Abstract
Genome-wide association studies (GWAS) have yielded significant insights into the genetic architecture of myocardial infarction (MI), although studies in non-European populations are still lacking. Saudi Arabian cohorts offer an opportunity to discover novel genetic variants impacting disease risk due to a high rate of consanguinity. Genome-wide genotyping (GWG), imputation and GWAS followed by meta-analysis were performed based on two independent Saudi Arabian studies comprising 3950 MI patients and 2324 non-MI controls. Meta-analyses were then performed with these two Saudi MI studies and the CardioGRAMplusC4D and UK BioBank GWAS as controls. Meta-analyses of the two Saudi MI studies resulted in 17 SNPs with genome-wide significance. Meta-analyses of all 4 studies revealed 66 loci with genome-wide significance levels of p < 5 × 10–8. All of these variants, except rs2764203, have previously been reported as MI-associated loci or to have high linkage disequilibrium with known loci. One SNP association in Shisa family member 5 (SHISA5) (rs11707229) was evident at a much higher frequency in the Saudi MI populations (> 12% MAF). In conclusion, our results replicated many MI associations, whereas in Saudi-only GWAS (meta-analyses), several new loci were implicated that require future validation and functional analyses.
Similar content being viewed by others
Introduction
Coronary artery disease (CAD) leading to myocardial infarction (MI) is a leading cause of mortality, and modifiable risk factors, including sedentary lifestyle, diet, and smoking, play major roles in disease risk1. While exogenous risk factors, including dyslipidaemia, type 2 diabetes (T2D), and hypertension, exacerbate disease progression, 40–60% of CAD susceptibility has been attributed to genetic factors2,3,4,5. Genome-wide association studies (GWAS) have yielded significant insights into the complex aetiology of CAD and MI, including the interplay of hundreds of genetic risk variants impacting phenotypic development, as well as CAD-independent variants that impact the risk of MI alone6. These genetic variants provide important insights into the molecular mechanisms underlying MI and can lead to potential downstream targets for therapeutic intervention. However, much work remains to be done to fully understand the complex interplay between genetic and environmental factors in the development and progression of CAD and MI.
Large international consortia, including the UK Biobank (UKBB), Million Veteran Program (MVP), Coronary ARtery DIsease Genome-Wide Replication and Meta-Analysis (CARDIoGRAM), and Coronary Artery Disease (C4D) Genetics Consortia studies, have provided large-scale population-based cohorts to study the genetic underpinnings of CAD and/or MI7,8,9,10,11,12. However, most study participants in these large consortia are of European ancestry. The need for improved diversity of populations in genomic studies has been recognized, and while some CAD-related GWAS meta-analyses in other ancestral groups have been performed13,14,15, further large-scale studies are needed to evaluate the frequencies and consistency of risk allele effect sizes across different ancestries and to assess linkage disequilibrium, which can vary substantially across genetic ancestries15.
Performing GWAS in Saudi Arabian populations offers a unique opportunity to discover novel genetic variants impacting disease risk, as there is a high rate of consanguinity among tribal pedigrees, leading to a higher frequency of rare genetic variants due to increased levels of shared ancestry. Furthermore, undetected or untreated CAD is a significant health and financial burden in Saudi Arabia, with community-based epidemiological studies reporting a prevalence of CAD of approximately 55 cases per 1000 individuals in 30- to 70-year-old adults16,17,18.
In this study, genome-wide genotyping (GWG), imputation and GWAS followed by meta-analysis were performed based on two independent Saudi Arabian studies comprising 3950 MI patients and 2324 non-MI controls. Meta-analyses were then performed with the two Saudi MI studies together with the CardioGRAMplusC4D and UK BioBank GWAS, which comprised an additional 56,278 MI patients and 577,716 non-MI controls.
Materials and methods
Patient sampling and phenotyping
Saudi MI Study 1
From 2019 to 2020, samples and data from consecutive subjects with MI visiting the Cardiology Clinics, King Fahd Hospital of the University, Al-Khobar, and King Fahd Hospital, Alhafof, Saudi Arabia, were collected for inclusion in this study. Participants ranged in age from 25 to 66 and were clinically diagnosed with MI at the time of recruitment. Clinical diagnosis of MI was derived according to the fourth universal definition of MI19. The phenotypic data of all subjects were reviewed by a cardiologist consultant to verify uniformity among sites and eligibility according to study criteria. Eligibility for each of the individual cases was reviewed by the consultant committee and assessed for inclusion. For secondary analyses, T2D and hypertension were defined using WHO criteria; LDL, HDL, total cholesterol and troponin I were determined using Direct LDL-, Ultra HDL-, Cholesterol- and STAT High Sensitive Troponin I-Alinity c Reagent kits (Abbott, Wiesbaden, Germany)20,21.
Saudi MI Study 2
Details of the MI patients and controls in this Saudi study are described in a 2016 GWAS of CAD/MI by Wakil et al.22. Patients with suspected CAD/MI based on coronary angiography and echocardiography (ECG) abnormalities at the Catheterization Centre of King Faisal Heart Institute, King Faisal Specialist Hospital and Research Centre, Riyadh (KFSHRC), Saudi Arabia, were evaluated and represented all five regions of the country. Changes in the biomarkers myoglobin, cardiac troponin T, pro-brain natriuretic peptide and pro-calcitonin were also assessed. Two experienced interventional cardiologists independently reviewed patient records for the presence of ischaemia as per recommendations of the Joint ESC/ACCF/AHA/WHF Task Force for the Redefinition of MI23. The exclusion criteria included major cardiac rhythm disturbances, history of cerebral vascular disease, neurological disorder, psychiatric illness, and substance abuse. Controls consisted of individuals from KFSHRC undergoing heart valvular disease surgery and subjects with chest pain but no significant coronary stenosis based on angiography. There were 3481 MI patients available after delineating MI from CAD-alone cases, with 2299 controls.
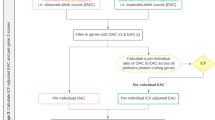
Details regarding the UK Biobank and CARDIoGRAMplusC4D Consortium GWAS MI patients (56,278 subjects), controls (577,716 non-MI subjects), phenotype ascertainment, and ancestry information are described elsewhere9. The study design for these analyses and details of how the datasets were combined is also depicted in the flowchart shown in Fig. 1.
For the Saudi MI Study 1, ethical approval was obtained from the Imam Abdulrahman Bin Faisal University Institutional Review Board (IRB) committee (IRB-2019-01-104), and the study was conducted according to the ethical principles of the Declaration of Helsinki and Good Clinical Practice guidelines. Informed written consent in English, with a verified translation in Arabic, was obtained from all participants in accordance with the IRB rules. The Saudi MI Study 2 protocol was approved by the Institutional Review Board (IRB) of the King Faisal Specialist Hospital and Research Centre. Summary-level GWAS datasets for the UK BioBank and CardioGRAMplusC4D were downloaded through a resource database outlined in Hartiala et al.9.
Generation of genotype data and imputation
Saudi MI Study 1
Peripheral blood samples were collected in EDTA tubes and stored at 4 °C before extraction of genomic DNA using Gentra Puregene Blood kits (Qiagen, Maryland, USA) according to the manufacturer’s protocol. DNA concentrations and purity were estimated by fluorometry using a NanoDrop 2000 Spectrophotometer (Thermo Fisher, MA, USA) and were diluted to 20 ng/µl. GWG was then performed using the Infinium Global Screening Array v3.0 (Illumina, CA, USA), which captures 654,027 SNPs or monomorphic/rare variants. Genotype data were clustered using Illumina GenomeStudio software, and standard quality control (QC) was performed using PLINK24. Normalized intensities for all samples were generated using optiCall clustering25. Raw genotypes were imputed using the 1000 Genomes Project (1KGP) v3 multiethnic reference panel through the Michigan Imputation Server26. The genotype data were subjected to QC with variants with < 90% missingness and consistency against the Haplotype Reference Consortium (HRC) reference panel for strand, reference/alternative alleles, SNP names and genome build positions. Furthermore, the imputed data were subjected to QC to retain variants with imputation INFO scores of R2 > 0.3 using Minimac, a 99% genotyping and sample call rate, and minor allele frequency (MAF) > 0.0127. Variants with a Hardy–Weinberg equilibrium (HWE) p value < 1 × 10−8 were excluded from the analyses. Principal component analyses (PCA) were computed using the fastPCA module in the eigensoft package28. The data points were then projected on the 1KGP populations29.
Saudi MI Study 2
DNA, GWG and QC are described in detail in Wakil et al.22. In brief, GWG was performed using Affymetrix Axiom Genome-Wide “ASI Array” (Asian population) with ~ 537,800 directly genotyped SNPs passing QC filtering. CARDIoGRAMplusC4D and UKBioBank GWAS data and imputation are fully described in Hartiala et al.9. This data was also imputed to 1000 Genomes dataset using Michigan Imputation server26.
Statistical analyses
Meta-analyses of GWAS: The variants passing QC for imputed dosage data were used to perform genome-wide association analyses for MI patients and controls. To account for the relatedness in the dataset, the analyses for Saudi studies 1 and 2 were performed using REGENIE30. Supplementary Fig. 1 illustrates the Manhattan and QQ plot for Saudi study 1 GWAS analyses. The associations were adjusted for age, sex, and the first 4 principal components. Two GWAS meta-analyses were performed to discover MI loci. First, a meta-analysis of Saudi MI studies 1 and 2 was conducted using PLINK 2.0 as shown in supplementary Fig. 2. Second, a meta-analysis of Saudi MI studies 1 and 2 was performed with the CARDIoGRAMplusC4D and UK Biobank MI datasets using PLINK 2.031.
Results
Study population characteristics
Table 1 summarizes the demographic characteristics of the two Saudi cohorts included in this study. In both cohorts, there were more subjects with MI represented compared to controls having no MI. Saudi MI Study 1 included 469 patients (95%) and 25 controls (5%), whereas Saudi MI Study 2 included 3481 (60%) patients and 2299 controls (40%). Overall, there were more men than women represented in the study; the male to female ratio in both cohorts was ~ 70% to 30%. Both sexes were equally represented in the control group of Study 2. Study 1 had a balanced median age of 55 (47, 63) years for the patients and 54 (44, 64) years for the controls, while Study 2 was represented by a larger distribution of ages with a median age of 60 (51, 69) for patients and 48 (35, 59) for controls. BMI measurements were not available in 4–10% of study subjects, but of those measured, the median BMI was slightly higher in Study 1 {29.3 (25.8, 32.7) for the patients and 30.1 (27.4, 35.3) for the controls} than in Study 2 {28.9 (25.6, 32.5) for the patients and 28.6 (24,5, 33.4) for the controls}. In Study 2, the patients with MI had much higher counts of hypertension (81%) than those in Study 1 (33%).
Replication of previously reported MI risk loci
GWAS meta-analyses of Saudi MI Studies 1 and 2 only
Meta-analyses of 3950 MI patients and 2324 controls from Saudi MI Study 1 and 2 resulted in 17 SNPs (6 loci) reaching genome-wide significance. The Manhattan plot for Saudi data meta-analyses is shown in Supplementary Fig. 2. Supplementary Table 1 shows the Quality control and Quality assurance metrics for the SNP filtering for: the two Saudi MI studies. The meta-analysis summary statistics of Study 1 and 2 signals for p < 0.001 are shown in Supplementary Table 2. We tested for replication of eight MI-associated SNPs from the Wakil et al. original GWAS paper from which Study 2 cases and controls were derived, of which 3 SNPs were of genome-wide significance and 5 additional SNPs had a suggestive p value of < 1 × 10–522. Seven out of eight SNPs from Wakil et al. were replicated in this study at the Bonferroni threshold (p value ≤ 0.05/8 = 0.006)22. The loci for these SNPs are linked to the genes RNF13 (rs41411047), PDZD2 (rs32793), ITGA1 (rs16880442), CDKN2A/B (rs2891168, rs10757274 and rs1333045), EIF4A3 (rs7211079), KCNE2 (rs998261), NDST2 (rs4691), and MRPS6 (rs28451064).
We also assessed the replication of 213 SNPs with genome-wide significance from the CARDIoGRAMplusC4D + UKBiobank meta-analysis by Hartiala et al.9. Three out of 213 SNPs from the Hartiala et al. study demonstrated replication in Saudi data 1 and data 2 meta-analyses9. Figure 2 also shows the three SNPs that were replicated from the 213 genome-wide significant SNPs from the Hartiala et al.9 meta-analysis. SNPs were considered significant for inclusion if they passed the Bonferroni calculation (p ≤ 0.05/213 = 0.0002).
Meta-analysis overview of Saudi MI Study 1 and 2 plus CARDIoGRAMplusC4D + UKBiobank GWAS: Synthesis view plot showing p values from the four analyses in the first panel and their odds ratio and confidence intervals for: Saudi MI Study 2 (Panel 2, blue); CARDIoGRAMplusC4D + UKBioBank (Panel 3, red); Saudi MI Study 1 + 2 (Panel 4, green) and Saudi MI Study 1 + 2 and CARDIoGRAMplusC4D + UKBioBank (Panel 5, yellow). The 10 replicated SNPs are shown on the y-axis.
GWAS meta-analyses of Saudi datasets + CardiogramplusC4D + UkBioBank
Figure 3 shows a Manhattan plot for 2523 association signals corresponding to 66 loci (mapping to 212 genes) observed above genome-wide significance (p < 5 × 10–8). The summary statistics of the Saudi MI Study 1 and 2 plus CARDIoGRA MplusC4D + UKBiobank GWAS for p < 0.001 are shown in Supplementary Table 3. The difference in the allele frequencies for all variants in these 66 loci among European and Saudi populations is reported in Supplementary Table 4. Fifteen variants showed a > 10% difference in allele frequencies, but the majority of the variants were common (> 10% MAF) in both populations. Notably, rs11707229 in SHISA5 has an MAF of 0.02 in European populations but an MAF of 0.12 in our Saudi MI populations. The results for all 66 significant genome-wide loci are reported in Table 2. Sixty-five out of 66 loci have been previously implicated to be significantly associated with MI based on the GWAS catalogue (downloaded on April 27, 2023). rs2764203 was previously identified to be nominally associated with MI (p = 1.0 × 10−7) but was found to be significantly associated with MI after the addition of the Saudi data in the meta-analyses (p = 2 × 10–8).
(A) Manhattan plot for MI genome-wide significant signals for the full meta-analysis comprising 3950 Saudi MI patients and 2324 controls and 56,278 MI patients and 577,716 controls from CARDIoGRAMplusC4D + UKBiobank. (B) Quantile‒Quantile (Q‒Q) plot for the meta-analyses (genomic inflation factor λ = 1.203). The horizontal red line indicates genome-wide significance (p value ≤ 5 × 10–8). SNPs coloured green have not been identified in previous studies.
Discussion
We performed GWG, imputation and GWAS on two independent Saudi Arabian studies comprising a total of 3950 MI patients and 2324 non-MI controls. Meta-analyses were performed with the two Saudi MI studies separately, resulting in 6 loci with genome-wide significance, and then combined with the CardioGRAMplusC4D and UK BioBank GWAS SNRPC studies, resulting in 66 loci with genome-wide significance. Our results replicated many MI associations, whereas in Saudi-only GWAS (meta-analyses), several new loci were implicated that require future validation and functional analyses.
The new genome-wide signal for MI from the meta-analyses of the four MI studies, rs2764203, is located approximately 4 kb from RP3-375P9.2 and ~ 20 kb from small nuclear ribonucleoprotein polypeptide C (SNRPC). Very little information is available from any previous studies of the long noncoding RNA RP3-375P9.2, apart from an association in a hepatocellular carcinoma (HCC) genomic and epigenomics study within early- and late-stage patients32. The RP3-375P9.2 lncRNA does not appear to be associated with MI in a recent pathway-based study33.
Small nuclear ribonucleoprotein polypeptide C (SNRPC) encodes one of the specific protein components of the U1 small nuclear ribonucleoprotein (snRNP) particle, which is needed for the formation of the spliceosome34,35. It is critical to the initiation and regulation of pre-mRNA splicing and is broadly expressed in most tissues, including heart tissues36. A recent study by Zhang et al. showed that SNRPC has the potential to promote the motility of hepatocellular carcinoma (HCC) cells via induction of epithelial-mesenchymal transition and to serve as a prognostic biomarker in HCC and predictor of immunotherapy responses37,38. SNRPC has also been shown to impact sex biases in systemic autoimmune diseases39.
The Shisa family member 5 (SHISA5) intronic association (rs11707229) in this MI study is interesting, as the observed minor allele frequency was > 12% in our overall Saudi population but has been reported to be approximately 2% in European populations, less than 1% in African populations and very rare in most Asian populations (http://www.ncbi.nlm.nih.gov/snp/rs11707229). SHISA5 is a member of the Shisa family, which is a single-transmembrane protein characterized by N-terminal cysteine-rich domains and proline-rich C-terminal regions. SHISA5 is located in the endoplasmic reticulum and the nuclear membrane and appears to have roles in numerous biological processes including regulation of autophagy, with involvement in p53-inducible pro-apoptosis in a caspase-dependent manner, is inducible by interferon and has an effect on the Wnt signalling pathway40,41,42,43. Associations of SHISA5 to date are largely limited to anthropometric, red cell characteristics and the glomerular filtration rate (GFR)44,45,46. Lakota and colleagues have previously described the upregulation of SHISA5 in mesenchymal stem cells (MSCs) transplanted into human subjects with ischaemic cardiomyopathy and controls and postulated that SHISA5 contributes to the death of cardiomyocytes via apoptosis after ischaemia–reperfusion injury47,48. Alternative splicing isoforms of different C-terminal isoforms of Shisa5 have been previously reported, and numerous variants impacting alternative splicing acceptor or donor sites appear likely to affect the specificity of its interactions41.
In conclusion, our study not only successfully replicated many known MI associations but also, through our Saudi-specific GWAS meta-analyses, identified several novel loci. These newly implicated loci, including RP3-375P9.2 lncRNA and the SNRPC gene, present exciting opportunities for future validation and functional analyses. Moreover, the association with SNPs in SHISA5, considering the distinct minor allele frequency differences between Saudi and European populations, offers potential insights into the high MI prevalence in Saudi Arabia. Such findings emphasize the critical need for genetic studies across diverse ancestral cohorts to ensure a holistic understanding of MI. This study has numerous limitations, including a limited number of MI controls, discordance in hypertension prevalence between the two Saudi MI studies and incomplete BMI measurements for a small number of the study subjects. Consanguineous populations such as the Saudi Arabian population offer an invaluable opportunity to explore rare and structural variants that are linked to disease. Future studies will involve more elegant methodologies to enhance the power of GWAS in consanguineous populations, inclusion of modifiable and nonmodifiable risk factors in predicting the risk of common diseases and strategic tools to analyse multiple genetic variants and exposure variables to uncover the hidden heritability of MI and concomitant comorbidities.
Data availability
The datasets generated during the current study are available in the European Variation Archive (EVA) repository (https://www.ebi.ac.uk/ebisearch/search?query=PRJEB59353&submit=Search&db=allebi&requestFrom=global-masthead) under the title "Genome-Wide Association Studies of Myocardial Infarction in Saudi Arabian Cohorts" with accession number PRJEB59353.
References
Benjamin, E. J. et al. Heart disease and stroke statistics-2019 update: A report from the American Heart Association. Circulation 139, e56–e528 (2019).
Yusuf, S. et al. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): Case-control study. Lancet 364, 937–952 (2004).
Marenberg, M. E., Risch, N., Berkman, L. F., Floderus, B. & de Faire, U. Genetic susceptibility to death from coronary heart disease in a study of twins. N. Engl. J. Med. 330, 1041–1046 (1994).
Mayer, B., Erdmann, J. & Schunkert, H. Genetics and heritability of coronary artery disease and myocardial infarction. Clin. Res. Cardiol. 96, 1–7 (2007).
Dai, X., Wiernek, S., Evans, J. P. & Runge, M. S. Genetics of coronary artery disease and myocardial infarction. World J. Cardiol. 8, 1–23 (2016).
Shah, S. et al. Genome-wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat. Commun. 11, 163 (2020).
Littlejohns, T. J., Sudlow, C., Allen, N. E. & Collins, R. UK Biobank: Opportunities for cardiovascular research. Eur. Heart J. 40, 1158–1166 (2019).
Gaziano, J. M. et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J. Clin. Epidemiol. 70, 214–223 (2016).
Hartiala, J. A. et al. Genome-wide analysis identifies novel susceptibility loci for myocardial infarction. Eur. Heart J. 42, 919–933 (2021).
Nikpay, M. et al. A comprehensive 1,000 genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 47, 1121–1130 (2015).
Consortium, C. A. D. C. D. G. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat. Genet. 43, 339–344 (2011).
Khera, A. V. et al. Genetic risk, adherence to a healthy lifestyle, and coronary disease. N. Engl. J. Med. 375, 2349–2358 (2016).
Lu, X. et al. Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nat. Genet. 44, 890–894 (2012).
Takeuchi, F. et al. Genome-wide association study of coronary artery disease in the Japanese. Eur. J. Hum. Genet. 20, 333–340 (2012).
Koyama, S. et al. Population-specific and trans-ancestry genome-wide analyses identify distinct and shared genetic risk loci for coronary artery disease. Nat. Genet. 52, 1169–1177 (2020).
Al-Nozha, M. M. et al. Coronary artery disease in Saudi Arabia. Saudi Med. J. 25, 1165–1171 (2004).
Osman, A. M., Alsultan, M. S. & Al-Mutairi, M. A. The burden of ischemic heart disease at a major cardiac center in Central Saudi Arabia. Saudi Med. J. 32, 1279–1284 (2011).
Walli-Attaei, M. et al. Variations between women and men in risk factors, treatments, cardiovascular disease incidence, and death in 27 high-income, middle-income, and low-income countries (PURE): A prospective cohort study. Lancet 396, 97–109 (2020).
Thygesen, K. et al. Fourth universal definition of myocardial infarction (2018). Circulation 138, e618–e651 (2018).
Hird, T. R. et al. Burden of diabetes and first evidence for the utility of HbA1c for diagnosis and detection of diabetes in urban black South Africans: The Durban Diabetes Study. PLoS ONE 11, e0161966 (2016).
Foldes, C. & Fisher, J. W. Hypertension. Ann. Intern. Med. 171, 525 (2019).
Wakil, S. M. et al. A genome-wide association study reveals susceptibility loci for myocardial infarction/coronary artery disease in Saudi Arabs. Atherosclerosis 245, 62–70 (2016).
Thygesen, K., Alpert, J. S., White, H. D., Infarction, J. E. A. A. W. T. F. f. t. R. o. M. Universal definition of myocardial infarction. J. Am. Coll. Cardiol. 50, 2173–2195 (2007).
Purcell, S. et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Shah, T. S. et al. optiCall: A robust genotype-calling algorithm for rare, low-frequency and common variants. Bioinformatics 28, 1598–1603 (2012).
Das, S. et al. Next-generation genotype imputation service and methods. Nat. Genet. 48, 1284–1287 (2016).
van Leeuwen, E. M. et al. Population-specific genotype imputations using minimac or IMPUTE2. Nat. Protoc. 10, 1285–1296 (2015).
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Auton, A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Mbatchou, J. et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet. 53, 1097–1103 (2021).
Chang, C. C. et al. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Kaur, H., Bhalla, S. & Raghava, G. P. S. Classification of early and late stage liver hepatocellular carcinoma patients from their genomics and epigenomics profiles. PLoS ONE 14, e0221476 (2019).
Ma, P., Li, J. & Cheng, M. Screening of biomarkers related to myocardial infarction based on the construction of a ceRNA regulation network. Anatol. J. Cardiol. 27, 274–281 (2023).
Yamamoto, K. et al. Isolation and characterization of a complementary DNA expressing human U1 small nuclear ribonucleoprotein C polypeptide. J. Immunol. 140, 311–317 (1988).
Libri, D., Duconge, F., Levy, L. & Vinauger, M. A role for the Psi-U mismatch in the recognition of the 5′ splice site of yeast introns by the U1 small nuclear ribonucleoprotein particle. J. Biol. Chem. 277, 18173–18181 (2002).
Fagerberg, L. et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell Proteomics 13, 397–406 (2014).
Zhang, Y. et al. SNRPC promotes hepatocellular carcinoma cell motility by inducing epithelial-mesenchymal transition. FEBS Open Bio 11, 1757–1770 (2021).
Cai, J., Zhou, M. & Xu, J. N6-methyladenosine (m6A) RNA methylation regulator SNRPC is a prognostic biomarker and is correlated with immunotherapy in hepatocellular carcinoma. World J. Surg. Oncol. 19, 241 (2021).
Lindén, M. et al. Sex influences eQTL effects of SLE and Sjögren’s syndrome-associated genetic polymorphisms. Biol. Sex Differ. 8, 34 (2017).
Lee, J. E., Kim, N., Jung, M., Mun, J. Y. & Yoo, J. Y. SHISA5/SCOTIN restrains spontaneous autophagy induction by blocking contact between the ERES and phagophores. Autophagy 18, 1613–1628 (2022).
Ergun, A. et al. Differential splicing across immune system lineages. Proc. Natl. Acad. Sci. USA 110, 14324–14329 (2013).
Bourdon, J. C., Renzing, J., Robertson, P. L., Fernandes, K. N. & Lane, D. P. Scotin, a novel p53-inducible proapoptotic protein located in the ER and the nuclear membrane. J. Cell Biol. 158, 235–246 (2002).
Kim, N. et al. Interferon-inducible protein SCOTIN interferes with HCV replication through the autolysosomal degradation of NS5A. Nat. Commun. 7, 10631 (2016).
Christakoudi, S., Evangelou, E., Riboli, E. & Tsilidis, K. K. GWAS of allometric body-shape indices in UK Biobank identifies loci suggesting associations with morphogenesis, organogenesis, adrenal cell renewal and cancer. Sci. Rep. 11, 10688 (2021).
Kichaev, G. et al. Leveraging polygenic functional enrichment to improve GWAS power. Am. J. Hum. Genet. 104, 65–75 (2019).
Liu, H. et al. Epigenomic and transcriptomic analyses define core cell types, genes and targetable mechanisms for kidney disease. Nat. Genet. 54, 950–962 (2022).
Lakota, J. Molecular mechanism of ischemia–reperfusion injury after myocardial infarction and its possible targeted treatment. Int. J. Cardiol. 220, 571–572 (2016).
Lakota, J., Dubrovcakova, M., Bohovic, R. & Goncalvesova, E. Intracoronary mesenchymal stem cell transplantation in patients with ischemic cardiomyopathy. Int. J. Cardiol. 176, 547–549 (2014).
Acknowledgements
The authors wish to thank King Abdulaziz City for Science and Technology for their financial support.
Funding
This work is supported by King Abdulaziz City for Science and Technology, Riyadh, Saudi Arabia, grant number 11-BIO1342-46. This is a government agency that had no role in the design, collection, analysis and interpretation of the data and writing of the manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization: A.K.A., A.M.A., R.A.A., M.A., M.A.J., A.A. and M.S.A. were involved in the design of the work, critical revision of the protocol, patient recruitment, data acquisition, experimental analysis, interpretation of the data and drafting of the manuscript. C.V., S.S.V., T.C., S.Y., G.M., B.P.K., B.K. and A.K.A. were involved in the design of the work, bioinformatic analysis, interpretation of the data and drafting of the manuscript. These authors contributed equally to this work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Al-Ali, A.K., Al-Rubaish, A.M., Alali, R.A. et al. Uncovering myocardial infarction genetic signatures using GWAS exploration in Saudi and European cohorts. Sci Rep 13, 21866 (2023). https://doi.org/10.1038/s41598-023-49105-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-49105-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.