Abstract
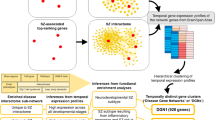
To identify potential candidate genes for future pharmacogenetic studies of antipsychotic (AP)-induced extrapyramidal symptoms (EPS), we used gene expression arrays to analyze changes induced by risperidone in mice strains with different susceptibility to EPS. We proposed a systems biology analytical approach that combined the identification of gene co-expression modules related to AP treatment, the construction of protein–protein interaction networks with genes included in identified modules and finally, gene set enrichment analysis of constructed networks. In response to risperidone, mice strain with susceptibility to develop EPS showed downregulation of genes involved in the mammalian target of rapamycin (mTOR) pathway and biological processes related to this pathway. Moreover, we also showed differences in the phosphorylation pattern of the ribosomal protein S6 (rpS6), which is a major downstream effector of mTOR. The present study provides new evidence of the involvement of the mTOR pathway in AP-induced EPS and offers new and valuable markers for pharmacogenetic studies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Divac N, Prostran M, Jakovcevski I, Cerovac N . Second-generation antipsychotics and extrapyramidal adverse effects. Biomed Res Int 2014; 2014: 656370.
Crowley JJ, Kim Y, Szatkiewicz JP, Pratt AL, Quackenbush CR, Adkins DE et al. Genome-wide association mapping of loci for antipsychotic-induced extrapyramidal symptoms in mice. Mamm Genome 2012; 23: 322–335.
Greenbaum L, Strous RD, Kanyas K, Merbl Y, Horowitz A, Karni O et al. Association of the RGS2 gene with extrapyramidal symptoms induced by treatment with antipsychotic medication. Pharmacogenet Genomics 2007; 17: 519–528.
Gassó P, Mas S, Bernardo M, Alvarez S, Parellada E, Lafuente A . A common variant in DRD3 gene is associated with risperidone-induced extrapyramidal symptoms. Pharmacogenomics J 2009; 9: 404–410.
Gassó P, Mas S, Oliveira C, Bioque M, Parellada E, Bernardo M et al. Searching for functional SNPs or rare variants in exonic regions of DRD3 in risperidone-treated patients. Eur Neuropsychopharmacol 2011; 21: 294–299.
Crescenti A, Mas S, Gassó P, Parellada E, Bernardo M, Lafuente A . Cyp2d6*3, *4, *5 and *6 polymorphisms and antipsychotic-induced extrapyramidal side-effects in patients receiving antipsychotic therapy. Clin Exp Pharmacol Physiol 2008; 35: 807–811.
Mas S, Gassò P, Alvarez S, Parellada E, Bernardo M, Lafuente A . Intuitive pharmacogenetics: spontaneous risperidone dosage is related to CYP2D6, CYP3A5 and ABCB1 genotypes. Pharmacogenomics J 2012; 12: 255–259.
Lencz T, Malhotra AK . Pharmacogenetics of antipsychotic-induced side effects. Dialogues Clin Neurosci 2009; 11: 405–415.
Llerena A, Berecz R, Peñas-Lledó E, Süveges A, Fariñas H . Pharmacogenetics of clinical response to risperidone. Pharmacogenomics 2013; 14: 177–194.
Almoguera B, Riveiro-Alvarez R, Lopez-Castroman J, Dorado P, Vaquero-Lorenzo C, Fernandez-Piqueras J et al. CYP2D6 poor metabolizer status might be associated with better response to risperidone treatment. Pharmacogenet Genomics 2013; 23: 627–630.
Remington G, Kapur S . Atypical antipsychotics: are some more atypical than others? Psychopharmacology (Berl) 2000; 148: 3–15.
Niculescu AB, Le-Niculescu H . Convergent Functional Genomics: what we have learned and can learn about genes, pathways, and mechanisms. Neuropsychopharmacology 2010; 35: 355–356.
Minguez P, Dopazo J . Functional genomics and networks: new approaches in the extraction of complex gene modules. Expert Rev Proteomics 2010; 7: 55–63.
Konradi C, Heckers S . Antipsychotic drugs and neuroplasticity: insights into the treatment and neurobiology of schizophrenia. Biol Psychiatry 2001; 50: 729–742.
Strand AD, Baquet ZC, Aragaki AK, Holmans P, Yang L, Cleren C et al. Expression profiling of Huntington's disease models suggests that brain-derived neurotrophic factor depletion plays a major role in striatal degeneration. J Neurosci 2007; 27: 11758–11768.
Korostynski M, Piechota M, Dzbek J, Mlynarski W, Szklarczyk K, Ziolkowska B et al. Novel drug-regulated transcriptional networks in brain reveal pharmacological properties of psychotropic drugs. BMC Genomics 2013; 14: 606.
Wei C, Li J, Bumgarner RE . Sample size for detecting differentially expressed genes in microarray experiments. BMC Genomics 2004; 5: 87.
Langfelder P, Horvath S . WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008; 9: 559.
Minguez P, Götz S, Montaner D, Al-Shahrour F, Dopazo J . SNOW, a web-based tool for the statistical analysis of protein-protein interaction networks. Nucleic Acids Res 2009; 37: W109–W114.
Mas S, Gassó P, Parellada E, Bernardo M, Lafuente A . Network analysis of gene expression in peripheral blood identifies mTOR and NF-κB pathways involved in antipsychotic induced extrapyramidal symptoms. Pharmacogenomics J, epub ahead of print 27 Jan 2015.10.1038/tpj.2014.84.
Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009; 25: 1091–1093.
Carretón O, Giralt A, Torres-Peraza JF, Brito V, Lucas JJ, Ginés S et al. Age-dependent decline of motor neocortex but not hippocampal performance in heterozygous BDNF mice correlates with a decrease of cortical PSD-95 but an increase of hippocampal TrkB levels. Exp Neurol 2012; 237: 335–345.
Giralt A, Carretón O, Lao-Peregrin C, Martín ED, Alberch J . Conditional BDNF release under pathological conditions improves Huntington's disease pathology by delaying neuronal dysfunction. Mol Neurodegener 2011; 6: 71.
Hales EC, Taub JW, Matherly LH . New insights into Notch1 regulation of the PI3K-AKT-mTOR1 signaling axis: targeted therapy of γ-secretase inhibitor resistant T-cell acute lymphoblastic leukemia. Cell Signal 2014; 26: 149–161.
Laplante M, Sabatini DM . mTOR signaling at a glance. J Cell Sci 2009; 122: 3589–3594.
Mas S, Gassó P, Malagelada C, Bernardo M, Lafuente A . Pharmacogenetic predictor of extrapyramidal symptoms induced by antipsychotics: multilocus interaction in the mTOR pathway. Eur Neuropsychopharmacol 2014; 25: 51–59.
Schmidt RH, Jokinen JD, Massey VL, Falkner KC, Shi X, Yin X et al. Olanzapine activates hepatic mammalian target of rapamycin: new mechanistic insight into metabolic dysregulation with atypical antipsychotic drugs. J Pharmacol Exp Ther 2013; 347: 126–135.
Shin SY, Lee KS, Choi YK, Lim HJ, Lee HG, Lim Y et al. The antipsychotic agent chlorpromazine induces autophagic cell death by inhibiting the Akt/mTOR pathway in human U-87MG glioma cells. Carcinogenesis 2013; 34: 2080–2089.
Bonito-Oliva A, Pallottino S, Bertran-Gonzalez J, Girault JA, Valjent E, Fisone G . Haloperidol promotes mTORC1-dependent phosphorylation of ribosomal protein S6 via dopamine- and cAMP-regulated phosphoprotein of 32 kDa and inhibition of protein phosphatase-1. Neuropharmacology 2013; 72: 197–203.
Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N . Translational control of long-lasting synaptic plasticity and memory. Neuron 2009; 61: 10–26.
Roux PP, Shahbazian D, Vu H, Holz MK, Cohen MS, Taunton J et al. RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J Biol Chem 2007; 282: 14056–14064.
Moore CE, Xie J, Gomez E, Herbert TP . Identification of cAMP-dependent kinase as a third in vivo ribosomal protein S6 kinase in pancreatic beta-cells. J Mol Biol 2009; 389: 480–494.
Valjent E, Bertran-Gonzalez J, Bowling H, Lopez S, Santini E, Matamales M et al. Haloperidol regulates the state of phosphorylation of ribosomal protein S6 via activation of PKA and phosphorylation of DARPP-32. Neuropsychopharmacology 2011; 36: 2561–2570.
Bowling H, Zhang G, Bhattacharya A, Pérez-Cuesta LM, Deinhardt K, Hoeffer CA et al. Antipsychotics activate mTORC1-dependent translation to enhance neuronal morphological complexity. Sci Signal 2014; 7: ra4.
Mannoury la Cour C, Salles MJ, Pasteau V, Millan MJ . Signaling pathways leading to phosphorylation of Akt and GSK-3β by activation of cloned human and rat cerebral D2 and D3 receptors. Mol Pharmacol 2011; 79: 91–105.
Beaulieu JM, Sotnikova TD, Marion S, Lefkowitz RJ, Gainetdinov RR, Caron MG . An Akt/beta-arrestin 2/PP2A signaling complex mediates dopaminergic neurotransmission and behavior. Cell 2005; 122: 261–273.
Seo MS, Scarr E, Lai CY, Dean B . Potential molecular and cellular mechanism of psychotropic drugs. Clin Psychopharmacol Neurosci 2014; 12: 94–110.
Dwivedi Y, Rizavi HS, Pandey GN . Differential effects of haloperidol and clozapine on [(3)H]cAMP binding, protein kinase A (PKA) activity, and mRNA and protein expression of selective regulatory and catalytic subunit isoforms of PKA in rat brain. J Pharmacol Exp Ther 2002; 301: 197–209.
Mas S, Gassó P, Bernardo M, Lafuente A . Functional analysis of gene expression in risperidone treated cells provide new insights in molecular mechanism and new candidate genes for pharmacogenetic studies. Eur Neuropsychopharmacol 2013; 23: 329–337.
Marder SR . Perspective: retreat from the radical. Nature 2014; 508: S18.
Hyman SE . Perspective: revealing molecular secrets. Nature 2014; 508: S20.
Acknowledgements
This study was supported by the Spanish Ministry of Health, Instituto de Salud Carlos III (FIS, Fondo de Investigacion Sanitaria PI10/02430) and the Catalan Innovation, Universities and Enterprise Authority (Grants DURSI 2014SGR436 and 2014SGR441). We thank the Language Advisory Service of the University of Barcelona, Spain for manuscript revision. The authors also thank Ana Meseguer for sample collection assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the The Pharmacogenomics Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Mas, S., Gassó, P., Boloc, D. et al. Network analysis of gene expression in mice provides new evidence of involvement of the mTOR pathway in antipsychotic-induced extrapyramidal symptoms. Pharmacogenomics J 16, 293–300 (2016). https://doi.org/10.1038/tpj.2015.48
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tpj.2015.48