Abstract
Triple negative breast cancers (TNBC) are aggressive tumors, with high rates of metastatic spread and targeted therapies are critically needed. We aimed to assess the prognostic and predictive value of a 3q 19-gene signature identified previously from lung cancer in a collection of 4,801 breast tumor gene expression data. The 3q gene signature had a strong association with features of aggressiveness such as high grade, hormone receptor negativity, presence of a basal-like or TNBC phenotype and reduced distant metastasis free survival. The 3q gene signature was strongly associated with lung metastasis only in TNBC (P < 0.0001, Hazard ratio (HR) 1.44, 95% confidence interval (CI), 1.31–1.60), significantly associated with brain but not bone metastasis regardless of TNBC status. The association of one 3q driver gene FXR1 with distant metastasis in TNBC (P = 0.01) was further validated by immunohistochemistry. In addition, the 3q gene signature was associated with better response to neoadjuvant chemotherapy in TNBC (P < 0.0001) but not in non-TNBC patients. Our study suggests that the 3q gene signature is a novel prognostic marker for lung and/or brain metastasis and a predictive marker for the response to neoadjuvant chemotherapy in TNBC, implying a potential role for 3q genes in the mechanism of organ-specific metastasis.
Similar content being viewed by others
Introduction
Breast cancer is the most frequent malignant disease in women worldwide. Patients with breast cancer are at risk of experiencing metastasis for their lifetime. It is not the primary tumor, but its metastases at distant sites such as lung, bone and liver that are the main cause of death in these patients1. Many gene expression studies have demonstrated that breast cancer is a clinically and molecularly heterogeneous disease comprising subtypes with distinct gene expression patterns and outcomes, making it difficult not only to cure this disease, but also to assess risk factors for metastasis2. A small number of expression profiling strategies have been successfully developed and validated for clinical use, some of which are now commercially available2. Nevertheless, uncertainty remains in the clinical use of many breast gene signatures. Moreover, new prognostic markers are urgently needed to identify patients who are at the highest risk for developing metastases in each subtype of breast cancer, which might enable oncologists to begin tailoring treatment strategies3.
Amplification of the chromosomal region 3q26-29 is the most frequent genomic alteration in primary squamous cell lung cancers and occurs in many other cancers including breast cancer4,5. Recent comprehensive genomic studies in breast cancer reveal that gene copy number (CN) changes correlated with mRNA subtype including characteristic loss of 5q and gain of 3q, 10p in basal-like cancers and gain of 1q and 16q loss in luminal tumors5. Earlier studies showed that gains of chromosome 3q, 9p, 11p and 11q and loss of 17p are associated with breast cancer recurrence6. In an effort to identify oncogenic drivers in lung cancer associated the 3q26-29 amplicon, we previously integrated genomic and gene expression analysis of 593 primary lung squamous carcinoma from seven independent datasets and identified 20 driver genes in this amplicon7. Some of these driver genes such as phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), fragile X mental retardation, autosomal homolog 1 (FXR1) and protein kinase C iota (PRKCI) have been implicated in the progression of lung or breast cancers8,9,10.
In this report, we interrogated the expression profiles of 4,801 breast tumors and report that this 3q gene expression signature is associated with poor outcomes in node negative breast cancer patients. We discovered that the 3q gene signature is strongly associated with the risk of developing lung and/or brain specific metastasis and the response to neoadjuvant chemotherapy in triple negative breast cancer (TNBC).
Results
3q-gene signature is associated with aggressive behavior of breast cancer
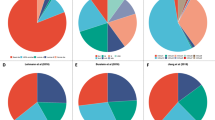
Among the 4,801 patients with breast cancer, we tested the association between the 3q 19-gene signature and established prognostic variables including age, grade, tumor size, lymph node status, and the expression status of ER, PR and HER2. The 3q gene signature was significantly associated with higher grade (P < 2.2e-16), larger tumor size (P = 0.005), ER- (P = 1.42e-08) and PR- status (P = 4.75e-10), but not associated with age (P = 0.07), HER2 status (P = 0.53) or lymph node involvement (P = 0.26) (Table 1 and Supplemental Fig. 1). The 3q gene signature was significantly associated with basal-like and luminal B subtypes of breast tumors (P < 2.2e-16, Fig. 1) or TNBC (P = 3.06e-12, Supplemental Fig. 1). Moreover, both univariable Cox analysis and a meta-analysis indicated that the high 3q gene signature was significantly associated with worse distant metastasis–free survival (DMFS) (P = 3.25e-05), but not recurrence-free survival (RFS) (P = 0.07), OS (P = 0.29) or DSS (P = 0.24) (Table 1 and Supplemental Figs 2 and 3).
3q gene signature is associated with distant metastasis in node-negative breast cancer
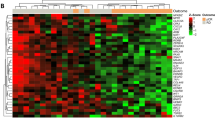
To identify determinants of recurrence independent of treatment, we focused on a subset of lymph node-negative breast tumor samples in three datasets (GSE11121, GSE2034 and TRANSBIG) obtained from 788 patients who did not receive systemic neoadjuvant or adjuvant treatment and had DMFS information available (Supplemental Table 2). The expression of the 3q gene signature was not different across three datasets (P = 0.99, Supplemental Figure 4) but was significantly associated with DMFS in all three cohorts (Supplemental Table 3). After adjustment for age, ER, PR and HER2 status, grade and/or tumor size, multivariable Cox analysis demonstrated that the 3q gene signature remained an independent prognostic marker for DMFS in GSE11121 (HR 1.83, 95% CI 1.41–2.37, P = 6.45e-06) and GSE2034 (HR 1.38, 95% CI 1.11–1.72, P = 0.004, Supplemental Table 4), respectively. In the TRANSBIG dataset, the 3q gene signature tended to be an independent prognostic marker for DMFS (HR 1.18, 95% CI 0.95–1.47, P = 0.13, Supplemental Table 4). In the combined two datasets (GSE11121 + TRANSBIG, P = 0.03) or three datasets (GSE2034 + GSE11121 + TRANSBIG, P = 0.0001), the association remained significant independent of variables available (Supplemental Table 4). The association also remained statistically significant when adjusted for PAM50 molecular subtypes (HR 1.38, 95% CI 1.23–1.55, P = 8.43e-08, Supplemental Table 5) or five well-established gene signatures such as the 70-gene, 76-gene, Genomic Grade Index (GGI), Oncotype Dx and proliferating cell nuclear antigen (PCNA)-117 gene signatures in pooled datasets (HR 1.24, 95% CI 1.14–1.36, P = 7.8e-07, Supplemental Table 6). Schmidt et al. reported a B-cell metagene as a prognostic factor for highly proliferating tumors, independent of the proliferation, ER, and T-cell metagenes in the same combined cohort11. Our study showed that the 3q gene signature was also significantly associated with worse DMFS independently of B-cell metagene (HR 1.26, 95% CI 1.17–1.36, P = 2.96e-09, Supplemental Table 7). Further analysis showed that the 3q gene signature was able to identify high risk patients in fast-proliferating tumors such as Basal-like and Luminal B subtypes of breast cancer (Fig. 2 and Supplemental Table 8), but not in HER2 or Normal-like subtypes after adjusting for B-cell and proliferation metagene signatures (Supplemental Table 9). Therefore, we concluded that high expression of the 3q genes is an independent prognostic factor for DMFS in Basal-like and Luminal B subtypes of node-negative breast cancer patients and has added prognostic value to known prognostic variables and known proliferation or immune response related gene signatures.
3q gene signature is an independent prognostic marker for triple negative breast cancer lung and brain metastasis
The ability to predict the risk and location of tumor recurrence could influence the surveillance for patients with a history of breast cancer. Therefore, we sought to ask whether the 3q gene signature (originally derived from lung cancer gene expression analysis) is associated with breast cancer metastasis to specific organ sites such as the lung. To test this hypothesis, we analyzed expression profiles of three independent cohorts of patients with breast cancer where the outcome information on lung, brain and bone specific metastasis was available (EMC344, GSE12276 and GSE2603, Supplemental Table 10). The univariable Cox analysis showed that the 3q gene signature was significantly associated with shorter lung metastasis–free survival in all three dataset (P = 0.0008, 8.58e-06, and 0.0032 for EMC344, GSE12276, and GSE2603, respectively), whereas only associated with shorter brain metastasis–free survival in two datasets (P = 0.00003 in EMC344 and P = 0.008 in GSE12276), and associated with shorter bone metastasis–free survival only in EMC344 (P = 0.013, Supplemental Table 11). When tested on the combined cohort (n = 618), the 3q gene signature was highly correlated to lung (P < 2.0e-16, HR 1.74, 95% CI 1.60 to 1.9) and brain metastasis (P = 4.79e-11, HR 1.99, 95% CI 1.62 to 2.45) but not bone metastasis (P = 0.71) (Supplemental Table 11). The multivariable Cox model further showed that the 3q gene signature was an independent predictor of metastases to lung (P < 2.0e-16, HR 1.58, 95% CI 1.42 to 1.76) or brain (P = 0.001, HR 1.61, 95% CI 1.21 to 2.13), after adjusting for age, ER, PR, HER2 and lymph node status, five common variables in the combined cohort (Supplemental Table 12). In addition, the 3q gene signature was significantly associated with lung metastases in basal-like (P < 2.0e-16), HER2 (P = 0.01) and luminal A (P = 0.01) subtypes, whereas was associated with brain metastases in basal-like (P < 2.0e-16) and luminal B (P = 1.94e-14) subtypes (Table 2). Basal-like breast cancer is known to have a large overlap with triple negative breast cancer, a subtype associated with distant metastasis. Further multivariable Cox model indicated that the 3q gene signature remained strongly associated with the risk of development of lung metastasis in TNBC (P = 8.48e-13, HR 1.44, 95% CI 1.31to 1.60) or basal like (P = 1.86e-07, HR 1.47, 95% CI 1.27 to 1.70) breast cancer patients after adjusting for age, node status and five known gene signatures (including two proliferating related signatures and three reported lung specific metastasis gene signatures), but not in non-TNBC breast cancer patients (P = 0.09, Table 2 and Supplemental Table 13). In contrast, the 3q gene signature was significantly associated with brain metastasis in both TNBC (P = 0.02) and non-TNBC patients (P = 8.46e-10, Table 2 and Supplemental Table 14). Together, these data suggest that the 3q gene signature is an independent prognostic marker for lung and brain specific metastasis in breast cancer, especially for lung metastasis in TNBC.
3q gene signature is associated with response to neoadjuvant chemotherapy
Among 4,801 patients, there are 1,058 patients in four datasets (GSE16446, GSE25066, GSE20194 and GSE20271) who had information on the response to neoadjuvant chemotherapy (Supplemental Table 15). All these patients received anthracycline (Epirubicin) monotherapy (GSE16446), taxane-anthracycline based (GSE25066) or paclitaxel followed by 5-fluorouracil, doxorubicin and cyclophosphamide (FAC) (GSE20194 and GSE20271) based neoadjuvant chemotherapy. In the combined dataset, multiple logistic regression analysis showed that the 3q gene signature was significantly associated with pathological complete response (pCR) after adjusting for clinical variables known to be associated with pCR including grade, status of ER, PR, HER2 and node (P < 0.0001, OR = 1.32, 95% CI 1.24 to 1.41, Supplemental Table 16). Further analysis showed that the 3q gene signature was associated with better pCR in TNBC (P < 0.0001, OR = 1.50, 95% CI 1.33 to 1.71) but worse pCR in non-TNBC patients (P = 0.35), after adjusting for known gene signatures and clinical variables (Supplemental Table 17). These data suggest that the 3q gene signature is an independent predictive biomarker for better response to neoadjuvant chemotherapy in TNBC.
FXR1 protein overexpression predicts distant metastasis in TNBC
We previously identified FXR1 as a novel cancer gene that is associated with poor outcomes in multiple human cancers including lung and breast9. FXR1 was most significantly associated with lung metastasis only in TNBC (Supplemental Table 18), therefore we elected to validate this finding using immunohistochemistry on 69 breast tumors (Fig. 3 and Supplemental Table 19). Univariable Cox analysis showed that elevated FXR1 expression was associated with DMFS in TNBC (P = 0.01, HR, 9.63, 95% CI, 1.7–43.96, Fig. 3B) but not in non-TNBC (Supplemental Table 20). Multivariable Cox analysis further indicates that FXR1 protein expression was associated with DMFS after adjusting for tumor stage in TNBC (P = 0.03, HR, 6.37, 95% CI, 1.2–33.7, Supplemental Table 20). Together, these results suggest that FXR1 is a novel prognostic biomarker for distant metastasis in TNBC. Nonetheless, organ specific metastasis information was not available on these patients and thus we were unable to further analyze this association.
Discussion
In a study including 27 publicly available gene expression datasets of clinically annotated breast cancer on a total of 4,801 patients, we found that the 3q 19-gene signature has strong association with breast cancer features of aggressiveness, reduced DMFS and response to neo-adjuvant chemotherapy, especially in triple negative breast cancer (TNBC). TNBC which are defined by a lack of ER, PR and HER2 expression, are still poorly characterized at the molecular level, and lack prognostic markers or targets for therapy12.
The majority of breast cancer deaths result from metastases rather than from direct effects of the primary tumor itself. The most common site of breast metastasis includes bone, lung and brain1. The prediction of a metastatic behavior remains a major challenge with only few metastasis-inducing proteins experimentally validated so far6. In a subset of 618 patients who had lung, brain and bone specific metastasis information available, the 3q 19-gene signature independently predicted breast cancer to metastasize to the lung or brain but not bone in TNBC. While gene signatures have been associated with breast outcome13, gene signatures that significantly associate with specific organ metastases are very limited6. Many gene signatures are not able to predict organ specific metastasis when adjusted for molecular subtype. For instance, the LMS 6-gene is highly correlated with basal subtype but its ability to predict lung metastasis is controversial14,15. Venet et al. showed that even randomly selected gene expression signatures could be significantly associated with breast cancer outcome, due to a high correlation with proliferation16. In contrast, the 3q gene signature is able to predict lung and brain metastasis in basal and luminal A/B. Furthermore, when compared with other lung metastasis signatures or well-studied proliferation signatures, the 3q gene signature maintained significance especially in TNBC. Likewise, in the setting of neoadjuvant chemotherapy, we found that the 3q gene signature was associated with pathological complete response (pCR) in TNBC patients. Together, the 3q gene signature provides additional prognostic information beyond the previously reported organ metastasis signatures, shedding new light on the possible biological processes relevant for predicting outcome or response to therapy in TNBC.
To date molecular mechanisms underlying breast cancer metastasis to the lungs and brain are still poorly understood. Our study highlights the molecular pathways possibly driven by 3q genes that might contribute to the organ preference of breast tumor cells for lung or brain. There have been studies showing that basal-like breast tumors represent a unique molecular entity strikingly similar to squamous cell lung cancers17, supporting clinical trials testing immune checkpoint agents in basal-like breast cancer as well as TNBC patients. We previously found the 3q gene signature is inversely associated with the suppressed immune response pathway in squamous carcinoma of the lung18. Whether and how 3q genes contribute to the regulation of immune pathway in human cancer warrants further study. Moreover, none of our 3q 19 genes overlaps with known gene signatures tested in this study, arguing a lack of efficacy of using these known gene signatures as prognostic factors in TNBC patients. Although the exact role of these 19 genes in breast cancer metastasis is unclear, 12 genes including FXR1 have been studied functionally or mechanistically in human cancers including lung and breast (Supplemental Table 21)9,19,20,21,22,23,24,25. For instance, PIK3CA is the most well-studied oncogene among our 3q gene signature due to its high mutation rate in human cancer, especially in breast cancer23. However, the prognostic value of PIK3CA mutation status in breast cancer is controversial23. PIK3CA mutation is associated with ER + and PR + breast cancer. A PIK3CA-mutated gene signature actually predicted better outcome in ER + breast cancer26. In contrast, our study showed that PIK3CA expression is significantly associated with lung or brain metastasis in TNBC (Supplemental Table 18). An analysis using TCGA breast cancer datasets showed that PIK3CA expression was indeed associated with copy number but not mutation (Supplemental Figure 5). It is PIK3CA amplification rather than PIK3CA mutation status that is associated with basal-like breast cancer (Supplemental Table 22). Early clinical trials also showed that PIK3CA mutations do not result in a dramatic responses to PI3K inhibitors23. Collectively, the significance of genotyping PIK3CA in clinical practice and targeted therapies based on the PIK3CA mutation in breast cancer remains unclear. Our study further argues that PIK3CA amplification or PIK3CA overexpression is more relevant than PIK3CA mutation status in breast cancer.
PRKCI is another well-known oncogene in human cancers and has been proposed to be a novel therapeutic target27. A recent study specifically demonstrated that PRKCI signaling promotes TNBC cell growth and metastasis through NF-κB pathway which could be regulated by TGFβ and IL1β25. We previously reported a new oncogenic role for RNA binding protein FXR1 linking to PRKCI and ECT2 in NSCLC9. In this study, multivariable cox analysis indicated that among 19 3q genes, FXR1 was actually most specifically associated with lung but not brain metastasis in TNBC (Supplemental Table 18). Therefore, we postulate that FXR1 is a novel prognostic biomarker specific for lung metastasis in TNBC and further validated the association of FXR1 protein with distant metastasis in TNBC using IHC on 69 breast tumors. We were not able to analyze organ specific metastasis due to the lack of such information on this small cohort of patients, a limitation we acknowledge. Future studies on a larger cohort are warranted.
Although the possible role of the 3q genes in specific steps of the metastatic process needs to be functionally validated, our study suggests an unrecognized role of certain 3q genes or gene interaction in the 3q amplicon in the basal cell type of human cancers and TNBC. A better knowledge of their function might lead to new insights into the mechanisms of disease progression and to the development of new predictive markers of metastatic behavior in breast cancer.
Methods
Breast tumor microarray datasets
We interrogated gene expression profiles from a total of 4,801 breast cancer patients in 27 publically available breast cancer datasets (Supplemental Table 1). 129 raw CEL files of E-TABM-158 (U133AAofAv2) were obtained from ArrayExpress and normalized using affy R package and Robust Multi-array Average (RMA) method28,29. Raw CEL files of GSE25066 (n = 508) were downloaded from the NCBI Gene Expression Omnibus (GEO) database and normalized using frozen Robust Multi-Array Analysis (fRMA) method, a procedure that allows one to pre-process microarrays individually or in small batches and to then combine the data into a single comparable dataset for further analyses30,31. The other 4,164 breast cancer gene expression profiles from 25 breast cancer datasets (GSE11121, GSE12093, GSE12276, GSE1456, GSE16391, GSE16446, GSE17705, GSE19615, GSE20194,GSE20271, GSE2034, GSE20685, GSE20711, GSE21653, GSE25066,GSE2603, GSE26971, GSE31519, GSE3494, GSE42568, GSE45255, GSE4922, GSE5327, GSE6532, GSE7390 and GSE9195) were obtained on the Affymetrix U133A or U133 Plus 2.0 expression array11,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57. These samples were collected from InsilicoDB database58 and normalized using fRMA method. A final combined dataset compiling 4,801 breast tumor samples was generated using inSilicoMerging R package and COMBAT algorithm59, an Empirical Bayes method to adjust for potential batch effects in the dataset. TRANSBIG (n = 731) cohort consists of samples from GSE3494, GSE4922, GSE6532 and GSE7390 and the replicates within the studies were removed49. EMC344 (n = 344) consists of GSE2034 and GSE5327. After integration of the datasets, all gene expression profiles were filtered to include 21,172 probes on the HG-U133A platform and were further collapsed to 13,129 gene symbols. TCGA breast mutation (wustl curated), copy number (gistic2_thresholded) and mRNA expression dataset (IllumninaHiSeq_RNASeqV2) were downloaded using the UCSC Cancer Genomics Browser60 as described before9.
In addition to the raw expression data, we also obtained clinical outcome data from a subset of the samples (Supplemental Table 1), which included data on overall survival (OS, n = 990), recurrence-free survival (RFS, n = 1,814), distant metastasis free survival (DMFS, n = 3,715), as well as disease specific survival (DSS, event of death from breast cancer, n = 614). Four datasets included data on response to neoadjuvant chemotherapy (n = 1,028). For samples not characterized by immunohistochemistry (IHC) in this cohort, the final calls for estrogen receptor (ER) progesterone receptor (PR) and epidermal growth factor receptor 2 (HER2) statuses was defined by analyzing mRNA expression bimodal cutoffs in pooled 4801 samples using a 2-component Gaussian mixture distribution model and parameters were estimated by maximum likelihood optimization in optim R package61. PAM50 (basal-like, luminal A, luminal B, HER2-enriched, and normal-like) molecular subtype was calculated using genefu R package62,63. Triple-negative breast cancer (TNBC) was defined as ER-, PR- and HER2- based on mRNA expression cutoff.
Breast tumor biospecimens
Breast cancer tissues were collected from surgical specimens through the Specialized Program of Research Excellence (SPORE) in breast at Vanderbilt University Medical Center in Nashville, Tennessee. All samples were reviewed by a pathologist (R.E). 69 breast cancer tissues contained in tumor tissue microarrays were used for the evaluation of FXR1 protein expression using immunohistochemistry as described before9. Clinical characteristics of these patients are described in Supplemental Table 19. All primary tumors were fresh-frozen, with efforts made to use samples with tumor content >70%. This study using human biospecimens was approved by the Vanderbilt University Internal Review Board and complied with all state, federal, and NIH regulations. Informed consent was obtained from all patients.
Immunohistochemistry study
Immunohistochemical staining and scoring were performed as previously described9. Briefly, the staining index was considered as the sum of the intensity score (0, no staining; 1 + , weak; 2 + , moderate; 3 + , strong) and the distribution score (0, no staining; 0.1, staining of 1%-9% of cells; 0.5, 10%-49% and 1 if >50% of cells). The final immunoreactivity H score was determined by multiplying the intensity and extent of positivity scores of stained cells, with the minimum score of 0 and a maximum score of 3. The median value of all the H scores was a priori chosen as the cutoff point for separating FXR1-high tumors from FXR1-low tumors.
Statistical analysis
Known gene signatures including MammaPrint (70-gene)64, Veridex (76-gene)51, GGI65 and Oncotype DX66 were calculated using genefu R package. 18-gene Lung metastasis signature (LMS) was defined as a linear combination of the gene-expression values weighted by their estimated regression coefficients obtained from univariable Cox proportional-hazard regression modeling as originally published46,52,67. The 3q 19-gene signature (19 out of 20 genes were mapped to the HG-U133A platform) and other published gene signatures including LMS 6-gene14, TGFβ pathway (152-gene)67 and PCNA (117-gene)16 were summarized to the mean expression within each sample and standardized to zero mean and unit variances before further analyses were performed. Pearson correlations between the signature indices were performed. Genes in signatures were mapped to the combined dataset by gene symbol.
Mann-Whitney-Wilcoxon test and Kruskal–Wallis one way analysis of variance (ANOVA) were used to compare the difference of gene signatures for groups of interest including dichotomized category clinical variables such as age (<50 or >50), tumor size (<2 cm or >2 cm), status of ER, PR, HER2 (positive vs negative), and lymph node (positive or negative), grade (grade III vs grade I/II), PAM50 subtypes, status of triple negative breast cancer (TNBC vs non-TNBC) and chemotherapy response (pathological complete response; pCR vs residual disease; RD). The distribution of gene signatures were visualized by using Box-and-whisker plots. Multiple logistic regression models were used to analyze the association between chemotherapy response and gene signatures as well as other clinical variables of interest. Cox proportional hazard (CPH) regression was used to analyze time to event data and the survival curve was calculated from Kaplan-Meier (KM) method. Robust sandwich covariance estimator was used for logistic and CPH regressions to account for the gene expression cluster (dataset) effect. The estimated odds ratio (OR), hazard ratio (HR) and 95% confidence intervals were provided to measure the effect of the association. In addition to the univariate CPH analysis, meta-analysis was conducted to confirm the finding in the univariate CPH regression. When 3q gene signature was used as dichotomized variable, the patients were divided into tertiles (high, intermediate and low) according to gene expression value. All p-values were based on two-sided tests and differences were considered statistically significant when p-value < 0.05. Analyses were performed using R version 3.3.1.
Additional Information
How to cite this article: Qian, J. et al. A 3q gene signature associated with triple negative breast cancer organ specific metastasis and response to neoadjuvant chemotherapy. Sci. Rep. 7, 45828; doi: 10.1038/srep45828 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Weigelt, B., Peterse, J. L. & van ‘t Veer, L. J. Breast cancer metastasis: markers and models. Nature reviews. Cancer 5, 591–602, doi: 10.1038/nrc1670 (2005).
Sotiriou, C. & Piccart, M. J. Taking gene-expression profiling to the clinic: when will molecular signatures become relevant to patient care? Nature reviews. Cancer 7, 545–553, doi: 10.1038/nrc2173 (2007).
Rodenhiser, D. I., Andrews, J. D., Vandenberg, T. A. & Chambers, A. F. Gene signatures of breast cancer progression and metastasis. Breast cancer research: BCR 13, 201, doi: 10.1186/bcr2791 (2011).
Qian, J. & Massion, P. P. Role of chromosome 3q amplification in lung cancer. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer 3, 212–215, doi: 10.1097/JTO.0b013e3181663544 (2008).
Cancer Genome. Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70, doi: 10.1038/nature11412 (2012).
Urquidi, V. & Goodison, S. Genomic signatures of breast cancer metastasis. Cytogenetic and genome research 118, 116–129, doi: 10.1159/000108292 (2007).
Wang, J. et al. Integrative genomics analysis identifies candidate drivers at 3q26-29 amplicon in squamous cell carcinoma of the lung. Clinical cancer research: an official journal of the American Association for Cancer Research 19, 5580–5590, doi: 10.1158/1078-0432.CCR-13-0594 (2013).
Karakas, B., Bachman, K. E. & Park, B. H. Mutation of the PIK3CA oncogene in human cancers. British journal of cancer 94, 455–459, doi: 10.1038/sj.bjc.6602970 (2006).
Qian, J. et al. The RNA binding protein FXR1 is a new driver in the 3q26-29 amplicon and predicts poor prognosis in human cancers. Proceedings of the National Academy of Sciences of the United States of America 112, 3469–3474, doi: 10.1073/pnas.1421975112 (2015).
Murray, N. R., Kalari, K. R. & Fields, A. P. Protein kinase Ciota expression and oncogenic signaling mechanisms in cancer. Journal of cellular physiology 226, 879–887, doi: 10.1002/jcp.22463 (2011).
Schmidt, M. et al. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer research 68, 5405–5413, doi: 10.1158/0008-5472.CAN-07-5206 (2008).
Penault-Llorca, F. & Viale, G. Pathological and molecular diagnosis of triple-negative breast cancer: a clinical perspective. Annals of oncology: official journal of the European Society for Medical Oncology 23 Suppl 6, vi19–22, doi: 10.1093/annonc/mds190 (2012).
Harrell, J. C. et al. Genomic analysis identifies unique signatures predictive of brain, lung, and liver relapse. Breast Cancer Res Treat 132, 523–535, doi: 10.1007/s10549-011-1619-7 (2012).
Landemaine, T. et al. A six-gene signature predicting breast cancer lung metastasis. Cancer research 68, 6092–6099, doi: 10.1158/0008-5472.CAN-08-0436 (2008).
Culhane, A. C. & Quackenbush, J. Confounding effects in “A six-gene signature predicting breast cancer lung metastasis”. Cancer research 69, 7480–7485, doi: 10.1158/0008-5472.CAN-08-3350 (2009).
Venet, D., Dumont, J. E. & Detours, V. Most random gene expression signatures are significantly associated with breast cancer outcome. PLoS computational biology 7, e1002240, doi: 10.1371/journal.pcbi.1002240 (2011).
Prat, A. et al. Genomic analyses across six cancer types identify basal-like breast cancer as a unique molecular entity. Scientific reports 3, 3544, doi: 10.1038/srep03544 (2013).
Qian, J., Zou, Y., Wang, J., Zhang, B. & Massion, P. P. Global gene expression profiling reveals a suppressed immune response pathway associated with 3q amplification in squamous carcinoma of the lung. Genomics data 5, 272–274, doi: 10.1016/j.gdata.2015.06.014 (2015).
Litviakov, N. V. et al. Deletions of multidrug resistance gene loci in breast cancer leads to the down-regulation of its expression and predict tumor response to neoadjuvant chemotherapy. Oncotarget 7, 7829–7841, doi: 10.18632/oncotarget.6953 (2016).
Gao, S. et al. Dsh homolog DVL3 mediates resistance to IGFIR inhibition by regulating IGF-RAS signaling. Cancer research 74, 5866–5877, doi: 10.1158/0008-5472.CAN-14-0806 (2014).
Zhao, J. et al. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene 32, 4814–4824, doi: 10.1038/onc.2012.494 (2013).
Cashman, R., Cohen, H., Ben-Hamo, R., Zilberberg, A. & Efroni, S. SENP5 mediates breast cancer invasion via a TGFbetaRI SUMOylation cascade. Oncotarget 5, 1071–1082, doi: 10.18632/oncotarget.1783 (2014).
Mukohara, T. PI3K mutations in breast cancer: prognostic and therapeutic implications. Breast cancer 7, 111–123, doi: 10.2147/BCTT.S60696 (2015).
Nait Achour, T. et al. Transcriptional repression of estrogen receptor alpha signaling by SENP2 in breast cancer cells. Molecular endocrinology 28, 183–196, doi: 10.1210/me.2013-1376 (2014).
Paul, A. et al. PKClambda/iota signaling promotes triple-negative breast cancer growth and metastasis. Cell death and differentiation 21, 1469–1481, doi: 10.1038/cdd.2014.62 (2014).
Cizkova, M. et al. PIK3CA mutation impact on survival in breast cancer patients and in ERalpha, PR and ERBB2-based subgroups. Breast cancer research: BCR 14, R28, doi: 10.1186/bcr3113 (2012).
Parker, P. J., Justilien, V., Riou, P., Linch, M. & Fields, A. P. Atypical protein kinase Ciota as a human oncogene and therapeutic target. Biochemical pharmacology 88, 1–11, doi: 10.1016/j.bcp.2013.10.023 (2014).
Chin, K. et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer cell 10, 529–541, doi: 10.1016/j.ccr.2006.10.009 (2006).
Gautier, L., Cope, L., Bolstad, B. M. & Irizarry, R. A. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315, doi: 10.1093/bioinformatics/btg405 (2004).
Hatzis, C. et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. Jama 305, 1873–1881, doi: 10.1001/jama.2011.593 (2011).
McCall, M. N., Bolstad, B. M. & Irizarry, R. A. Frozen robust multiarray analysis (fRMA). Biostatistics 11, 242–253, doi: 10.1093/biostatistics/kxp059 (2010).
Zhang, Y. et al. The 76-gene signature defines high-risk patients that benefit from adjuvant tamoxifen therapy. Breast Cancer Res Treat 116, 303–309, doi: 10.1007/s10549-008-0183-2 (2009).
Bos, P. D. et al. Genes that mediate breast cancer metastasis to the brain. Nature 459, 1005–1009, doi: 10.1038/nature08021 (2009).
Pawitan, Y. et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast cancer research: BCR 7, R953–964, doi: 10.1186/bcr1325 (2005).
Desmedt, C. et al. The Gene expression Grade Index: a potential predictor of relapse for endocrine-treated breast cancer patients in the BIG 1-98 trial. BMC medical genomics 2, 40, doi: 10.1186/1755-8794-2-40 (2009).
Desmedt, C. et al. Multifactorial approach to predicting resistance to anthracyclines. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 29, 1578–1586, doi: 10.1200/JCO.2010.31.2231 (2011).
Symmans, W. F. et al. Genomic index of sensitivity to endocrine therapy for breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 28, 4111–4119, doi: 10.1200/JCO.2010.28.4273 (2010).
Sircoulomb, F. et al. Genome profiling of ERBB2-amplified breast cancers. BMC cancer 10, 539, doi: 10.1186/1471-2407-10-539 (2010).
Silver, D. P. et al. Efficacy of neoadjuvant Cisplatin in triple-negative breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 28, 1145–1153, doi: 10.1200/JCO.2009.22.4725 (2010).
Li, Y. et al. Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nature medicine 16, 214–218, doi: 10.1038/nm.2090 (2010).
Popovici, V. et al. Effect of training-sample size and classification difficulty on the accuracy of genomic predictors. Breast cancer research: BCR 12, R5, doi: 10.1186/bcr2468 (2010).
Tabchy, A. et al. Evaluation of a 30-gene paclitaxel, fluorouracil, doxorubicin, and cyclophosphamide chemotherapy response predictor in a multicenter randomized trial in breast cancer. Clinical cancer research: an official journal of the American Association for Cancer Research 16, 5351–5361, doi: 10.1158/1078-0432.CCR-10-1265 (2010).
Kao, K. J., Chang, K. M., Hsu, H. C. & Huang, A. T. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: implications for treatment optimization. BMC cancer 11, 143, doi: 10.1186/1471-2407-11-143 (2011).
Dedeurwaerder, S. et al. DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO molecular medicine 3, 726–741, doi: 10.1002/emmm.201100801 (2011).
Sabatier, R. et al. A gene expression signature identifies two prognostic subgroups of basal breast cancer. Breast Cancer Res Treat 126, 407–420, doi: 10.1007/s10549-010-0897-9 (2011).
Minn, A. J. et al. Genes that mediate breast cancer metastasis to lung. Nature 436, 518–524, doi: 10.1038/nature03799 (2005).
Filipits, M. et al. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clinical cancer research: an official journal of the American Association for Cancer Research 17, 6012–6020, doi: 10.1158/1078-0432.CCR-11-0926 (2011).
Rody, A. et al. A clinically relevant gene signature in triple negative and basal-like breast cancer. Breast cancer research: BCR 13, R97, doi: 10.1186/bcr3035 (2011).
Nagalla, S. et al. Interactions between immunity, proliferation and molecular subtype in breast cancer prognosis. Genome biology 14, R34, doi: 10.1186/gb-2013-14-4-r34 (2013).
Loi, S. et al. Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC genomics 9, 239, doi: 10.1186/1471-2164-9-239 (2008).
Wang, Y. et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365, 671–679, doi: 10.1016/S0140-6736(05)17947-1 (2005).
Minn, A. J. et al. Lung metastasis genes couple breast tumor size and metastatic spread. Proceedings of the National Academy of Sciences of the United States of America 104, 6740–6745, doi: 10.1073/pnas.0701138104 (2007).
Hoppe, S., Bier, F. F. & von Nickisch-Rosenegk, M. Rapid identification of novel immunodominant proteins and characterization of a specific linear epitope of Campylobacter jejuni. PloS one 8, e65837, doi: 10.1371/journal.pone.0065837 (2013).
Miller, L. D. et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proceedings of the National Academy of Sciences of the United States of America 102, 13550–13555, doi: 10.1073/pnas.0506230102 (2005).
Ivshina, A. V. et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer research 66, 10292–10301, doi: 10.1158/0008-5472.CAN-05-4414 (2006).
Loi, S. et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 25, 1239–1246, doi: 10.1200/JCO.2006.07.1522 (2007).
Desmedt, C. et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clinical cancer research: an official journal of the American Association for Cancer Research 13, 3207–3214, doi: 10.1158/1078-0432.CCR-06-2765 (2007).
Taminau, J. et al. inSilicoDb: an R/Bioconductor package for accessing human Affymetrix expert-curated datasets from GEO. Bioinformatics 27, 3204–3205, doi: 10.1093/bioinformatics/btr529 (2011).
Taminau, J. et al. Unlocking the potential of publicly available microarray data using inSilicoDb and inSilicoMerging R/Bioconductor packages. BMC bioinformatics 13, 335, doi: 10.1186/1471-2105-13-335 (2012).
Zhu, J. et al. The UCSC Cancer Genomics Browser. Nature methods 6, 239–240, doi: 10.1038/nmeth0409-239 (2009).
Karn, T. et al. Data-driven derivation of cutoffs from a pool of 3,030 Affymetrix arrays to stratify distinct clinical types of breast cancer. Breast Cancer Res Treat 120, 567–579, doi: 10.1007/s10549-009-0416-z (2010).
Parker, J. S. et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 27, 1160–1167, doi: 10.1200/JCO.2008.18.1370 (2009).
Gendoo, D. M. et al. Genefu: an R/Bioconductor package for computation of gene expression-based signatures in breast cancer. Bioinformatics 32, 1097–1099, doi: 10.1093/bioinformatics/btv693 (2016).
van ‘t Veer, L. J. et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415, 530–536, doi: 10.1038/415530a (2002).
Sotiriou, C. et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. Journal of the National Cancer Institute 98, 262–272, doi: 10.1093/jnci/djj052 (2006).
Paik, S. et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. The New England journal of medicine 351, 2817–2826, doi: 10.1056/NEJMoa041588 (2004).
Padua, D. et al. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell 133, 66–77, doi: 10.1016/j.cell.2008.01.046 (2008).
Acknowledgements
We thank Drs Carlos L. Arteaga, Melinda Sanders, Jennifer M. Giltnane for assisting the breast cancer tissue acquisition. We are also grateful for the datasets publically available made by scientific community including the TCGA Research Network (http://cancergenome.nih.gov/). The work is supported by the NIH grants R01 CA102353 and U01CA152662 to PPM.
Author information
Authors and Affiliations
Contributions
J.Q., H.C. and P.P.M. contributed to study design. J.Q. and J.X. contributed to data collection. R.E contributed to IHC study. J.Q. and H.C. contributed to data analysis. J.Q., H.C., A.B.C, I.A.M. and P.P.M. contributed to manuscript preparation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Qian, J., Chen, H., Ji, X. et al. A 3q gene signature associated with triple negative breast cancer organ specific metastasis and response to neoadjuvant chemotherapy. Sci Rep 7, 45828 (2017). https://doi.org/10.1038/srep45828
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep45828
This article is cited by
-
Basal–epithelial subpopulations underlie and predict chemotherapy resistance in triple-negative breast cancer
EMBO Molecular Medicine (2024)
-
The emerging role of RNA N6-methyladenosine methylation in breast cancer
Biomarker Research (2021)
-
Recurrence biomarkers of triple negative breast cancer treated with neoadjuvant chemotherapy and anti-EGFR antibodies
npj Breast Cancer (2021)
-
The RNA-binding protein FXR1 modulates prostate cancer progression by regulating FBXO4
Functional & Integrative Genomics (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.