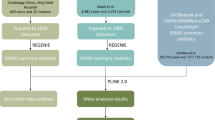
Abstract
The robustness of genome-wide association study (GWAS) results depends on the genotyping algorithms used to establish the association. This paper initiated the assessment of the impact of the Corrected Robust Linear Model with Maximum Likelihood Classification (CRLMM) genotyping quality on identifying real significant genes in a GWAS with large sample sizes. With microarray image data from the Wellcome Trust Case–Control Consortium (WTCCC), 1991 individuals with coronary artery disease (CAD) and 1500 controls, genetic associations were evaluated under various batch sizes and compositions. Experimental designs included different batch sizes of 250, 350, 500, 2000 samples with different distributions of cases and controls in each batch with either randomized or simply combined (4:3 case–control ratios) or separate case–control samples as well as whole 3491 samples. The separate composition could create 2–3% discordance in the single nucleotide polymorphism (SNP) results for quality control/statistical analysis and might contribute to the lack of reproducibility between GWAS. CRLMM shows high genotyping accuracy and stability to batch effects. According to the genotypic and allelic tests (P<5.0 × 10−7), nine significant signals on chromosome 9 were found consistently in all batch sizes with combined design. Our findings are critical to optimize the reproducibility of GWAS and confirm the genetic role in the pathophysiology of CAD.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Risch N, Merikangas K . The future of genetic studies of complex human diseases. Science 1996; 273: 1516–1517.
Dong S, Wang E, Hsie L, Cao Y, Chen X, Gingeras TR . Flexible use of high-density oligonucleotide arrays for single-nucleotide polymorphism discovery and validation. Genome Res 2001; 11: 1418–1424.
Lin S, Chakravarti A, Cutler DJ . Exhaustive allelic transmission disequilibrium tests as a new approach to genome-wide association studies. Nat Genet 2004; 36: 1181–1188.
Cutler DJ, Zwick ME, Carrasquillo MM, Yohn CT, Tobin KP, Kashuk C et al. High-throughput variation detection and genotyping using microarrays. Genome Res 2001; 11: 1913–1925.
Liu WM, Di X, Yang G, Matsuzaki H, Huang J, Mei R et al. Algorithms for large-scale genotyping microarrays. Bioinformatics 2003; 19: 2397–2403.
Di X, Matsuzaki H, Webster TA, Hubbell E, Liu G, Dong S et al. Dynamic model based algorithms for screening and genotyping over 100K SNPs on oligonucleotide microarrays. Bioinformatics 2005; 21: 1958–1963.
Rabbee N, Speed TP . A genotype calling algorithm for Affymetrix SNP arrays. Bioinformatics 2006; 22: 7–12.
BRLMM: an improved genotype calling method for the GeneChip human mapping 500K array set. http://media.affymetrix.com/support/technical/whitepapers/brlmm_whitepaper.pdf.
Carvalho B, Bengtsson H, Speed TP, Irizarry RA . Exploration, normalization, and genotype calls of high-density oligonucleotide SNP array data. Biostatistics 2007; 8: 485–499.
TheWellcome Trust Case Control Consortium. Genome-wide association study of 14 000 cases of seven common diseases and 3000 shared controls. Nature 2007; 447: 661–678.
Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003; 4: 249–264.
Lin S, Carvalho B, Cutler DJ, Arking DE, Chakravarti A, Irizarry RA . Validation and extension of an empirical Bayes method for SNP calling on Affymetrix microarrays. Genome Biol 2008; 9: R63.
Armitage P . Tests for linear trends in proportions and frequencies. Biometrics 1971; 11: 375–386.
Hong H, Su Z, Ge W, Shi L, Perkins R, Fang H et al. Assessing batch effects of genotype calling algorithm BRLMM for the Affymetrix GeneChip Human Mapping 500K array set using 270 HapMap samples. BMC Bioinformatics 2008; 9 (Suppl 9): S17.
Carvalho B, Louis TA, Irizarry RA . Quantifying uncertainty in genotype calls. Bioinformatics 2010; 26: 242–249.
Acknowledgements
We thank the Wellcome Trust Case–Control Consortium (WTCCC), UK, for providing us the original Affymetix 500 data with CAD patients and controls. We thank Benilton Carvalho and Rafael A Irizarry (Johns Hopkins University) for providing us advice on setting up CRLMM on our computer server. The views presented in this article do not necessarily reflect those of the US Food and Drug Administration.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Zhang, L., Yin, S., Miclaus, K. et al. Assessment of variability in GWAS with CRLMM genotyping algorithm on WTCCC coronary artery disease. Pharmacogenomics J 10, 347–354 (2010). https://doi.org/10.1038/tpj.2010.27
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tpj.2010.27
Keywords
This article is cited by
-
A TBX5 3′UTR variant increases the risk of congenital heart disease in the Han Chinese population
Cell Discovery (2017)
-
Variability in GWAS analysis: the impact of genotype calling algorithm inconsistencies
The Pharmacogenomics Journal (2010)