Abstract
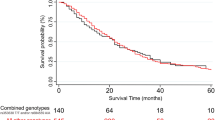
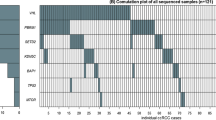
The 5-year survival rate for gastric adenocarcinoma (GA) remains only 40% and biomarkers to identify patients at high risk of tumor recurrence are urgently needed. Secreted protein acidic and rich in cysteine (SPARC) is an extracellular matrix glycoprotein that mediates cell matrix interactions, and upregulation of SPARC can promote tumor progression and metastasis. This study investigated whether single-nucleotide polymorphisms (SNPs) in SPARC impact the prognosis of GA. Blood or formalin-fixed, paraffin-embedded tissues were obtained from 137 GA patients at the University of Southern California and Memorial Sloan-Kettering Cancer Center medical facilities. DNA was isolated and five SNPs in the SPARC 3′-untranslated region (UTR) were evaluated by DNA sequencing or PCR-restriction fragment length polymorphism. Associations between SNPs and time to tumor recurrence (TTR) were analyzed using Kaplan–Meier curves, log-rank tests, and likelihood-ratio test within logistic or Cox regression model as appropriate. Patients carrying at least one G allele of the SPARC rs1059829 polymorphism (GG, AG) showed a median TTR of 3.7 years compared with 2.1 years TTR for patients with AA (hazard ratio (HR) 0.57; P=0.033). In a multivariate analysis adjusted for T and N category as covariates and stratified by race, hospital and chemotherapy, patients with at least one SPARC rs1059829 G allele (GG, AG) remained significantly associated with superior TTR than patients with AA genotype (adjusted P=0.026). In addition, patients harboring the G-A-A haplotype had the highest risk of tumor recurrence (HR 1.892; adjusted P=0.016). Our findings suggest that SPARC 3′-UTR SNPs may be useful in predicting GA patients at increased risk of recurrence.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others

References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D . Global cancer statistics. CA Cancer J Clin 2011; 61: 212–236.
Cunningham D, Allum WH, Stenning SP, Thompson JN, Van de Velde CJ, Nicolson M et al. Perioperative chemotherapy versus surgery alone for resectable gastroesophageal cancer. N Engl J Med 2006; 355: 11–20.
Hartgrink HH, van de Velde CJ, Putter H, Bonenkamp JJ, Klein Kranenbarg E, Songun I et al. Extended lymph node dissection for gastric cancer: who may benefit? Final results of the randomized Dutch gastric cancer group trial. J Clin Oncol 2004; 22: 2069–2077.
Sage H, Johnson C, Bornstein P . Characterization of a novel serum albumin-binding glycoprotein secreted by endothelial cells in culture. J Biol Chem 1984; 259: 3993–4007.
Porter PL, Sage EH, Lane TF, Funk SE, Gown AM . Distribution of SPARC in normal and neoplastic human tissue. J Histochem Cytochem 1995; 43: 791–800.
Bradshaw AD, Sage EH . SPARC, a matricellular protein that functions in cellular differentiation and tissue response to injury. J Clin Invest 2001; 107: 1049–1054.
Raines EW, Lane TF, Iruela-Arispe ML, Ross R, Sage EH . The extracellular glycoprotein SPARC interacts with platelet-derived growth factor (PDGF)-AB and -BB and inhibits the binding of PDGF to its receptors. Proc Natl Acad Sci USA 1992; 89: 1281–1285.
Robert G, Gaggioli C, Bailet O, Chavey C, Abbe P, Aberdam E et al. SPARC represses E-cadherin and induces mesenchymal transition during melanoma development. Cancer Res 2006; 66: 7516–7523.
Shi Q, Bao S, Maxwell JA, Reese ED, Friedman HS, Bigner DD et al. Secreted protein acidic, rich in cysteine (SPARC), mediates cellular survival of gliomas through AKT activation. J Biol Chem 2004; 279: 52200–52209.
Yunker CK, Golembieski W, Lemke N, Schultz CR, Cazacu S, Brodie C et al. SPARC-induced increase in glioma matrix and decrease in vascularity are associated with reduced VEGF expression and secretion. Int J Cancer 2008; 122: 2735–2743.
Briggs J, Chamboredon S, Castellazzi M, Kerry JA, Bos TJ . Transcriptional upregulation of SPARC, in response to c-Jun overexpression, contributes to increased motility and invasion of MCF7 breast cancer cells. Oncogene 2002; 21: 7077–7091.
Atorrasagasti C, Malvicini M, Aquino JB, Alaniz L, Garcia M, Bolontrade M et al. Overexpression of SPARC obliterates the in vivo tumorigenicity of human hepatocellular carcinoma cells. Int J Cancer 2010; 126: 2726–2740.
Tang MJ, Tai IT . A novel interaction between procaspase 8 and SPARC enhances apoptosis and potentiates chemotherapy sensitivity in colorectal cancers. J Biol Chem 2007; 282: 34457–34467.
Koukourakis MI, Giatromanolaki A, Brekken RA, Sivridis E, Gatter KC, Harris AL et al. Enhanced expression of SPARC/osteonectin in the tumor-associated stroma of non-small cell lung cancer is correlated with markers of hypoxia/acidity and with poor prognosis of patients. Cancer Res 2003; 63: 5376–5380.
Wang CS, Lin KH, Chen SL, Chan YF, Hsueh S . Overexpression of SPARC gene in human gastric carcinoma and its clinic-pathologic significance. Br J Cancer 2004; 91: 1924–1930.
Yin J, Chen G, Liu Y, Liu S, Wang P, Wan Y et al. Downregulation of SPARC expression decreases gastric cancer cellular invasion and survival. J Exp Clin Cancer Res 2010; 29: 59.
Zhao ZS, Wang YY, Chu YQ, Ye ZY, Tao HQ . SPARC is associated with gastric cancer progression and poor survival of patients. Clin Cancer Res 2010; 16: 260–268.
Podhajcer OL, Benedetti LG, Girotti MR, Prada F, Salvatierra E, Llera AS . The role of the matricellular protein SPARC in the dynamic interaction between the tumor and the host. Cancer Metastasis Rev 2008; 27: 691–705.
Jeung HC, Rha SY, Im CK, Shin SJ, Ahn JB, Yang WI et al. A randomized phase 2 study of docetaxel and S-1 versus docetaxel and cisplatin in advanced gastric cancer with an evaluation of SPARC expression for personalized therapy. Cancer 2011; 117: 2050–2057.
Mendell JT, Dietz HC . When the message goes awry: disease-producing mutations that influence mRNA content and performance. Cell 2001; 107: 411–414.
Zhou X, Tan FK, Reveille JD, Wallis D, Milewicz DM, Ahn C et al. Association of novel polymorphisms with the expression of SPARC in normal fibroblasts and with susceptibility to scleroderma. Arthritis Rheum 2002; 46: 2990–2999.
Delany AM, McMahon DJ, Powell JS, Greenberg DA, Kurland ES . Osteonectin/SPARC polymorphisms in Caucasian men with idiopathic osteoporosis. Osteoporos Int 2008; 19: 969–978.
Lee PH, Shatkay H . F-SNP: computationally predicted functional SNPs for disease association studies. Nucleic Acids Res 2008; 36 (Database issue): D820–D824.
Lee PH, Shatkay H . An integrative scoring system for ranking SNPs by their potential deleterious effects. Bioinformatics 2009; 25: 1048–1055.
Molinaro AM, Simon R, Pfeiffer RM . Prediction error estimation: a comparison of resampling methods. Bioinformatics 2005; 21: 3301–3307.
Generali D, Buffa FM, Berruti A, Brizzi MP, Campo L, Bonardi S et al. Phosphorylated ERalpha, HIF-1alpha, and MAPK signaling as predictors of primary endocrine treatment response and resistance in patients with breast cancer. J Clin Oncol 2009; 27: 227–234.
Ghadimi BM, Grade M, Difilippantonio MJ, Varma S, Simon R, Montagna C et al. Effectiveness of gene expression profiling for response prediction of rectal adenocarcinomas to preoperative chemoradiotherapy. J Clin Oncol 2005; 23: 1826–1838.
Hu-Lieskovan S, Vallbohmer D, Zhang W, Yang D, Pohl A, Labonte MJ et al. EGF61 polymorphism predicts complete pathologic response to cetuximab-based chemoradiation independent of KRAS status in locally advanced rectal cancer patients. Clin Cancer Res 2011; 17: 5161–5169.
Stephenson AJ, Scardino PT, Eastham JA, Bianco Jr FJ, Dotan ZA, DiBlasio CJ et al. Postoperative nomogram predicting the 10-year probability of prostate cancer recurrence after radical prostatectomy. J Clin Oncol 2005; 23: 7005–7012.
Rothman KJ . No adjustments are needed for multiple comparisons. Epidemiology 1990; 1: 43–46.
Lurje G, Husain H, Power DG, Yang D, Groshen S, Pohl A et al. Genetic variations in angiogenesis pathway genes associated with clinical outcome in localized gastric adenocarcinoma. Ann Oncol 2010; 21: 78–86.
Winder T, Bohanes P, Zhang W, Yang D, Power DG, Ning Y et al. GRP78 promoter polymorphism rs391957 as potential predictor for clinical outcome in gastric and colorectal cancer patients. Ann Oncol 2011; 22: 2431–2439.
Winder T, Ning Y, Yang D, Zhang W, Power DG, Bohanes P et al. Germline polymorphisms in genes involved in the CD44 signaling pathway are associated with clinical outcome in localized gastric adenocarcinoma. Int J Cancer 2011; 129: 1096–1104.
Maeng HY, Song SB, Choi DK, Kim KE, Jeong HY, Sakaki Y et al. Osteonectin-expressing cells in human stomach cancer and their possible clinical significance. Cancer Lett 2002; 184: 117–121.
Wewer UM, Albrechtsen R, Fisher LW, Young MF, Termine JD . Osteonectin/SPARC/BM-40 in human decidua and carcinoma, tissues characterized by de novo formation of basement membrane. Am J Pathol 1988; 132: 345–355.
Gilles C, Bassuk JA, Pulyaeva H, Sage EH, Foidart JM, Thompson EW . SPARC/osteonectin induces matrix metalloproteinase 2 activation in human breast cancer cell lines. Cancer Res 1998; 58: 5529–5536.
Thomas R, True LD, Bassuk JA, Lange PH, Vessella RL . Differential expression of osteonectin/SPARC during human prostate cancer progression. Clin Cancer Res 2000; 6: 1140–1149.
Tremble PM, Lane TF, Sage EH, Werb Z . SPARC, a secreted protein associated with morphogenesis and tissue remodeling, induces expression of metalloproteinases in fibroblasts through a novel extracellular matrix-dependent pathway. J Cell Biol 1993; 121: 1433–1444.
Sankpal NV, Moskaluk CA, Hampton GM, Powell SM . Overexpression of CEBPbeta correlates with decreased TFF1 in gastric cancer. Oncogene 2006; 25: 643–649.
El-Rifai W, Frierson Jr HF, Moskaluk CA, Harper JC, Petroni GR, Bissonette EA et al. Genetic differences between adenocarcinomas arising in Barrett′s esophagus and gastric mucosa. Gastroenterology 2001; 121: 592–598.
Desai N, Trieu V, Damascelli B, Soon-Shiong P . SPARC Expression Correlates with Tumor Response to Albumin-Bound Paclitaxel in Head and Neck Cancer Patients. Transl Oncol 2009; 2: 59–64.
Inoue M, Senju S, Hirata S, Ikuta Y, Hayashida Y, Irie A et al. Identification of SPARC as a candidate target antigen for immunotherapy of various cancers. Int J Cancer 2010; 127: 1393–1403.
Acknowledgements
This work was funded by the NIH Grant 5 P30CA14089-27I and the Dhont Family Foundation. It was performed in the Sharon A. Carpenter Laboratory at USC/Norris Comprehensive Cancer Center and in memory of David Donaldson.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Previous Presentation: ASCO 2010 (J Clin Oncol 28:15 s, 2010 (suppl; abstr 4054))
PowerPoint slides
Rights and permissions
About this article
Cite this article
Winder, T., Wilson, P., Yang, D. et al. An individual coding polymorphism and the haplotype of the SPARC gene predict gastric cancer recurrence. Pharmacogenomics J 13, 342–348 (2013). https://doi.org/10.1038/tpj.2012.11
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tpj.2012.11