silver Enhanced In Situ Hybridization and Fluorescence In Situ Hybridization
From 95 cases entered in this study 90 cases could be evaluated for both, silver enhanced in situ hybridization and fluorescence in situ hybridization. Three cases were excluded due to the lack of signals (two with no fluorescence in situ hybridization/silver enhanced in situ hybridization signals and one with absent silver enhanced in situ hybridization signals). One of the excluded cases was the frozen tissue section. Two cases had to be excluded due to small sample size without any detectable tumour tissue on consecutive sections. In all, 93 of 95 samples were informative for fluorescence in situ hybridization, with an amplification rate of 30% (Figure 2a and b). No case showed an ambiguous EGFR gene status. In comparison, 90 of 95 samples were informative for silver enhanced in situ hybridization, with an amplification rate of 28% (Figure 2c and d). One case showed an EGFR gene ratio of 1.87 and was consequently termed ‘ambiguous for EGFR gene amplification’.
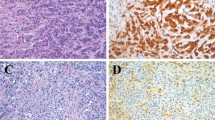
Representative images of fluorescence in situ hybridization and silver enhanced in situ hybridization EGFR in two glioblastoma tumour samples. In fluorescence in situ hybridization (a,b), green signals represent the centromere of chromosome 7; red signals represent the EGFR gene locus on chromosome 7p12. In silver enhanced in situ hybridization (c,d), black signals represent the EGFR gene locus. (a) and (c) are EGFR not amplified cases; (b) and (d) are EGFR amplified cases. In (d), endothelial cells serve as internal negative control (arrow). Magnification: (a,b) 1000 × oil objective; (c,d): 630 × air objective.
Discrepancy between the fluorescence in situ hybridization and silver enhanced in situ hybridization occurred in only 2 of 90 cases, both positive for fluorescence in situ hybridization and negative for silver enhanced in situ hybridization. A case-by-case analysis revealed the following:
Case 1 (151/07b): The silver enhanced in situ hybridization ratio was negative for EGFR gene amplification (ratio of 1.54) The fluorescence in situ hybridization yielded a positive result (ratio of 2.39). In both methods, the EGFR gene signals were increased (fluorescence in situ hybridization=4.90; silver enhanced in situ hybridization=7.45) but the number of centromere signals was different (fluorescence in situ hybridization=2.05; silver enhanced in situ hybridization=4.83). It is therefore assumed that this discrepancy was caused by a polysomy of chromosome 7, which had not been recognized in the fluorescence in situ hybridization slide. This might have been due to weak green (centromere 7) fluorescence in situ hybridization signals, which were presumably overlooked by the observer.
Case 2 (285/08a): The silver enhanced in situ hybridization ratio was 1.78 (negative for EGFR gene amplification but very close to the ambiguous category). The fluorescence in situ hybridization yielded a clear positive result with a ratio of 8.49. This discrepancy had been caused by heterogeneity of the signal distribution. Both methods identified amplified single cells, but the percentage of these cells differed in variable areas of the slide. This heterogeneity might have lead to a drifting result.
As a conclusion, the concordance rate for silver enhanced in situ hybridization and fluorescence in situ hybridization set at cut-off point 2.2 was 98% (κ=0.95, 95% CI [0.87,1.0]). The Pearson correlation coefficient for the detailed data of silver enhanced in situ hybridization and fluorescence in situ hybridization was 0.85 (P<0.0001).
Immunohistochemistry
Altogether, 10 of 91 glioblastomas (11%) were scored negatively (staining intensity 0) on EGFR immunohistochemistry. From 91 glioblastoma 81 cases (89%) revealed a weak, medium or strong (staining intensity 1–3) EGFR protein expression. Applying the immunohistochemistry score (0–149 negative, 150–300 positive) 39 of 91 glioblastomas (43%) were positive.
Correlation of Silver Enhanced In Situ Hybridization with Immunohistochemistry
In all, 89 cases were evaluable for both, immunohistochemistry and silver enhanced in situ hybridization. All cases (27/91) with EGFR gene amplification were also tested positive with immunohistochemistry. In all, 26 of the 27 glioblastomas with amplified EGFR showed staining intensity of three and one tumour demonstrated a weak-staining intensity (Table 1). In contrast, 29 of 64 cases without amplification exhibited high EGFR expression (staining intensity 3). EGFR protein expression was significantly associated with gene amplification. The correlation between staining intensity and fluorescence in situ hybridization was r=0.402 (P<0.0001). The correlation for immunohistochemistry score (staining intensity × percentage stained cells) and fluorescence in situ hybridization was r=0.570 (P<0.0001).
EGFR Expression by Polymerase Chain Reaction-Based Assay and Silver Enhanced In Situ Hybridization
EGFR DNA expression by a polymerase chain reaction-based assay was highly associated with EGFR silver enhanced in situ hybridization (Table 2). All semiquantitative polymerase chain reaction-based assay positive samples (n=10) were highly amplified with silver enhanced in situ hybridization (mean ratio=10.7). All semiquantitative polymerase chain reaction-based assay negative cases (n=12) were not amplified with silver enhanced in situ hybridization (mean ratio=1.03). The correlation for semiquantitative polymerase chain reaction-based assay and silver enhanced in situ hybridization was r=0.79 (P<0.0001). In the ambiguous semiquantitative polymerase chain reaction-based assay group (n=4) all silver enhanced in situ hybridization results were negative. The mean value for EGFR gene ratio in this group was slightly higher than in the nonamplified group (2.95 signals per cell vs 2.68 signals per cell) but this was statistically not significant (P=0.49). The mean silver enhanced in situ hybridization ratio differences between the semiquantitative polymerase chain reaction-based assay positive and negative group were highly significant (P<0.001) but not between ambiguous cases and nonamplified cases (P=0.57) (Figure 3).
Discussion
The number of clinical studies using EGFR-based therapies in addition to the conventional radio-chemotherapy is growing constantly. For treatment of glioblastoma, there are several clinical protocols including Erlotinib, Gefitinib or Cetuximab in addition to the standard treatment regime.16, 17, 18, 19, 20 Accurate assessment of EGFR status is a prerequisite to identify subgroups of patients, which will benefit most likely from this treatment. Mellinghoff et al21 supported this concept by demonstrating that glioblastomas with coexpression of EGFRvIII (a mutant form of EGFR) and PTEN showed better clinical response when treated with erlotinib or gefitinib. Thus, need for rapid and reliable tests to determine EGFR status seems to be likely. The current genomic tests usually include fluorescence in situ hybridization. Although there are several articles demonstrating chromogenic in situ hybridization as a viable alternative, fluorescence in situ hybridization is still the gold standard and widely used.10, 22, 23, 24
In this study, we compared the novel method silver enhanced in situ hybridization with fluorescence in situ hybridization for the assessment of EGFR amplification in a large series of 90 human glioblastomas. Both methods showed low failure rates (fluorescence in situ hybridization 2%; silver enhanced in situ hybridization 3%). The incidence of EGFR amplification detected with silver enhanced in situ hybridization was 28% (fluorescence in situ hybridization 30%), according to previous literature.25, 26 The concordance rate between fluorescence in situ hybridization and silver enhanced in situ hybridization was 98% (κ=0.95, 95% CI), demonstrating that silver enhanced in situ hybridization is a valid alternative testing method for the determination of EGFR-amplification status in glioblastoma. These results are consistent with previous studies reporting on concordance rates for HER2 silver enhanced in situ hybridization vs. fluorescence in situ hybridization of 96% (κ=0.75, 95% CI) and 99%.13, 27
Additionally, we assessed the EGFR gene status with a semiquantitative polymerase chain reaction-based assay and compared the results with silver enhanced in situ hybridization. Semiquantitative polymerase chain reaction-based assay results concurred excellent with silver enhanced in situ hybridization findings (P<0.001) and showed high association with EGFR silver enhanced in situ hybridization. This result is consistent with a previous study illustrating a good correlation of polymerase chain reaction tested EGFR gene doses with fluorescence in situ hybridization.28
All cases that yielded ambiguous results with semiquantitative polymerase chain reaction-based assay (n=4) showed negative EGFR amplification with silver enhanced in situ hybridization. The slightly higher EGFR gene ratio in this group might have been caused by trisomy 7 as often seen in glioblastoma.29 However, this result was statistically not significant, due to the small number.
We demonstrated a significant association of EGFR protein overexpression with gene amplification according to previous literature.26, 30, 31 Although EGFR protein overexpression was highly associated with gene amplification, there were 29 of 64 cases that showed a high EGFR protein level and no EGFR amplification by fluorescence in situ hybridization or silver enhanced in situ hybridization. All cases with gene amplification showed immunohistochemistry positivity. The observation that the frequency of elevated EGFR immunoreactivity far exceeded that of EGFR gene amplification is consistent with prior results in adult high-grade gliomas.32, 33
Although fluorescence in situ hybridization is globally accepted as a standard method for determining EGFR status, pathologists have been reluctant to adopt fluorescence in situ hybridization for routine clinical laboratory testing. The reasons for this include the technical challenges associated with fluorescence in situ hybridization assays,34 the expenses, the required time for interpretation35 and the relative lack of community-based experience with this technology. Although the material costs are slightly higher for silver enhanced in situ hybridization than for fluorescence in situ hybridization (approximately costs per each test for silver enhanced in situ hybridization: 200$, for fluorescence in situ hybridization: 130$), the technician working time for silver enhanced in situ hybridization is much shorter compared with fluorescence in situ hybridization (pure working time for silver enhanced in situ: 20 min, for fluorescence in situ hybridization 3 h). In addition, fluorescence in situ hybridization requires access to a cost-intensive fluorescent microscope. However, the fluorescence in situ hybridization method comprises one advantage: one hybridization is sufficient for diagnostic purpose compared with SISH, which needs to be performed on two slides per case (one for the EGFR probe and one for chromosome 7).
The excellent concordance rate of fluorescence in situ hybridization and silver enhanced in situ hybridization proves equivalent diagnostic value. Silver enhanced in situ hybridization offers several advantages. It is performed as a 6-h automated protocol and can be completed within one working day. Without the need of specialized microscope equipments, the pathologist can easily integrate the tests in the routine day work. It allows simultaneous demonstration of interesting results for teaching purposes by using routinely available multiheaded microscopes. Silver enhanced in situ hybridization generates a stable chromogenic reaction product, which allows the permanent storage of the slides. This point is particularly important when the tests are used in routine diagnostics when proper documentation and archival storage is required for certification and quality management. Furthermore, the possibility to work with low microscopic magnifications allows a much easier handling of difficulties due to tumour heterogeneity. Most of the silver enhanced in situ hybridization signals are already recognizable at 200 × magnification (for fluorescence in situ hybridization a magnification of 400 × up to 1000 × is often necessary) and the intensity and sharpness of signals in silver enhanced in situ hybridization is much better than in chromogenic in situ hybridization.
Many pathologists are experienced in evaluating silver enhanced in situ hybridization-like peroxidase-based immunostainings but not familiar with fluorescence microscopy. Therefore, the time and effort needed for aquisition of silver enhanced in situ hybridization is presumably shorter than for fluorescence in situ hybridization.
In conclusion, the present results convincingly demonstrate that silver enhanced in situ hybridization is a powerful tool for the detection of EGFR amplification in archival paraffin-embedded glioblastoma samples. The universal need to standardize, simplify and accelerate the diagnostic process renders silver enhanced in situ hybridization a cost-effective alternative to fluorescence in situ hybridization.
References
Krex D, Klink B, Hartmann C, et al. Long-term survival with glioblastoma multiforme. Brain 2007;130:2596–2606.
Frederick L, Wang XY, Eley G, et al. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res 2000;60:1383–1387.
Shinojima N, Tada K, Shiraishi S, et al. Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res 2003;63:6962–6970.
Donato V, Papaleo A, Castrichino A, et al. Prognostic implication of clinical and pathologic features in patients with glioblastoma multiforme treated with concomitant radiation plus temozolomide. Tumori 2007;93:248–256.
Viana-Pereira M, Lopes JM, Little S, et al. Analysis of EGFR overexpression, EGFR gene amplification and the EGFRvIII mutation in Portuguese high-grade gliomas. Anticancer Res 2008;28:913–920.
Baselga J . Why the epidermal growth factor receptor? The rationale for cancer therapy. Oncologist 2002;7 (Suppl 4):2–8.
Dancey JE, Freidlin B . Targeting epidermal growth factor receptor--are we missing the mark? Lancet 2003;362:62–64.
Clinical laboratory assays for HER-2/neu amplification and overexpression: quality assurance, standardization, and proficiency testing. Arch Pathol Lab Med 2002;126:803–808.
Bilous M, Dowsett M, Hanna W, et al. Current perspectives on HER2 testing: a review of national testing guidelines. Mod Pathol 2003;16:173–182.
Fischer I, de la Cruz C, Rivera AL, et al. Utility of chromogenic in situ hybridization (CISH) for detection of EGFR amplification in glioblastoma: comparison with fluorescence in situ hybridization (FISH). Diagn Mol Pathol 2008;17:227–230.
Kleihues P, Sobin LH . World Health Organization classification of tumors. Cancer 2000;88:2887.
Hirsch FR, Varella-Garcia M, Bunn Jr PA, et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol 2003;21:3798–3807.
Dietel M, Ellis IO, Hofler H, et al. Comparison of automated silver enhanced in situ hybridisation (SISH) and fluorescence ISH (FISH) for the validation of HER2 gene status in breast carcinoma according to the guidelines of the American Society of Clinical Oncology and the College of American Pathologists. Virchows Arch 2007;451:19–25.
Korshunov A, Sycheva R, Golanov A . Molecular stratification of diagnostically challenging high-grade gliomas composed of small cells: the utility of fluorescence in situ hybridization. Clin Cancer Res 2004;10:7820–7826.
Waha A, Rollbrocker B, Wiestler OD, et al. A polymerase chain reaction-based assay for the rapid detection of gene amplification in human tumors. Diagn Mol Pathol 1996;5:147–150.
Combs SE, Heeger S, Haselmann R, et al. Treatment of primary glioblastoma multiforme with cetuximab, radiotherapy and temozolomide (GERT)--phase I/II trial: study protocol. BMC Cancer 2006;6:133.
Brown PD, Krishnan S, Sarkaria JN, et al. Phase I/II trial of erlotinib and temozolomide with radiation therapy in the treatment of newly diagnosed glioblastoma multiforme: North Central Cancer Treatment Group Study N0177. J Clin Oncol 2008;26:5603–5609.
de Groot JF, Gilbert MR, Aldape K, et al. Phase II study of carboplatin and erlotinib (Tarceva, OSI-774) in patients with recurrent glioblastoma. J Neurooncol 2008;90:89–97.
Franceschi E, Cavallo G, Lonardi S, et al. Gefitinib in patients with progressive high-grade gliomas: a multicentre phase II study by Gruppo Italiano Cooperativo di Neuro-Oncologia (GICNO). Br J Cancer 2007;96:1047–1051.
Doherty L, Gigas DC, Kesari S, et al. Pilot study of the combination of EGFR and mTOR inhibitors in recurrent malignant gliomas. Neurology 2006;67:156–158.
Mellinghoff IK, Wang MY, Vivanco I, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med 2005;353:2012–2024.
Marquez A, Wu R, Zhao J, et al. Evaluation of epidermal growth factor receptor (EGFR) by chromogenic in situ hybridization (CISH) and immunohistochemistry (IHC) in archival gliomas using bright-field microscopy. Diagn Mol Pathol 2004;13:1–8.
Miyanaga T, Hirato J, Nakazato Y . Amplification of the epidermal growth factor receptor gene in glioblastoma: an analysis of the relationship between genotype and phenotype by CISH method. Neuropathology 2008;28:116–126.
Quezado M, Ronchetti R, Rapkiewicz A, et al. Chromogenic in situ hybridization accurately identifies EGFR amplification in small cell glioblastoma multiforme, a common subtype of primary GBM. Clin Neuropathol 2005;24:163–169.
Lee JC, Vivanco I, Beroukhim R, et al. Epidermal growth factor receptor activation in glioblastoma through novel missense mutations in the extracellular domain. PLoS Med 2006;3:e485.
Tripp SR, Willmore-Payne C, Layfield LJ . Relationship between EGFR overexpression and gene amplification status in central nervous system gliomas. Anal Quant Cytol Histol 2005;27:71–78.
Carbone A, Botti G, Gloghini A, et al. Delineation of HER2 gene status in breast carcinoma by silver in situ hybridization is reproducible among laboratories and pathologists. J Mol Diagn 2008;10:527–536.
Coulibaly B, Nanni I, Quilichini B, et al. Immunohistochemistry, gene amplification and FISH: Which is the best method to assess EGFR status inglioblastomas? ASCO Molecular Markers 2008, Abstract No: 74.
Lopez-Gines C, Cerda-Nicolas M, Gil-Benso R, et al. Association of chromosome 7, chromosome 10 and EGFR gene amplification in glioblastoma multiforme. Clin Neuropathol 2005;24:209–218.
Layfield LJ, Willmore C, Tripp S, et al. Epidermal growth factor receptor gene amplification and protein expression in glioblastoma multiforme: prognostic significance and relationship to other prognostic factors. Appl Immunohistochem Mol Morphol 2006;14:91–96.
Tohma Y, Gratas C, Biernat W, et al. PTEN (MMAC1) mutations are frequent in primary glioblastomas (de novo) but not in secondary glioblastomas. J Neuropathol Exp Neurol 1998;57:684–689.
Bredel M, Pollack IF, Hamilton RL, et al. Epidermal growth factor receptor expression and gene amplification in high-grade non-brainstem gliomas of childhood. Clin Cancer Res 1999;5:1786–1792.
Waha A, Baumann A, Wolf HK, et al. Lack of prognostic relevance of alterations in the epidermal growth factor receptor-transforming growth factor-alpha pathway in human astrocytic gliomas. J Neurosurg 1996;85:634–641.
Bartlett JM . Fluorescence in situ hybridization: technical overview. Methods Mol Med 2004;97:77–87.
Yaziji H, Goldstein LC, Barry TS, et al. HER-2 testing in breast cancer using parallel tissue-based methods. JAMA 2004;291:1972–1977.
Acknowledgements
We thank Diana Jaeger, Department of Neuropathology, University Hospital Heidelberg, for excellent technical assistance in silver in situ hybridization and immunohistochemistry. Furthermore, the authors thank Georges Marchal, Ventana Application Laboratory in Strasbourg, for the EGFR silver enhanced in situ hybridization protocol setup and Maria R. Becker, National Institutes of Health in Bethesda, for critical reading of the manuscript. CONFIRM, INFORM, BENCHMARK and ultraVIEW are trademarks of Roche. EGFR pharmDx is a registered trademark of Dako. LSI and Vysis are trademarks of Abbott.
Author information
Authors and Affiliations
Corresponding author
Additional information
Conflict of interest
All authors declare no conflict of interest. No conflict exists for drugs or devices used in this study. Authors thank Ventana for generously providing the silver enhanced in situ hybridization probe and reagents.
Rights and permissions
About this article
Cite this article
Gaiser, T., Waha, A., Moessler, F. et al. Comparison of automated silver enhanced in situ hybridization and fluorescence in situ hybridization for evaluation of epidermal growth factor receptor status in human glioblastomas. Mod Pathol 22, 1263–1271 (2009). https://doi.org/10.1038/modpathol.2009.86
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2009.86
Keywords
This article is cited by
-
In-situ-Hybridisierung in der klinischen Pathologie
Der Pathologe (2012)