Abstract
Immunoglobulin A (IgA) has a critical role in immune defense particularly at the mucosal surfaces, and is equipped to do so by the unique structural attributes of its heavy chain and by its ability to polymerize. Here, we provide an overview of human IgA structure, describing the distinguishing features of the IgA1 and IgA2 subclasses and mapping the sites of interaction with host receptors important for IgA's functional repertoire. Remarkably, these same interaction sites are targeted by binding proteins and proteases produced by various pathogens as a means to subvert the protective IgA response. As interest in the prospect of therapeutic IgA-based monoclonal antibodies grows, the emerging understanding of the relationship between IgA structure and function will be invaluable for maximizing the potential of these novel reagents.
Similar content being viewed by others
Introduction
Genetic sequence analysis and functional comparisons have shown that immunoglobulin A (IgA) is present in all mammals (placental, marsupials, and monotremes) and birds. Mammals, except for rabbits and certain primates, have a single Cα gene encoding the α heavy-chain constant region that defines the IgA antibody class. Rabbits (Lagomorphs) possess 13 Cα genes, whereas humans, chimpanzees, gorillas, and gibbons have 2 Cα genes encoding distinct IgA subclasses, termed IgA1 and IgA2.1, 2 Orangutans, possessing only a single IgA that resembles IgA1, have presumably lost their IgA2. IgA1 molecules are marked out by the length of their hinge regions, flexible stretches of polypeptide at the antibody's core that separate the regions responsible for antigen binding and effector capability. The elongated hinge regions of IgA1 are lacking both in IgA2 molecules and in the single IgAs present in the majority of mammals. This striking feature seems to have evolved relatively recently through an insertion event, and represents one of the important distinctions that exist between IgA molecules from different species. Here, the focus will be on human IgA.
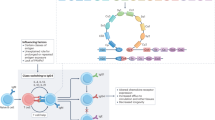
Like all Igs, IgA molecules are made up of pairings of two identical heavy chains (α-chains in the case of IgA) and two identical light chains. In humans, the IgA in serum is chiefly monomeric, comprising ∼90% IgA1 and 10% IgA2 (Figure 1a and b). Further heterogeneity arises because both subclasses can form dimers. These are stabilized through disulfide linkages between the carboxy-terminal 18 amino-acid extension (tailpiece) of one of the heavy chains of each monomer and a 15-kDa joining or J chain (Figure 1d). Secretory IgA (S-IgA), the predominant Ig in milk, colostrum, tears, saliva, and the secretions that bathe mucosal surfaces such as our respiratory, gastrointestinal, and genitourinary tracts, is chiefly derived from local synthesis and is mainly dimeric (Figure 1e), although low levels of some larger polymers, particularly tetramers, are also present. The subclass proportions vary with mucosal site, but typically range from 80 to 90% IgA1 in nasal and male genital secretions, through 60% IgA1 in saliva, to 60% IgA2 in colonic and female genital secretions.
Human IgA structure. Schematic diagrams of a, IgA1; b, IgA2; d, dimeric IgA1; e, secretory IgA1; and f, polymeric immunoglobulin receptor. IgA heavy-chain domains are shown in pink, light-chain domains in light blue, J chain in yellow, and pIgR domains (D1–D5) in dark blue. N- and O-linked oligosaccharides are shown in red and green, respectively. In panel b, the IgA2m(1) allotype is depicted. In panel f, the arrow indicates the cleavage point to yield secretory component. In c and g, molecular models of IgA1, IgA2m(1), and secretory component based on X-ray and neutron scattering are shown (accession numbers 1IGA, 1R70, and 2OCW, respectively). The inset in panel g shows the X-ray crystal structure for domain 1 of pIgR (accession number 1XED). The loops implicated in binding to dIgA are shown in green.
IgA Structure
The basic monomer unit of IgA, in common with all antibodies, is arranged into two identical Fab regions which bind antigen, linked through the hinge region to the Fc region, which mediates effector mechanisms (Figure 1a and b). Both heavy and light chains are folded into globular domains, four in each heavy chain (from the N-terminus VH, Cα1, Cα2, and Cα3) and two in each light chain (VL and CL). Each domain adopts the characteristic “immunoglobulin fold”, comprising a 110 residue β-sheet sandwich of anti-parallel strands arranged around a stabilizing internal disulfide bond.
Apart from the Cα2 domains, there is close pairing of domains between neighboring chains (VH with VL, Cα1 with CL, and Cα3 with Cα3). Inter-chain disulfide bridges further stabilize the structure. These are found between heavy chains in the Cα2 domain of IgA1 and IgA2 (and not in the hinge region as seen in IgG). Available X-ray crystal structures3, 4 implicate three (or four) cysteines on each heavy chain (Cys242, Cys299, Cys301, and possibly Cys241) in linkages across the upper parts of the Cα2 domains. The precise arrangements differ in the solved structures for IgA1 Fc complexes with different ligands, suggesting that a degree of disulfide interchange may be possible. Disulfide bridges also occur between the heavy and light chains in IgA1 and in one allotypic version of IgA2 termed IgA2m(2). In the other allotype of IgA2, known as IgA2m(1), there are generally no heavy–light chain disulfide bonds but instead there is a disulfide bridge between the light chains (Figure 1b).
The IgA1 hinge features a 16 amino-acid insertion, lacking in IgA2, comprising a repeat of 8 amino acids rich in proline, serine, and threonine, and is decorated with 3–5, or occasionally 6, O-linked oligosaccharides5, 6, 7 (Figure 1b). These sugars are generally small and heterogeneous, composed chiefly of N-acetyl galactosamine, galactose, and sialic acid. Nuclear magnetic resonance (NMR) analysis of synthetic IgA1 hinge peptides suggests that O-linked sugars are likely to affect the structure adopted by the hinge, decreasing its conformational variability.8
Both IgA subclasses carry N-linked oligosaccharides. In IgA1, these glycans are attached to Asn263 in the Cα2 domain and to Asn459 in the tailpiece (Figure 1a). In addition to these, IgA2m(1) has two further glycans attached to Asn166 in the Cα1 domain and to Asn337 in the Cα2 domain (Figure 1b), whereas IgA2m(2) has yet a further Cα1 domain sugar attached at Asn211. Analysis of serum IgA1 and S-IgA N-linked oligosaccharides revealed that the carbohydrates consist of a family of structures based on a mannosyl chitobiose core, which display marked heterogeneity in the type and number of terminal sugar residues (galactose and sialic acid) and the level of fucosylation.5, 6, 9 The majority of N-linked glycans are digalactosylated biantennary complex-type oligosaccharides, but some triantennary and a small percentage of tetra-antennary structures were detected.
In dimeric IgA (dIgA), the Fc regions of the two monomers are linked end to end through disulfide bridges to the J chain10 (Figure 1d). In particular, the penultimate residue of the tailpiece, Cys471, of one of the heavy chains of each monomer forms a disulfide bridge to the J chain.11 Both Fc domains also contribute to efficient dimer formation.12, 13
The J chain itself is an extremely highly conserved polypeptide believed to adopt either a single β-barrel-like domain14 or a two-domain structure.15 In polymeric Igs such as dIgA, larger IgA polymers, or pentameric IgM, a single J-chain molecule is incorporated. It has eight Cys residues, six of which form intrachain disulfide bridges. The remaining two, Cys14 and Cys68, form covalent links to the tailpiece Cys471 in dIgA.10, 16 The J chain has an N-linked oligosaccharide, chiefly of biantennary complex structure,6 attached to Asn48, which along with the oligosaccharide attached to the IgA tailpiece contributes to correct dimer formation.10, 11
dIgA is transported into mucosal secretions by the polymeric Ig receptor (pIgR) expressed on the basolateral surface of cells making up the mucosal epithelial barrier (Figure 1f). pIgR specifically binds and transports polymeric Igs. As dIgA is the predominant polymeric Ig at the mucosal surface, it forms the receptor's chief ligand and cargo. Upon binding, the pIgR–dIgA complex is internalized and transferred across the cell through a series of vesicles to the apical (lumenal) surface. The extracellular portion of pIgR, now linked by a disulfide bridge to dIgA, is cleaved to yield a fragment known as secretory component (SC) (Figure 1f and g). The complex of dIgA and SC, termed S-IgA (Figure 1e), is released into the secretions that bathe the mucosal surface.
The ligand-binding extracellular portion of pIgR folds into five Ig-like domains, similar to antibody variable domains, named D1 to D5 from the N terminus, and a further less globular region containing the cleavage site that releases SC (Figure 1f and g). In its unbound state, SC is believed to adopt a structure that curves back upon itself17 (Figure 1g). Interaction with dIgA requires the first three domains (D1–D3) of pIgR. D4 and D5 seem to contribute indirectly to the affinity of interaction.18 D1 in particular has a critical role in binding dIgA, through surface loops analogous to the complementarity-determining regions (CDRs) of variable domains.19, 20 Residues of D1 implicated in the interaction (Thr27–Thr33) lie in a conserved helical turn in CDR1, whereas the close-lying CDR2 loop (Glu53–Gly54) may also contribute to binding21 (inset to Figure 1g). Available evidence is consistent with these non-covalent interactions of D1 of pIgR and the Fc region of IgA initiating the interaction, with subsequent formation of a disulfide bond between Cys467 in D5 of SC and Cys311 of the IgA Cα2 domain.22, 23 In addition, direct interaction of the J chain with pIgR is required.16
Electron microscopy (EM) studies of colostral S-IgA show a double Y-shaped configuration similar to that seen for dIgA. Attachment of SC does not seem to change the overall length of the joined Fc regions. The idea that the domains of SC interact chiefly with the Fc regions of the dimer (Figure 1e) is supported by models based on solution structure analysis.24, 25 However, it should be noted that these low-resolution models fail to account for the interaction sites on IgA Fc that have been defined by mutagenesis studies in several laboratories (see below).
Interaction of IgA With Host Receptors
Between them, serum and secretory forms of IgA interact with various host receptors. For some of these (FcαRI, pIgR, Fcα/μR), an understanding of their mode of interaction with IgA has been developed and will be discussed here. For other receptors that have been described, including the M-cell receptor, the eosinophil receptor specific for SC and S-IgA, and the transferrin receptor CD71, the mode of interaction with IgA ligand is less clear.
Interaction with FcαRI
A key mediator of IgA effector function is FcαRI, also known as CD89.26 IgA molecules clustered on the surface of a pathogenic target can trigger various elimination processes through engagement of FcαRI present on neutrophils, monocytes, eosinophils and some macrophages and dendritic cells. These mechanisms include phagocytosis, release of cytokines and activated oxygen species, and antibody-dependent cell-mediated cytotoxicity.
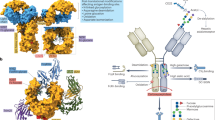
The Fc regions of both IgA1 and IgA2 bind to FcαRI through the receptor's membrane distal domain, one of two Ig-like domains making up the extracellular portion of its ligand-binding chain. An interaction site on IgA lying at the interface of the Cα2 and Cα3 domains proposed on the basis of mutagenesis analyses27, 28 was confirmed by resolution of the X-ray crystal structure of the complex of IgA1 Fc and the extracellular domains of FcαRI.3 The interaction surface comprises a central hydrophobic core of IgA residues Leu257, Leu258, Met433, Leu441, Ala442, Phe443, and the aliphatic portion of Arg382, and FcαRI residues Tyr53, Leu54, Phe56, Gly84, His85, and the aliphatic portion of Lys55, with additional contributions from a number of surrounding charged residues (Figure 2a). Despite homology between both ligands and receptors, the FcαRI site on IgA differs markedly from that on IgG and IgE for their specific Fc receptors.29 The latter receptors bind at a hinge-proximal site in the Fc regions of their respective ligands. One can speculate that, in the case of FcαRI, the Fc interdomain region was favored over a hinge proximal site because of steric restriction imposed by the IgA hinge and the interheavy chain disulfide bridges between the upper parts of the Cα2 domains. Phylogenetic and diversity analysis of IgA and FcαRI suggests that the FcαRI–IgA interaction has evolved under pressure from pathogen IgA-binding proteins that act as decoys30 (see below).
Views of IgA1 Fc X-ray crystal structure (accession code 1OW0) showing the interaction sites, as defined by X-ray crystallography or mutagenesis studies, for a, FcαRI, b, pIgR, and c, Fcα/μR. One IgA1 Fc heavy chain is shown in light blue, the other in yellow. Interaction sites are highlighted in contrasting colors. pIgR, polymeric immunoglobulin receptor.
Interaction with pIgR
In the non-covalent interaction of domain 1 of pIgR with IgA Fc, the Cα3 domain of IgA is considered to have a major role.13, 31 An exposed loop formed by residues 402–410 that lies at the upper reaches of the Cα3 domain has been particularly implicated,31, 32 with residues Phe411, Val413, and Thr414 having key roles, and the close-lying residue Lys377 and the interface loop Pro440–Phe443 making some contribution33 (Figure 2b). Interestingly, the latter loop is also involved in binding FcαRI, which may offer an explanation for the reported inability of S-IgA to trigger phagocytosis.34 However, D1 of pIgR binds to only one of the two IgA Fc regions in S-IgA, potentially leaving the other one available for FcαRI engagement, if the correct orientation is adopted. Indeed, S-IgA is reported to bind FcαRI when the integrin Mac-1 is co-engaged,35 and to be capable of triggering respiratory bursts in neutrophils.36
Interaction with Fcα/μR
Human Fcα/μR, a receptor that binds IgA and IgM,37 is expressed in follicular dendritic cells in tonsil,38 and on macrophages, plasma cells, and Paneth cells in the lamina propria and germinal centers of the intestine.39 Although the function of human Fcα/μR is not yet completely clear, these locations are consistent with a role in coordinating the immune response, at least in certain mucosal tissues. Fcα/μR maps close to pIgR on human chromosome 1. Moreover, its N-terminal Ig-like domain shares homology with domain 1 of pIgR, and the presence of certain conserved residues suggests that these domains share a similar structure.21 In particular, the CDR-like loops of both domains share similarity, implying that they may share a similar mode of interaction with dIgA. Fcα/μR interacts only with polymeric forms of IgA and IgM,40 but the presence of the J chain does not seem to be essential.41 The N-linked glycans of IgA are not required for interaction,41 and based on mapping mutagenesis experiments, the Cα2–Cα3 domain interface of the IgA heavy chain seems critical for the interaction40 (Figure 2c). The putative site overlaps with those that bind FcαRI and pIgR (Figure 2a–c).
Other Protective Functions of IgA
Glycan-mediated protective activity
The glycans of IgA have been shown to interact with sugar-dependent receptors or fimbriae on bacterial surfaces42, 43 (reviewed in the study by Mestecky and Russell44) and thereby inhibit attachment of several species of bacteria to mucosal surfaces. The significance of this interaction in vivo has been debated, and in some instances, the availability of appropriate blocking sugar residues may depend on their exposure by cleavage of glycan chains with glycosidases or on the variable composition of the glycan chains themselves. Potentially, however, S-IgA can serve as an effector molecule of both innate and adaptive immunity, through its constitutive blocking of lectin-like receptors on microbes and the specific recognition of antigens by its CDR, respectively.6
Interaction with complement
IgA lacks the residues identified in the Fc regions of IgG or IgM that bind to C1q, and consequently IgA does not activate the classical complement pathway. Although several papers have reported activation of the alternate pathway by heat-aggregated, denatured, or recombinantly generated IgA, this seems to be essentially artifactual, and intact native IgA antibodies complexed with antigen inhibit complement activation induced by IgG or IgM antibodies.45, 46, 47 This effect is also replicated by Fabα fragments generated by cleavage of IgA1 antibodies with IgA1 protease.45, 48 It is telling that mixed aggregates of heat-denatured IgG and IgA activate the alternate pathway in proportion to the content of IgG, and that C3b becomes covalently linked to the IgG heavy chains, not to IgA.49 Intriguing reports that IgA antibodies promote complement-dependent lysis or opsonization of encapsulated bacteria probably also arise from facilitation of alternate pathway activation by bacterial polysaccharides.50, 51
In addition, S-IgA has been shown to activate the lectin pathway of complement activation, by binding the mannose-binding lectin.52 However, this depends on the availability of terminal mannose (or N-acetylglucosamine) residues, which have been reported on some glycan chains expressed by IgA molecules.53 The in vivo significance of activation of this pathway by S-IgA (or other molecular forms of IgA) remains uncertain.
Hydrophilicity
IgA is the most hydrophilic of the Igs, which probably reflects its relatively high level of glycosylation. S-IgA is even more heavily glycosylated because of the glycans in SC, with an abundance of sugar residues surrounding the Fc–SC region (Figure 1e). This may also account for the long-known ability of S-IgA to associate with mucus.54, 55 It is also intriguing to speculate whether S-IgA could have unique functions in the development or control of biofilm formation, in which its hydrophilicity might be expected to facilitate penetration into the water channels. S-IgA is known to occur in dental plaque, a complex multispecies biofilm that develops on tooth surfaces. “Classical” functions of S-IgA antibodies include the inhibition of adherence of microbes to mucosal surfaces, inhibition of enzymes including the streptococcal glucosyltransferases that synthesize extracellular glucans from sucrose, and blocking of the glucan-binding proteins of oral streptococci, all of which contribute to the formation of dental plaque (reviewed in the study by Russell and Kilian56).
Pertubation of IgA Function by Pathogenic Microorganisms
The fact that numerous pathogens have evolved means to specifically evade or subvert the IgA immune response is an indicator of the importance of IgA in protection against infection. These evasion mechanisms include proteins that bind specifically to IgA and SC and inhibit their actions, and specific proteases that inactivate IgA through cleavage.
IgA-binding proteins
Akin to the well-known IgG-binding bacterial proteins used widely in research, certain pathogenic bacteria express surface proteins that bind specifically to IgA. For example, Staphylococcus aureus produces the SSL7 toxin, which binds to the Cα2–Cα3 interface of IgA Fc4, 57 (Figure 3a). Binding of SSL7 to IgA competitively inhibits FcαRI binding.57 Such blockade presumably helps bacteria evade the elimination mechanisms normally elicited by IgA through interaction with FcαRI.
Interaction sites on IgA Fc for bacterial proteins that perturb IgA function. One IgA1 Fc heavy chain is shown in light blue, the other in yellow. Interaction sites (highlighted in contrasting colors) for a, SSL7 protein from Staphylococcus aureus from the solved crystal structure of the IgA Fc–SSL7 complex; b, IgA-binding proteins from Streptococcus groups A and B based on mutagenesis analysis; c, type 2 IgA1 protease from Neisseria meningitidis implicated in by mutagenesis analysis; d, type 2 IgA1 protease from Haemophilus influenzae implicated by mutagenesis analysis.
Certain streptococci similarly express proteins that bind specifically to human IgA. Important examples are Arp4 and Sir22 on group A Streptococcus58, 59 and the unrelated β-protein on group B Streptococcus.60 These streptococcal proteins also interact with the IgA Cα2–Cα3 domain interface61 (Figure 3b). Again, they inhibit binding of IgA to FcαRI, as well as the triggering of FcαRI-mediated respiratory bursts,61 suggesting that this evasion strategy is used by a number of different bacteria. In fact, binding of specific bacterial proteins to the interdomain region of Ig Fc regions is a common theme crossing both host and bacterial species.62
SC binding by CbpA
Streptococcus pneumoniae uses binding of a surface protein CbpA (also termed “SpsA” or “PspC”) to pIgR on epithelial cells as a means to initiate colonization of the nasopharynx. Domains 3 and 4 of pIgR are involved in complex formation with CbpA.63, 64 This evasion strategy is tempered by the ability of free SC and S-IgA to bind CbpA and inhibit binding of the bacterium to cell-surface pIgR.
Bacterial IgA1 proteases
Certain clinically important pathogenic bacteria including Neisseria meningiditis, Neisseria gonorrhoeae, Haemophilus influenzae, S. pneumoniae, and Streptococcus sanguis produce proteolytic enzymes that are highly specific for the hinge region of IgA1. These enzymes, termed IgA1 proteases, cleave at specific Pro-Thr or Pro-Ser bonds within the IgA1 hinge. As the susceptible sequence is lacking in IgA2, this subclass is resistant to cleavage. The action of the enzymes on IgA1 results in release of the Fc region, leaving the Fab regions free to bind antigens on the bacteria (and to impede access of other Igs) but unable to trigger any Fc-mediated clearance mechanisms. Thus, the bacteria are able to evade the protective action normally provided by intact IgA. The bacteria that produce IgA1 proteases frequently colonize or gain a foothold at mucosal surfaces, and are responsible for major oral cavity and genital tract infections, as well as life-threatening meningitis. The ability to circumvent the IgA response seems important to their ability to breach mucosal defenses.
IgA1 proteases form a rather diverse enzyme group—e.g., some are serine proteases, whereas others are metalloproteinases—and it is therefore difficult to generalize on their substrate recognition and cleavage requirements. However, the context of the IgA1 hinge within the IgA protein and the sequence of the hinge have been shown to be important for recognition and cleavage by these proteases.65, 66, 67, 68, 69 Notably, there is a requirement for the susceptible bond to be suitably positioned relative to the Fc.69 For some IgA1 proteases, cleavage of IgA1 requires the presence of critical elements within the Fc region.67, 70 For example, the Neisseria meningitidis type 2 enzyme seems to require residues Pro440–Phe443, an interdomain loop also involved in the interaction with FcαRI, pIgR, and Fcα/μR70 (Figure 3c). The H. influenzae type 2 enzyme, meanwhile, seems to require Cα3 residues also implicated in binding to pIgR70 (Figure 3d). The solved X-ray crystal structure of an H. influenzae IgA1 protease is consistent with Fc involvement, and it has been proposed that binding of Fc by the protease may stabilize a conformation in which the IgA1 hinge peptide can access the enzyme's active site resulting in cleavage.71
Concluding Remarks
A striking theme to emerge from consideration of the interaction sites on IgA for different proteins is that many of these sites overlap or coincide. Remarkably, several host receptors and a number of pathogen molecules interact with the same region at the interface of the Cα2 and Cα3 domains in the Fc region. The same Fc interdomain region serves as a multifunctional interaction site on other Igs too. In IgG, the FcRn receptor that controls IgG turnover and transport binds here, as does the staphylococcal protein A and streptococcal protein G. Presumably, in each case, the interdomain region is intrinsically suited to protein–protein interactions and has been conserved as a site for key host receptors. Various pathogens have then targeted this region with their own proteins that often block interaction of host receptors, thereby subverting the protective antibody response.
While sharing characteristics with other Ig classes, the distinct structural properties of IgA lend this antibody class a unique array of functional capabilities. These arise in part from the ability to polymerize, which bestows affinity for additional receptors such as pIgR and Fcα/μR, and in part from the particular structural features of the IgA heavy chain. For example, the extended hinge seen in human IgA1 would seem likely to afford a greater reach between its two antigen-binding sites,72 allowing this subclass to achieve high-avidity interaction with more distantly spaced antigens than other Ig isotypes. Presumably, this may result in more efficient recognition of certain pathogens that express immunodominant antigens spaced out along their surface. Furthermore, the long hinge of IgA1 and the location of the FcαRI interaction site at the Fc interdomain site may provide for particularly efficient bridging between antigen on a target cell and FcαRI on an effector cell.29
These advantageous characteristics fit well with the increasing interest in the possibility of using IgA monoclonal antibodies therapeutically.73, 74, 75 The efficacy of IgA-based antibodies against bacterial pathogens including S. pneumoniae,51, 76 N. meningitidis,47 Bordetella pertussis,77, 78 and Mycobacterium tuberculosis79 is certainly encouraging, while the evidence for the value of FcαRI-directed therapies in cancer treatment continues to grow.80, 81, 82, 83, 84, 85 The detailed understanding of structure–function relationships in IgA that is now emerging will be invaluable in informing the design of a new generation of IgA-based monoclonal antibodies.
References
Burnett, R.C., Hanly, W.C., Zhai, S.K. & Knight, K.L. The IgA heavy chain gene family in rabbit: cloning and sequence analysis of 13 Cα genes. EMBO J. 8, 4041–4047 (1989).
Kawamura, S., Saitou, N. & Ueda, S. Concerted evolution of the primate immunoglobulin α-gene through gene conversion. J. Biol. Chem. 267, 7359–7367 (1992).
Herr, A.B., Ballister, E.R. & Bjorkman, P.J. Insights into IgA-mediated immune responses from the crystal structures of human FcαRI and its complex with IgA1-Fc. Nature. 423, 614–620 (2003).
Ramsland, P.A. et al. Structural basis for evasion of IgA immunity by Staphylococcus aureus revealed in the complex of SSL7 with Fc of human IgA1. Proc. Natl Acad. Sci. USA 104, 15051–15056 (2007).
Mattu, T.S. et al. The glycosylation and structure of human serum IgA1, Fab and Fc regions and the role of N-glycosylation on Fcα receptor interactions. J. Biol. Chem. 273, 2260–2272 (1998).
Royle, L. et al. Secretory IgA N- and O-glycans provide a link between the innate and adaptive immune systems. J. Biol. Chem. 278, 20140–20153 (2003).
Tarelli, E., Smith, A.C., Hendry, B.M., Challacombe, S.J. & Pouria, S. Human serum IgA1 is substituted with up to six O-glycans as shown by matrix assisted laser desorption ionisation time-of-flight mass spectrometry. Carbohydrate Res. 339, 2329–2335 (2004).
Narimatsu, Y. et al. Effect of glycosylation on the cis/trans isomerization of prolines in IgA1-hinge peptide. J. Am. Chem. Soc. 132, 5548–5549 (2010).
Field, M.C., Amatayakul-Chantler, S., Rademacher, T.W., Rudd, P.M. & Dwek, R.A. Structural analysis of the N-glycans from human immunoglobulin A1: comparison of normal human serum immunoglobulin A1 with that isolated from patients with rheumatoid arthritis. Biochem. J. 299, 261–275 (1994).
Krugmann, S., Pleass, R.J., Atkin, J.D. & Woof, J.M. Structural requirements for assembly of dimeric IgA probed by site-directed mutagenesis of J chain and a cysteine residue of the α chain CH2 domain. J. Immunol. 159, 244–249 (1997).
Atkin, J.D., Pleass, R.J., Owens, R.J. & Woof, J.M. Mutagenesis of the human IgA1 heavy chain tailpiece that prevents dimer assembly. J. Immunol. 157, 156–159 (1996).
Yoo, E.M. et al. Structural requirements for polymeric immunoglobulin assembly and association with J chain. J. Biol. Chem. 274, 33771–33777 (1999).
Braathen, R., Sorensen, V., Brandtzaeg, P., Sandlie, I. & Johansen, F.E. The carboxyl-terminal domains of IgA and IgM direct isotype-specific polymerization and interaction with the polymeric immunoglobulin receptor. J. Biol. Chem. 277, 42755–42762 (2002).
Zikan, J. et al. Secondary structure of the immunoglobulin J chain. Proc. Natl Acad. Sci. USA 82, 5905–5909 (1985).
Frutiger, S., Hughes, G.J., Paquet, N., Luthy, R. & Jaton, J.C. Disulfide bond assignment in human J chain and its covalent pairing with immunoglobulin M. Biochemistry. 31, 12643–12647 (1992).
Johansen, F.E., Braathen, R. & Brandtzaeg, P. The J chain is essential for polymeric Ig receptor-mediated epithelial transport of IgA. J. Immunol. 167, 5185–5192 (2001).
Bonner, A., Perrier, C., Corthesy, B. & Perkins, S.J. Solution structure of human secretory component and implications for biological function. J. Biol. Chem. 282, 16969–16980 (2007).
Norderhaug, I.N., Johansen, F.E., Krajci, P. & Brandtzaeg, P. Domain deletions in the human polymeric Ig receptor disclose differences between its dimeric and pentameric IgM interaction. Eur. J. Immunol. 29, 3401–3409 (1999).
Bakos, M.A., Kurosky, A., Cwerwinski, E.W. & Goldblum, R.M. A conserved binding site on the receptor for polymeric Ig is homologous to CDR1 of Ig V kappa domains. J. Immunol. 151, 1346–1352 (1993).
Coyne, R.S., Siebrecht, M., Peitsch, M.C. & Casanova, J.E. Mutational analysis of polymeric immunoglobulin receptor/ligand interactions. Evidence for the involvement of multiple complementarity determining region (CDR)-like loops in receptor domain I. J. Biol. Chem. 269, 31620–31625 (1994).
Hamburger, A.E., West, A.P. Jr . & Bjorkman, P.J. Crystal structure of a polymeric immunoglobulin binding fragment of the human polymeric immunoglobulin receptor. Structure (Camb) 12, 1925–1935 (2004).
Underdown, B.J., DeRose, J. & Plaut, A. Disulfide bonding of secretory component to a single monomer subunit in human secretory IgA. J. Immunol. 118, 1816–1821 (1977).
Fallgren-Gebauer, E. et al. The covalent linkage of the secretory component to IgA. Adv. Exp. Med. Biol. 371A, 625–628 (1995).
Bonner, A., Almogren, A., Furtado, P.B., Kerr, M.A. & Perkins, S.J. Location of secretory component on the Fc edge of dimeric IgA1 reveals insight into the role of secretory IgA1 in mucosal immunity. Mucosal Immunol. 2, 74–84 (2009).
Bonner, A., Almogren, A., Furtado, P.B., Kerr, M.A. & Perkins, S.J. The nonplanar secretory IgA2 and near planar secretory IgA1 solution structures rationalize their different mucosal immune responses. J. Biol. Chem. 284, 5077–5087 (2009).
Otten, M.A. & van Egmond, M. The Fc receptor for IgA (FcαRI, CD89). Immunol. Lett. 92, 23–31 (2004).
Carayannopoulos, L., Hexham, J.M. & Capra, J.D. Localization of the binding site for the monocyte immunoglobulin (Ig) A-Fc receptor (CD89) to the domain boundary between Cα2 and Cα3 in human IgA1. J. Exp. Med. 183, 1579–1586 (1996).
Pleass, R.J., Dunlop, J.I., Anderson, C.M. & Woof, J.M. Identification of residues in the CH2/CH3 domain interface of IgA essential for interaction with the human Fcα receptor (FcαR) CD89. J. Biol. Chem. 274, 23508–23514 (1999).
Woof, J.M. & Burton, D.R. Human antibody-Fc receptor interactions illuminated by crystal structures. Nat. Rev. Immunol. 4, 89–99 (2004).
Abi-Rached, L., Dorighi, K., Norman, P.J., Yawata, M. & Parham, P. Episodes of natural selection shaped the interactions of IgA-Fc with FcαRI and bacterial decoy proteins. J. Immunol. 178, 7943–7954 (2007).
Hexham, J.M. et al. A human immunoglobulin (Ig)A Cα3 domain motif directs polymeric Ig receptor-mediated secretion. J. Exp. Med. 189, 747–752 (1999).
White, K.D. & Capra, J.D. Targeting mucosal sites by polymeric immunoglobulin receptor-directed peptides. J. Exp. Med. 196, 551–555 (2002).
Lewis, M.J., Pleass, R.J., Batten, M.R., Atkin, J.D. & Woof, J.M. Structural requirements for the interaction of human IgA with the human polymeric Ig receptor. J. Immunol. 175, 6694–6701 (2005).
van Egmond, M. et al. FcαRI-positive liver Kuppfer cells: reappraisal of the function of immunoglobulin A in immunity. Nat. Med. 6, 680–685 (2000).
van Spriel, A.B., Leusen, J.H., Vilé, H. & van de Winkel, J.G. Mac-1 (CD11b/CD18) as accessory molecule for FcαR (CD89) binding of IgA. J. Immunol. 169, 3831–3836 (2002).
Stewart, W.W. & Kerr, M.A. The specificity of the human neutrophil IgA receptor (FcαR) determined by measurement of chemiluminescence induced by serum or secretory IgA1 or IgA2. Immunology. 71, 328–334 (1990).
Shibuya, A. & Honda, S.I. Molecular and functional characteristics of the Fcα/μR, a novel Fc receptor for IgM and IgA. Springer Semin. Immun. 28, 377–382 (2006).
Kikuno, K. et al. Unusual biochemical features and follicular dendritic cell expression of human Fcα/μ receptor. Eur. J. Immunol. 37, 3540–3550 (2007).
Wang, R., Fu, Y., Zhao, Q., Pan, L. & Zhang, W. Human Fcα/μR and pIgR distribute differently in intestinal tissues. Biochem. Biophys. Res. Commun. 381, 148–152 (2009).
Ghumra, A. et al. Structural requirements for the interaction of human IgM and IgA with the human Fcα/μ receptor. Eur. J. Immunol. 39, 1147–1156 (2009).
Yoo, E.M., Trinh, K.R., Lim, H., Wims, L.A. & Morrison, S.L. Characterization of IgA and IgM binding and internalization by surface-expressed human Fcα/μ receptor. Mol. Immunol. 48, 1818–1826 (2011).
Wold, A. et al. Secretory immunoglobulin A carries oligosaccharide receptors for Escherichia coli type 1 fimbrial lectin. Infect. Immun. 58, 3073–3077 (1990).
Ruhl, S., Sandberg, A.L., Cole, M.F. & Cisar, J.O. Recognition of immunoglobulin A1 by oral actinomyces and streptococcal lectins. Infect. Immun. 64, 5421–5424 (1996).
Mestecky, J. & Russell, M.W. Specific antibody activity, glycan heterogeneity and polyreactivity contribute to the protective activity of S-IgA at mucosal surfaces. Immunol. Lett. 124, 57–62 (2009).
Russell, M.W., Reinholdt, J. & Kilian, M. Anti-inflammatory activity of human IgA antibodies and their Fabα fragments: inhibition of IgG-mediated complement activation. Eur. J. Immunol. 19, 2243–2249 (1989).
Griffiss, J.M. & Goroff, D.K. IgA blocks IgM and IgG-initiated immune lysis by separate molecular mechanisms. J. Immunol. 130, 2882–2885 (1983).
Vidarsson, G. et al. Activity of human IgG and IgA subclasses in immune defense against Neisseria meningitidis serogroup B. J. Immunol. 166, 6250–6256 (2001).
Jarvis, G.A. & Griffiss, J.M. Human IgA1 blockade of IgG-initiated lysis of Neisseria meningitidis is a function of antigen-binding fragment binding to the polysaccharide capsule. J. Immunol. 147, 1962–1967 (1991).
Waldo, F.B. & Cochran, A.M. Mixed IgA-IgG aggregates as a model of immune complexes in IgA nephropathy. J. Immunol. 142, 3841–3846 (1989).
Jarvis, G.A. & Griffiss, J.M. Human IgA1 initiates complement-mediated killing of Neisseria meningitidis. J. Immunol. 143, 1703–1709 (1989).
Janoff, E.N. et al. Killing of Streptococcus pneumoniae by capsular polysaccharide-specific polymeric IgA, complement, and phagocytes. J. Clin. Invest. 104, 1139–1147 (1999).
Roos, A. et al. Human IgA activates the complement system via the mannan-binding lectin pathway. J. Immunol. 167, 2861–2868 (2001).
Endo, T., Mestecky, J., Kulhavy, R. & Kobata, A. Carbohydrate heterogeneity of human myeloma proteins of the IgA1 and IgA2 subclasses. Mol. Immunol. 31, 1415–1422 (1994).
Magnusson, K.E. & Stjernström, I. Mucosal barrier systems. Interplay between secretory IgA (SIgA), IgG and mucins on the surface properties and association of salmonellae with intestine and granulocytes. Immunology. 45, 239–248 (1982).
Biesbrock, A.R., Reddy, M.S. & Levine, M.J. Interaction of a salivary mucin-secretory immunoglobulin A complex with mucosal pathogens. Infect. Immun. 59, 3492–3497 (1991).
Russell, M.W. & Kilian, M. Biological activities of IgA. In Mucosal Immunol. (Mestecky, J., Bienenstock, J., Lamm, M.E., Mayer, L., Strober, W., McGhee, J.R., eds) 267–289 ( Elsevier/Academic Press, Amsterdam, 2005 ).
Wines, B.D., Willoughby, N., Fraser, J.D. & Hogarth, P.M. A competitive mechanism for staphylococcal toxin SSL7 inhibiting the leukocyte IgA receptor, FcαRI, is revealed by SSL7 binding at the Cα2/Cα3 interface of IgA. J. Biol. Chem. 281, 1389–1393 (2006).
Frithz, E., Héden, L.O. & Lindahl, G. Extensive sequence homology between IgA receptor and M proteins in Streptococcus pyogenes. Mol. Microbiol. 3, 1111–1119 (1989).
Stenberg, L., O'Toole, P.W., Mestecky, J. & Lindahl, G. Molecular characterization of protein Sir, a streptococcal cell surface protein that binds both immunoglobulin A and immunoglobulin G. J. Biol. Chem. 269, 13458–13464 (1994).
Héden, L.O., Frithz, E. & Lindahl, G. Molecular characterization of an IgA receptor from group B streptococci: sequence of the gene, identification of a proline-rich region with unique structure and isolation of N-terminal fragments with IgA-binding capacity. Eur. J. Immunol. 21, 1481–1490 (1991).
Pleass, R.J., Areschoug, T., Lindahl, G. & Woof, J.M. Streptococcal IgA-binding proteins bind in the Cα2-Cα3 interdomain region and inhibit binding of IgA to human CD89. J. Biol. Chem. 276, 8197–8204 (2001).
Lewis, M.J., Meehan, M., Owen, P. & Woof, J.M. A common theme in interaction of bacterial immunoglobulin-binding proteins with immunoglobulins illustrated in the equine system. J. Biol. Chem. 283, 17615–17623 (2008).
Lu, L., Lamm, M.E., Li, H., Corthesy, B. & Zhang, J.R. The human polymeric immunoglobulin receptor binds to Streptococcus pneumoniae via domains 3 and 4. J. Biol. Chem. 278, 48178–48187 (2003).
Elm, C. et al. Ectodomains 3 and 4 of human polymeric immunoglobulin receptor (hpIgR) mediate invasion of Streptococcus pneumoniae into the epithelium. J. Biol. Chem. 279, 6296–6304 (2004).
Senior, B.W., Dunlop, J.I., Batten, M.R., Kilian, M. & Woof, J.M. Cleavage of a recombinant human immunoglobulin A2 (IgA2)-IgA1 hybrid antibody by certain bacterial IgA1 proteases. Infect. Immun. 68, 463–469 (2000).
Batten, M.R., Senior, B.W., Kilian, M. & Woof, J.M. Amino acid sequence requirements in the hinge of human immunoglobulin A1 (IgA1) for cleavage by streptococcal IgA1 proteases. Infect. Immun. 71, 1462–1469 (2003).
Chintalacharuvu, K.R. et al. Cleavage of the human immunoglobulin A1 (IgA1) hinge region by IgA1 proteases requires structures in the Fc region of IgA. Infect. Immun. 71, 2563–2570 (2003).
Senior, B.W. & Woof, J.M. Effect of mutations in the human immunoglobulin A1 (IgA1) hinge on its susceptibility to cleavage by diverse bacterial IgA1 proteases. Infect. Immun. 73, 1515–1522 (2005).
Senior, B.W. & Woof, J.M. The influences of hinge length and composition on the susceptibility of human IgA to cleavage by diverse bacterial IgA1 proteases. J. Immunol. 174, 7792–7799 (2005).
Senior, B.W. & Woof, J.M. Sites in the CH3 domain of human IgA1 that influence sensitivity to bacterial IgA1 proteases. J. Immunol. 177, 3913–3919 (2006).
Johnson, T.A., Qiu, J., Plaut, A.G. & Holyoak, T. Active-site gating regulates substrate selectivity in a chymotrypsin-like serine protease the structure of Haemophilus influenzae immunoglobulin A1 protease. J. Mol. Biol. 389, 559–574 (2009).
Boehm, M.K., Woof, J.M., Kerr, M.A. & Perkins, S.J. The Fab and Fc fragments of IgA1 exhibit a different arrangement from that in IgG: a study by X-ray and neutron solution scattering and homology modelling. J. Mol. Biol. 286, 1421–1447 (1999).
Dechant, M. & Valerius, T. IgA antibodies for cancer therapy. Crit. Rev. Oncol. Hematol. 39, 69–77 (2001).
Corthésy, B. Recombinant secretory immunoglobulin A in passive immunotherapy: linking immunology and biotechnology. Curr. Pharm. Biotechnol. 4, 51–67 (2003).
Bakema, J.E. & van Egmond, M. Immunoglobulin A: a next generation of therapeutic antibodies? MAbs. 3, 352–361 (2011).
van der Pol, W., Vidarsson, G., Vile, H.A., van de Winkel, J.G. & Rodriguez, M.E. Pneumococcal capsular polysaccharide-specific IgA triggers efficient neutrophil effector functions via FcαRI (CD89). J. Infect. Dis. 182, 1139–1145 (2000).
Rodriguez, M.E. et al. Fc receptor-mediated immunity against Bordetella pertussis. J. Immunol. 167, 6545–6551 (2001).
Hellwig, S.M.M., van Spriel, A.B., Schellekens, J.F.P., Mooi, F.R. & van de Winkel, J.G.J. Immunoglobulin A-mediated protection against Bordetella pertussis infection. Infect. Immun. 69, 4846–4850 (2001).
Balu, S. et al. A novel human IgA monoclonal antibody protects against tuberculosis. J. Immunol. 186, 3113–3119 (2011).
Stockmeyer, B. et al. Triggering Fcα-receptor I (CD89) recruits neutrophils as effector cells for CD20-directed antibody therapy. J. Immunol. 165, 5954–5961 (2000).
van Egmond, M. et al. Enhancement of polymorphonuclear cell-mediated tumor cell killing on simultaneous engagement of FcγRI (CD64) and FcαRI (CD89). Cancer Res. 61, 4055–4060 (2001).
Dechant, M. et al. Chimeric IgA antibodies against HLA class II effectively trigger lymphoma cell killing. Blood. 100, 4574–4580 (2002).
Otten, M.A. et al. Immature neutrophils mediate tumor cell killing via IgA but not IgG Fc receptors. J. Immunol. 174, 5472–5480 (2005).
Lohse, S. et al. Recombinant dimeric IgA antibodies against the epidermal growth factor receptor mediate effective tumor cell killing. J. Immunol. 186, 3770–3778 (2011).
Bakema, J.E. et al. Targeting FcαRI on polymorphonuclear cells induces tumor cell killing through autophagy. J. Immunol. 187, 726–732 (2011).
Acknowledgements
We acknowledge funding from Asthma UK (JMW) and USPHS grants AI074791 and AI092348 (MWR).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declared no conflict of interest.
Rights and permissions
About this article
Cite this article
Woof, J., Russell, M. Structure and function relationships in IgA. Mucosal Immunol 4, 590–597 (2011). https://doi.org/10.1038/mi.2011.39
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mi.2011.39
This article is cited by
-
Increased Intestinal Permeability: An Avenue for the Development of Autoimmune Disease?
Exposure and Health (2024)
-
Elevated serum IgA following vaccination against SARS-CoV-2 in a cohort of high-risk first responders
Scientific Reports (2022)
-
Quantitative proteomics analysis of lysine 2-hydroxyisobutyrylation in IgA nephropathy
Clinical Proteomics (2021)
-
Immunoglobulin subtype-coated bacteria are correlated with the disease activity of inflammatory bowel disease
Scientific Reports (2021)
-
The immune landscape of IgA induction in the gut
Seminars in Immunopathology (2021)