Abstract
Photosynthetic activity is indispensable for plant growth and survival and it depends on the synthesis of plastidial isoprenoids as chlorophylls and carotenoids. In the non-mevalonate pathway (MEP), the 1-deoxy-D-xylulose-5-phosphate synthase 1 (DXS1) enzyme has been postulated to catalyze the rate-limiting step in the formation of plastidial isoprenoids. In tomato, the function of DXS1 has only been studied in fruits, and hence its functional relevance during plant development remains unknown. Here we report the characterization of the wls-2297 tomato mutant, whose severe deficiency in chlorophylls and carotenoids promotes an albino phenotype. Additionally, growth of mutant seedlings was arrested without developing vegetative organs, which resulted in premature lethality. Gene cloning and silencing experiments revealed that the phenotype of wls-2297 mutant was caused by 38.6 kb-deletion promoted by a single T-DNA insertion affecting the DXS1 gene. This was corroborated by in vivo and molecular complementation assays, which allowed the rescue of mutant phenotype. Further characterization of tomato plants overexpressing DXS1 and comparative expression analysis indicate that DXS1 may play other important roles besides to that proposed during fruit carotenoid biosynthesis. Taken together, these results demonstrate that DXS1 is essentially required for the development and survival of tomato plants.
Similar content being viewed by others
Introduction
Photosynthesis is a vital process for plant development and survival which is precisely regulated at several genetic, molecular and physiological levels during the plant growth cycle1. Photosynthetic pigments (chlorophylls and carotenoids) are a group of molecules that belong to a large class of compounds of different biochemical nature, the isoprenoids. Isoprenoids comprise a chemically and functionally wide heterogeneous group including more than 30,000 molecules2. In addition to photosynthesis, plants produce many isoprenoids whose functions are essential in developmental processes such as growth regulation (gibberellins, cytokinins, brassinosteroids and abscisic acid), defense mechanism (phytoalexins), membrane structure (sterols) and redox chemistry (plastoquinone, ubiquinone). Despite their diversity, all isoprenoids are formed from two common precursors, isopentenyl diphosphate (IPP) and its isomer dimethylallyl diphosphate (DMAPP)3. In higher plants, there are two distinct cellular compartmentalized pathways for the biosynthesis of IPP and DMAPP4,5,6,7: the cytosolic mevalonate (MVA) pathway, which is shared with fungi, yeast, and animals8,9, and the non-mevalonate pathway designated as the 2-C-methyl-D-erythritol-4-Phosphate (MEP) pathway, located in plastids and also occurring in eubacteria, green algae and protozoa10,11,12,13,14. The initial step of the MEP pathway, a transketolase-type condensation of pyruvate with D-glyceraldhyde-3-phosphate to form 1-deoxy-D-xylulose-5-phosphate (DXP), is catalyzed by DXP synthase (DXS)12. The cloning and characterization of the DXS gene was first described in Escherichia coli15. DXS homologous genes were subsequently cloned from several plant species, including peppermint16, pepper17, Arabidopsis18 and tomato19. Since first reported, the DXS gene has been studied in diverse plant species in order to gain deeper knowledge of the DXS family, and functional diversification has been suggested in this family due to the existence of at least three functionally specialized types of DXS, i.e. DXS1, DXS2 and DXS320,21,22. Despite high amino acid sequence homology among DXS proteins, mainly between DXS1 and DXS2, not all DXS-like genes have the same expression pattern during plant development. DXS1 has been proposed to be specifically involved in the synthesis of essential isoprenoids (such as photosynthetic pigments and phytohormones) in chloroplasts18,19. In turn, DXS2 is thought to be located in non-photosynthetic plastids, involved in the synthesis of specific isoprenoids as secondary metabolites related to mycorrhiza23. In addition, it has been suggested that DXS3 might participate in the synthesis of essential isoprenoids required at low levels, such as phytohormones20.
Many isoprenoids, carotenoids among them, have gained importance as biotechnological tools as they show beneficial effects on human health24,25. Since DXS1 is the key enzyme that limits output from the MEP pathway in plants19,26,27, changes in DXS1 expression may directly influence the content of photosynthetic pigments. Many experiments have therefore been carried out to enhance the level of some isoprenoids in model and crop species, mainly by the generation of DXS overexpression transgenic lines28,29. Indeed, constitutive expression of a bacterial DXS gene under the control of the CaMV 35S promoter in transgenic tomato lines did alter the isoprenoid end products in fruits, but not in leaves28. Similarly, the overexpression of DXS1 in Arabidopsis26 and the constitutive expression of the soybean DXS1 gene in tobacco transgenic plants29 produced increases in various isoprenoids, including total chlorophylls and carotenoids. However, results from DXS1 overexpression experiments differed among plant species, including tomato, indicating a complex regulation of DXS activity28. Additionally, characterization of DXS mutants has shed light on the key function of DXS proteins in plant development26,30. In Arabidopsis, DXS1 plays an essential role in chloroplast development during leaf cell maturation30,31, while DXS2 seems to have no DXS activity, but it has acquired another unknown biochemical function32. In tomato, the DXS1 function is required for carotenoid biosynthesis during fruit ripening19, while tomato DXS2 participates in the biosynthesis of isoprenoids in trichomes16,22,33 and of secondary metabolites16. Such results suggest that differences in the biochemical roles of DXS proteins seem to be related with variable functions in plant development and growth.
In this work, we report the identification and molecular characterization of the tomato white lethal seedling-2297 (wls-2297) mutant, which was isolated from the screening of a T-DNA mutant collection generated by an enhancer trapping gene construct. Seedlings of the wls-2297 mutant showed albino phenotype and were unable to develop any true leaves from the shoot apical meristem; as a consequence, albino seedlings died after reaching the fully expanded cotyledon stage. Expression and functional analyses demonstrated that these phenothypic alterations were caused by the loss-of-function of the DXS1 gene, indicating that, besides its role in fruit carotenoid biosynthesis, DXS1 activity is required at early stages of plant development and determines plant survival.
Results
The wls-2297 mutant showed significant alterations in pigment content and plant development
A tomato T-DNA mutant collection generated using an enhancer-trapping construct was screened for developmental traits in TG2 plants grown in soil. As a result, the wls-2297 mutant was selected due to the noteworthy albino phenotype of its cotyledons. After seed germination, mutant seedlings expanded cotyledons, but they did not fully elongate and never showed the characteristic green color associated with chlorophyll biosynthesis (Fig. 1a,b). Development of the shoot apical meristem was also arrested in the wls-2297 mutant, as a few days after the cotyledons emerged seedlings experienced premature senescence and died without developing any true leaves. In general, albino phenotypes commonly result from alterations in the photosynthetic process, either directly affecting chloroplast development and functionality, or indirectly, as a consequence of mutations that promote significant defects in plant development or metabolism31. In a first step toward determining the metabolic pathway affected in the wls-2297 mutant, seedlings were grown in vitro using a standard Murashige and Skoog (MS) culture medium supplemented with a high sugar concentration. Under these conditions, wls-2297 mutant seedlings survived 3–5 days longer than in soil but finally died; during this period, they developed slower than wild type (WT) seedlings and developed no true leaves (Fig. 1c,d).
One-week-old wild type (left) and wls-2297 seedlings (right) viewed from the side (a) and from above (b). Three-week-old wild type and mutant seedlings cultured in vitro on MS medium viewed from the side (c) and from above (d). Wild type (e) and mutant (f) cotyledons visualized using bright-field microscopy proving the lack of chloroplast in the mutant cells. (g) Chlorophyll and carotenoid content in wild type and wls-2297 seedlings. Error bars are the standard error value from two biological replicates. Asterisk indicates significant differences between wild type control plants and mutant plants (Fisher’s Least Significant Difference test, P < 0.05).
With a view to verifying whether the wls-2297 mutation is involved in chloroplast development, fresh cotyledon tissues were observed under bright field microscopy, which revealed that cells of wls-2297 cotyledons had an apparent lower density of chloroplasts than WT ones. This suggested decreased chlorophyll content of mutant cotyledons, in line with the pale green pigmentation of wls-2297 chloroplasts as compared with WT ones (Fig. 1e,f). Therefore, chlorophyll and carotenoid contents were measured in WT and mutant cotyledons. As expected, results showed that levels of both chlorophylls and carotenoids were significantly lower in wls-2297 mutant cotyledons (Fig. 1g), proving that the albino mutant phenotype was caused by a severe deficiency in photosynthetic activity.
Molecular cloning and characterization of the wls-2297 mutation
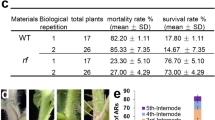
Genetic analysis carried out on a segregating TG2 progeny of 121 plants indicated that the wls-2297 mutant phenotype was inherited as a monogenic recessive trait (3:1 ratio, 84 WT:37 mutant; χ2 = 2.01; P = 0.16). This result agreed with that of Southern blot hybridization, which showed that a single T-DNA copy segregated in the wls-2297 TG2 family here analyzed (Fig. 2a). Moreover, 88 TG2 seedling plants were screened in vitro for kanamycin resistance, and 22 of them displayed a wls-2297 mutant phenotype. A complete co-segregation was found between wls-2297 mutant and kanamycin resistance phenotypes in this segregating population (1:2:1 ratio; 22 mutant and kanamycin resistant:41 WT and kanamycin resistant:25 WT and kanamycin susceptible; χ2 = 0.61; P = 0.74), indicating that a single T-DNA insertion was responsible for the albino phenotype showed by the wls-2297 mutant.
(a) Southern-blot hybridization of wild type (WT) and the wls-2297 TG1 plant using the FA-NPTII probe. (b) Genomic re-organization caused by the T-DNA integration on the wls-2297 genome. (c) PCR assays designed to detect the presence in wild type plant or the deletion in the wls-2297 mutant of the three POX genes, the DXS1 gene or (d) the T-DNA insertion in WT and the wls-2297 mutant. (e) Co-segregation analysis performed in a TG2 family segregating for the wls-2297 mutant phenotype. Cropped gels/blots are displayed in panels (a), (c), (d) and (e). Full-length gels/blots are included in Fig. S1.
The genomic region flanking the T-DNA insertion was cloned and the sequences obtained were compared with the reference sequence of tomato genome (https://solgenomics.net). Results showed that the T-DNA is located on chromosome 1, the right border (RB) of the T-DNA being located at position 76834.3 kb, 1528 bp downstream of a gene encoding a putative PEROXIDASE enzyme (POX, Solyc01g067850). In addition, the left border (LB) of the T-DNA is placed at position 76872.9 kb, 217 bp upstream from the DXS1 gene (Solyc01g067890) encoding for 1-deoxy-D-xylulose-5-phosphate synthase. According to this in silico analysis, the T-DNA insertion causes a 38.6 kb-deletion in the genome of the wls-2297 mutant, which affects four genes, three putative genes encoding for POX enzymes (POX24-Solyc01g067860, POX24-Solyc01g067870 and POX27-Solyc01g067880) and the DXS1 gene (Fig. 2b).
In order to demonstrate that the 38.6 kb-deletion was the only mutation promoted by the T-DNA insertion and that no other chromosome rearrangements (e.g. translocation) occurred during mutagenesis, specific primers of the four genes located on the 38.6 kb genomic region were designed and used in PCR experiments to confirm their absence in wls-2297 mutant plants. PCR products specific to the four reported genes from WT had the expected size and their sequences coincided with those reported in the tomato genome database (https://solgenomics.net), whereas no amplification products were obtained from the wls-2297 mutant DNA (Fig. 2c). On the other hand, a long-PCR assay was performed to amplify the region between the two previously cloned T-DNA flanking regions. The long-PCR product obtained from wls-2297 genomic DNA had the expected size of 5.2 kb (Fig. 2d) and its sequence confirmed the presence of a single T-DNA insertion causing a 38.6 kb-deletion in the genome of the wls-2297 mutant. A co-segregation analysis performed using allele-specific primers designed from both the T-DNA and the genomic flanking sequences tagged in the wls-2297 mutant revealed that the T-DNA insertion co-segregated with the wls-2297 mutant phenotype (Fig. 2e). Taken together, these results provide strong evidence as to the genetic and molecular nature of the wls-2297 mutant phenotype, which may be due to a 38.6 kb-deletion promoted by a single copy T-DNA insertion that affects three POX genes and the DXS1 gene.
Gene silencing experiments
Given the role of the DXS1 enzyme in isoprenoid biosynthesis through the MEP pathway18 and the pigment measurements in the present work, the DXS1 gene was considered the best candidate gene to be responsible for the wls-2297 mutant phenotype. In order to demonstrate this hypothesis, we generated DXS1 silencing lines in the cultivar Moneymaker using an RNA interference strategy (RNAi). After ploidy level determination, nine RNAi DXS1 diploid lines showing a significant reduction of DXS1 transcript levels (Fig. 3a) were further characterized under in vitro conditions. Interestingly, all DXS1 repressed lines showed an albino phenotype, and even developed young leaves when sucrose was added to the MS medium, allowing explant development (Fig. 3b,c). Moreover, neither RNAi DXS1 explants nor seedlings could be propagated since they died at early developmental stages. Therefore, the silencing of DXS1 leads to similar developmental changes to wls-2297 mutation, which provides strong evidence that the wls-2297 mutant phenotype was due to the lack of DXS1 gene function.
(a) Expression analysis of the DXS1 gene in the control (MM) and the RNAi DXS1 lines. (b) Top view and (c) lateral view of albino RNAi transformants compared to wild type (WT) non-transformed plants. Error bars show the standard error value of two biological and two technical replicates. Asterisk indicates significant differences between control and RNAi DXS1 lines (Fisher’s Least Significant Difference test, P < 0.05).
In addition, RNAi lines simultaneously silencing the three POX genes located in the deleted region were generated by using a gene construct that included a consensus sequence shared by the three POX messengers (Fig. S2b). Fifteen POX silencing diploid lines were selected for further analysis as they showed a significant decrease of POX transcripts ranging from 63 to 82% with respect to the control background (Fig. 4a). All of the RNAi POX lines developed normally, although plant vigor was slightly affected most likely due to peroxidase deficiency (Fig. 4b). Most importantly, none of these RNAi lines showed phenotype changes resembling those of the wls-2297 mutant (Fig. 1a–d), which indicates that POX genes affected by the T-DNA insertion are not involved in the mutant phenotype here reported.
(a) Expression analysis of the POX genes in the wild type control (MM, cv. Moneymaker) and two representative RNAi POX lines (RNAi POX 13 and RNAi POX 23). (b) Phenotype shown by wild type (WT) control non-transformed plants (left) and transgenic plant silencing the POX genes by RNAi (right). Error bars show the standard error value of two biological and two technical replicates. Asterisk indicates significant differences between control and RNAi POX lines (Fisher’s Least Significant Difference test, P < 0.05).
Expression patterns of the DXS gene family
Tomato DXS1 gene, which is deleted in the wls-2297 mutant, is located on chromosome 1; according to the tomato genome database (https://solgenomics.net), it has a genome size of 4.2 kb and includes 10 exons and 9 introns. In addition to DXS1, two other tomato genes belonging to the DXS family have been identified, i.e. DXS2, located on chromosome 11 (Solyc11g010850) and DXS3 located on chromosome 8 (Solyc08g066950), both also composed of 10 exons and 9 introns. The ORFs from the three genes, DXS1, DXS2, and DXS3 code for proteins of 719, 714 and 709 amino acid residues, respectively (Fig. S3). Phylogenetic analysis performed with the three tomato DXS proteins as well as representative DXS protein of diverse plant species showed three independent groups of DXS proteins conserved among plants, i.e. DXS1, DXS2 and DXS3 clades (Fig. S4), a result which agrees with those previously described by Hofberger et al.34. In addition, the overall similarity among the three DXS tomato proteins ranges between 59 and 72%, DXS3 being the most divergent. The three proteins have conserved the three characteristic domains of the DXS family, these are an N-terminal thiamine pyrophosphate (TPP)-binding module, a pyrimidine binding domain and a transketolase C-terminal domain (Fig. S3).
With a view to analyzing the spatial expression pattern of the DXS1 tomato gene, as well as the other two members of the DXS gene family, quantitative RT-PCR experiments have been carried out (Fig. 5a). A preliminary picture showed that all three DXS genes are expressed in a wide range of tissues, but a very low level of DXS3 transcripts was found in comparison to DXS1 and DXS2 transcripts. DXS1 expression was detected in both vegetative, i.e. root and leaf, and reproductive organs, including floral buds, flowers and fruits at several stages of development (Fig. 5a). DXS1 transcript accumulation was high during flower development and later decreased sharply in fully developed green fruits (MG stage; Fig. 5a). Expression of DXS2 was also found in leaf and flowers at several developmental stages, and particularly in flowers at anthesis; however, no DXS2 transcripts were detected in roots, and only a very low number in MG fruits (Fig. 5a). These last two features, i.e. lack of expression in roots and a significant reduction of transcript level in MG fruits, also characterized the expression pattern shown by the DXS3 gene (Fig. 5a).
(a) The spatial expression of DXS1, DXS2 and DXS3 genes was analyzed by qRT-PCR in different organs of tomato plants. Expression analysis of the DXS1 (b), DXS2 (c) and DXS3 (d) genes in wild type and wls-2297 mutant seedlings, as well as two RNAi DXS1 lines. R: root, L: leaf, FB0: Floral bud 0-3mm, FB1: Floral bud 4–7 mm, FB2: Floral bud 8–10 mm, AD: Anthesis day, AD + 3: Three days after anthesis, MG: Mature green fruit. Error bars show the standard error value of three biological and two technical replicates. Asterisk indicates significant differences between wild type control plants and mutant and silencing plants (Fisher’s Least Significant Difference test, P < 0.05).
The analysis of DXS transcript levels in the seedlings of the wls-2297 mutant and in the DXS1 RNAi silencing lines showed that DXS1 expression was undetectable in the former, while it was silenced in the two RNAi lines analyzed (Fig. 5b). Though DXS2 expression was not significantly affected in the wls-2297 mutant, it decreased in the RNAi DXS1 lines (Fig. 5c), despite the fact that the RNAi construct was specifically designed for silencing the DXS1 gene (Fig. S2a). Finally, DXS3 transcript levels were not significantly altered in the RNAi DXS1 lines, but were slightly increased in wls-2297 mutant seedlings (Fig. 5d).
In vivo and molecular complementation of the wls-2297 mutant phenotype
To corroborate the functional role of DXS1 gene in determining plant development and survival, two complementation experiments were performed. Thus, in vivo complementation of the wls-2297 mutant phenotype was attempted by treatment of seedling plants grown under in vitro conditions with 1-Deoxy-D-xylulose-5-phosphate (DXP), the product of the DXS enzyme activity. When DXP was added to the in vitro culture media, mutant seedlings not only did not recover the wild type phenotype, rather they died even faster than wls-2297 mutant ones (Fig. 6a,b). On the contrary, daily treatment with DXP on the shoot apical meristem led the mutant seedlings to develop green live tissues from those cells which incorporated DXP (Fig. 6c), and they were even able to develop small green leaves (Fig. 6e,f), suggesting that DXP is essential for photosynthetic activity of plant tissues. Nevertheless, it is noteworthy that plant development ceased and seedlings died a few days after DXP treatment ceased (Fig. 6d).
Two-week-old cotyledons of the wsl-2297 mutant without any treatment (a) or with DXP present in the media (b) became necrotic and the plant died. (c) wsl-2297 mutant seedlings treated daily with DXP on the shoot apical meristem became green but the plant died when the treatment with DXP was interrupted (d). (e,f) wsl-2297 seedlings treated with DXP daily were able to develop small green leaves. (g) Explants from the wsl-2297 mutant (left) and from wsl-2297 mutant lines overexpressing DXS1 (right).
In addition, the DXS1 gene was constitutively expressed under a 35S promoter in wls-2297 mutant seedlings to ascertain whether increased DXS1 transcript levels were able to rescue the wild type phenotype. Results showed that transgenic explants develop green tissues from albino tissue sections (Fig. 6g), confirming that the lack of photosynthetic activity displayed by the wls-2297 albino mutant can be firstly reverted by DXS1 expression. However, as the overexpressing DXS1 explants grew, tissues turned yellow and finally acquired an albino phenotype and died.
Effects of DXS1 overexpression on tomato fruit pigmentation
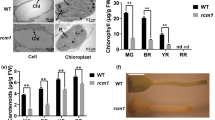
It has been proposed that the protein coded by DXS1, the plastidic form of 1-Deoxy-d-xylulose-5-phosphate synthase, is the limiting enzyme for isoprenoid biosynthesis26. In order to determine whether the DXS1 gene is a limiting step in the MEP pathway of tomato plants, nine diploid transgenic lines overexpressing DXS1 cDNA (OX lines) were generated in the cultivar Moneymaker using a constitutive 35S promoter gene construct, and subsequently characterized in greenhouse conditions (Fig. 7). With the exception of the OX7 line, no changes in the colour and lycopene concentration were observed in the fruits of 35S::DXS1 lines even though they expressed the DXS1 gene at a higher level than control plants (Fig. 7a). Contrary to expectations, tomato fruits yielded by the OX7 line showed a pale colour during the whole ripening process (Fig. 7b). The lack of red colour in mature fruits was most probably due to the low level of lycopene pigment detected in the OX7 transgenic line (Fig. 7c). Analysis of DXS1 relative expression carried out in tomato fruits at breaker (BR) and red ripe (RR) stages of development showed that DXS1 was not overexpressed, but rather it was repressed in these OX7 fruits, suggesting that the pale colour and low lycopene concentration displayed by this line was caused by the post-transcriptional gene silencing (PTGS) of the DXS1 gene (Fig. 7d). Indeed, PTGS triggered by the 35S promoter have widely been reported in transgenic plants35. However, the fact that T-DNA integration site in the OX7 line might be responsible for the altered phenotype cannot be definitively ruled out.
(a) DXS1 transcript levels in young leaves from 35S::DXS1 lines with respect to wild type untransformed cv. Moneymaker plants. (b) Fruit phenotypes at four developmental stages, i.e. mature green (MG), breaker (BR), 4 days after breaker (BR + 4) and red ripe (RR) of 35S::DXS1 line OX7 (above) and wild type (WT) (below). (c) Lycopene content of fruits from WT and 35S::DXS1 lines OX4 and OX7. (d) Relative expression of DXS1 in fruits (BR: breaker; RR: Red Ripe) of OX7 line and wild type. Error bars show the standard error value of two biological and two technical replicates. Asterisk indicates significant differences between control and 35S::DXS1 lines (Fisher’s Least Significant Difference test, P < 0.05).
Influence of DXS1 overexpression and silencing on the MEP pathway
The MEP pathway operates through the participation of eight consecutive enzymes to generate IPP and DMAPP, which serve as the basis for the biosynthesis of all isoprenoid compounds36,37. To investigate the effect of DXS1 overexpression and silencing on the MEP pathway, the expression of the genes participating in this pathway was analyzed by qRT-PCR in the three transgenic lines showing either the highest or lowest DXS1 expression levels (Fig. S5a).
Within the MEP pathway, DXS generates DXP by the transketolase-type condensation of pyruvate and D-glyceraldehyde 3-phosphate, which is converted to MEP by the enzyme coded by the DXP reductoisomerase gene (DXR, Solyc03g114340), whose expression profile remained quite similar in wild type and DXS1 overexpression lines (Fig S4b). However, DXS1 RNAi lines showed a decreased tendency in DXR transcript level compared to wild type plants (Fig S4b). Subsequently, MEP is converted into IPP and DMAPP by the consecutive action of five independent enzymes: i.e. 2-C-methyl-D-erythritol 4-phosphate cytidyltransferase (MCT, Solyc01g102820), 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase (CMK, Solyc01g009010), 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (MDS, Solyc08g081570), 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase (HDS, Solyc11g069380), and 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (HDR, Solyc01g109300). Expression analysis carried out in DXS1 overexpression lines showed similar expression profiles between wild type and transgenic lines in the transcript levels of the genes coding for these five enzymes (Fig. S5c–g). On the other hand, the transcript levels of MCT, CMK and MDS genes were remarkably reduced in DXS1 RNAi lines (Fig. S5c–g). However, HDS and HDR genes showed certain variability in expression levels across these silencing lines Fig. S5c–g).
In the final step of the MEP pathway, geranylgeranyl diphosphate (GGPP), a common precursor for the synthesis of phyllochinones, tocopherols, plastoquinones, chlorophylls, gibberellins and carotenoids, is generated by geranylgeranyl diphosphate synthase (GGPS, Solyc11g011240) that catalyzes the condensation of three IPP and one DMAPP units. Expression levels of the GGPS gene in DXS1 RNAi lines showed variable levels of expression (Fig. S5h), similar to those observed for HDS and HDR genes; whereas slight differences were found in DXS1 overexpression lines compared to wild type plants (Fig. S5h).
Discussion
In recent decades, insertional mutant collections have been particularly useful for identifying and characterizing genes of interest in different model species such as Arabidopsis, rice and tomato38,39,40. This work reports the isolation and functional analysis of the tomato wls-2297 T-DNA mutant, whose characterization led us to demonstrate once again the value of T-DNA insertional mutant libraries for gene discovery in a species of agronomic interest such as tomato. The wls-2297 mutant was selected based on its albino phenotype and the arrested development at expanded cotyledon stage. Molecular analyses performed on the wls-2297 mutant revealed that the observed albino phenotype is due to a single T-DNA insertion, whose integration in the tomato genome caused a 38.6 kb-deletion at chromosome 1 that resulted in the loss of DXS1 and three POX genes. Phenotypic and expression analyses of RNAi silencing lines led us to conclude that wls-2297 is a knock-out mutant affected in the DXS1 gene, and that the loss of DXS1 activity is responsible for the wls-2297 mutant phenotype. Indeed, this result was further confirmed by means of both in vivo complementation assays with DXP and DXS1 overexpression on the wls-2297 mutant, which partially rescued the albino phenotype. At early stages of plant development, the wls-2297 phenotype was completely rescued; however, over time the leaves of the wls-2297 mutant plants overexpressing the DXS1 gene turned yellow, became albino and eventually died prematurely. Functional assays based on DXS1 overexpression showed dissimilar results in different plant species, ranging from no changes in isoprenoid content28,41 to unequal increases in various isoprenoid compounds including total chlorophylls and carotenoids26,29. Overall, these previous works have proved that constitutive DXS1 overexpression does not necessarily mean high levels of isoprenoid end products. Indeed, it has been found that DXS1 transcript levels in overexpression lines, and therefore isoprenoid levels, are usually not so high as might be expected26,28,29,41. In this study, expression analysis of the MEP pathway genes suggested that DXS1 overexpression did not significantly alter transcript profiles of the MEP pathway since gene expression levels in DXS1 overexpressing lines were quite similar to those found in wild type plants (Fig. S5). However, different expression profiles of the MEP pathway genes were found in the DXS1 silencing lines. Thus, the expression levels of DXR, MCT, CMK and MDS genes were significantly reduced in albino RNAi transformants compared to wild type non-transformed plants (Fig. S5b–e), suggesting a role for DXS in transcriptional regulation of the first steps of the MEP pathway. On the other hand, expression profiles of HDS, HDR and GGPS genes showed an evident variation among different DXS1 RNAi lines (Fig. S5f–h). Most likely, the explanation for these results relies on the complex regulation of the MEP pathway, which occurs at both transcriptional and post-transcriptional levels42 to ensure its proper functioning. Due to this pathway complexity, it has been suggested that elevated levels of DXS1 transcripts can only exert an influence on the MEP pathway when adequate precursors from intermediary metabolism are accessible28. Given that levels of the different isoprenoids do not change equally in DXS1 overexpressing lines26,28,29,41, the fact that overexpression of DXS1 did not completely rescue the albino phenotype of the wls-2297 mutant might be due to disproportionately altered isoprenoid levels. However, the possibility that some wls-2297 DXS1 overexpressing lines here reported may undergo co-suppression effects of DXS1 expression cannot be discarded, as it is a commonly reported phenomenon in genetic transformation experiments using strong constitutive promoters35. Unfortunately, we were not able to perform expression studies to corroborate this hypothesis, as 35S::DXS1 plants died prematurely.
In Arabidopsis, the DXS1 knock-out mutants cla1-131 and temperature-sensitive allele chs530 also showed an albino phenotype as well as altered plant development. When both mutants were grown in a nutrient medium supplemented with glucose, they were able to sustain their development while maintaining their albino phenotype. Thus, chs5 plants developed slowly but reached a similar size to wild type plants30, whereas the cla1-1 mutant even generated floral buds, which did not however manage to produce normal flowers31. In this study, tomato RNAi DXS1 lines developed three-four albino leaves, probably as a consequence of residual DXS1 expression; however, the wls-2297 mutant did not develop true leaves, but rather its growth was arrested at the fully expanded cotyledon stage or slightly later when grown in a nutrient medium supplemented with a high sugar concentration. These results suggest a significant difference between DXS1 action mechanisms in Arabidopsis and tomato. Araki et al.30 suggested that a plant with a loss of DXS activity did not survive past the seedling stage due to the lack of thiamine, one of the first branches of the MEP pathway43. They therefore proposed that the partial development of albino chs5 plants was facilitated by other DXS genes30. Arabidopsis DXS1 and DXS2 genes show high homology and are both included in the subfamily DXS134, suggesting that they would have redundant functions. However, the Arabidopsis DXS2 gene has recently been renamed as DXL1, which might not have DXS activity, but rather has acquired a novel, as yet unknown biochemical function32. In contrast, the Arabidopsis DXS3 gene has been grouped within a clade composed of highly divergent protein sequences20, which lack many of the highly conserved residues known to be essential for DXS activity44. Nonetheless, marginal, though significant, DXS activity has recently been reported for the corresponding ortholog DXS3 in maize20. Hence, if the DXS3 function was conserved in other species, the survival of Arabidopsis chs5 and cla1-1 mutants might be explained by the DXS activity of the DXS3 enzyme. As in Arabidopsis, the tomato DXS family also consists of three genes, but in contrast to Arabidopsis, tomato DXS genes code for proteins belonging to each one of the proposed DXS clades (Fig. S4), and in addition they have different expression patterns. The tomato DXS1 gene is ubiquitously expressed and its highest transcript levels are observed during fruit ripening34,45. However, DXS2 transcripts are abundant in only few tissues, including young leaves and flowers, as well as isolated trichomes34,45. On the other hand, DXS3 transcript levels are extremely low compared to the other two tomato DXS genes according to previous results34. The roles of tomato DXS1 and DXS2 have been described without overlapping function in modulating isoprenoid metabolism16,22,33,45. Our results are in agreement with this hypothesis, since DXS2 and DXS3 activities did not compensate for the loss of DXS1 function in the wls-2297 mutant, indicating that DXS1 plays a vital role in the development of tomato plants.
Since the non-mevalonate pathway of isoprenoid biosynthesis was described, the exchange of isoprenoid intermediate compounds between MVA and MEP pathways has been a matter of some dispute. Certain evidence of cross-talk between MVA pathway in the cytosol and MEP pathway in the plastid has been reported in different plant species, including Arabidopsis and a solanaceous species like tobacco46,47,48. However, it has also been proposed that a tightly regulated cellular compartmentalization of MVA and MEP pathways occurs during isoprenoid biosynthesis28. In addition, a tissue-dependent regulation of tomato MEP genes has recently been reported supporting the complexity of this pathway49. Characterization of the wls-2297 mutant, which displays a complete inhibition of DXS activity due to a loss-of-function of the DXS gene, showed a drastic reduction in chlorophyll and carotenoid biosynthesis (Fig. 1g), a feature that should not be detected if there were an active exchange of isoprenoid intermediates between both pathways. Therefore, our results not only support that the MEP pathway is essential for photosynthetic pigment biosynthesis, but also that functional cross-talk with the MVA pathway may not occur at least during the first steps of isoprenoid biosynthesis. Although periodic supplementation of wsl-2297 mutant seedlings with DXP during several days promoted minor development, it did not avoid death of the seedlings once the treatment ceased. This result suggests that if an exchange of isoprenoid intermediates such as IPP and DMAPP occurs in tomato chloroplasts, it must be at a rate that is insufficient to allow plant growth. In addition, this hypothesis agrees with the blocking of plastidial isoprenoid biosynthesis observed after treatment with fosmidomycin, a specific inhibitor of DXP reductoisomerase, the enzyme catalyzing the previous step to that of DXS in the isoprenoid biosynthetic pathway50. Taken as a whole, these results regarding the lethality of the wls-2297 mutation, also found in DXS1 silencing lines, prove that the DXS1 gene is a crucial genetic factor for plant organ growth and survival.
Methods
Plant material and growth conditions
The wls-2297 mutant was isolated from a T-DNA insertional collection, which was generated from the tomato (Solanum lycopersicum L.) cultivar Moneymaker through infection with Agrobacterium strain LBA4404 containing the enhancer trap vector pD99151 (kindly supplied by Dr. Thomas Jack; Department of Biological Sciences, Dartmouth College, USA). The TG1 line was self-pollinated to obtain the TG2 progeny, which was grown in soil conditions (a mixture of peat and coconut fiber (1:1) and, after sowing, a cover of vermiculite). Due to the fact that the wls-2297 mutant homozygous plants did not survive enough to achieve fruit set, wls-2297 line were perpetuated by self-pollination of wild type heterozygous plants growth in soil under greenhouse conditions using standard practices with regular addition of fertilizers. Seedlings analysed in this project were grown under standard conditions (16 h light/8 h darkness, ≈ 25 °C, 60% RH) in a growth chamber. Co-segregation studies of the mutant phenotype with kanamycin resistance conferred by the NEOMYCIN PHOSPHOTRANSFERASE III (NPTII) marker gene was carried out by sowing seeds on Murashige and Skoog52 (MS) agar medium supplemented with sucrose (10 g l−1) and kanamycin (100 mg l−1). The sodium salt of 1-Deoxy-D-xylulose-5-phosphate (DXP, Sigma-Aldrich) resuspended in water (1 mg/mL) was used to perform in vivo complementation of the albino phenotype in the wls-2297 mutant.
Microscopy analysis of chloroplasts and pigment measurement
For observation of chloroplasts/plastids in living leaf tissues, a section of tomato cotyledon was excised and its cuticle removed manually. Then, these cotyledons were examined using a bright field microscope Nikon Optiphot-2 equipped with Nikon Digital Camera. The chlorophylls and carotenoids concentrations were determined by UV-VIS Spectroscopy following the method described by Lichtenthaler and Buschmann53.
DNA isolation and DNA-blot hybridization
Genomic DNA was isolated from 100 mg of young leaves using Plant DNAzol Reagent (Invitrogen) following the instructions provided by the manufacturer. In order to determine the number of T-DNA insertions introduced in the wls-2297 mutant, a DNA-blot hybridization was carried out from 15 μg of genomic DNA digested by restriction enzymes EcoRI and HindIII, electrophoresed through-out a 0.8% agarose-TBE gel, blotted onto Hybond N+ membrane (Amersham) and hybridized with a FA-NPTII radioactive probe, which was labelled with 32P by random priming with the Rediprime II Random Prime Labelling System (Amersham). Nylon membranes hybridized were exposed to Hyperfilms (Amersham). The FA-NPTII probe was synthetized including the complete coding sequence of the tomato FALSIFLORA (FA) gene, which was employed as hybridization positive control, and the NPTII gene used as a selective marker in the T-DNA.
Cloning of T-DNA flanking sequences and PCR genotyping
Sequences flanking the wls-2997 T-DNA insertion were cloned by amplification with a single-side specificity (anchor-PCR) assay described by Loh54. With this end, genomic DNA was digested with restriction endonucleases that generate blunt ends, i.e. AluI, DraI, EcoRV, HincII, PvuII, ScaI, StuI and SmaI (Takara Bio company). After digestion, adaptors were linked to the digested genomic DNA restriction fragments. The fragments linked to adaptors were then amplified by three assembled PCR with specific primers for the Right Border sequence (Anchor Right-1, Anchor Right-2 and Anchor Right-3; Table S1) or for the Left Border (Anchor Left-1, Anchor Left-1 and Anchor Left-1; Table S1) from the T-DNA, combined with adaptor specific primers Adaptor-1, Adaptor-2 and Adaptor-3 (Table S1). PCR amplification products were purified using GenElute PCR Clean-up Kit (SIGMA Aldrich) and sequenced in an Applied Biosystems 3500 Genetic Analyzer. The cloned sequences were compared with SGN Database (https://solgenomics.net) to assign the T-DNA insertion site on tomato genome. Co-segregation of the T-DNA insertion site with the wls-2997 phenotype was evaluated by PCR using i) the specific genomic forward and reverse primers to amplify the wild type allele (without T-DNA insertion) and ii) one specific genomic primer and the specific T-DNA primer to amplify the mutant allele (carrying the T-DNA insertion). The sequence of genotyping primers used is listed in Table S1.
Specific primers for DXS1 (Solyc01g067890) and the three POX (Solyc01g067860, Solyc01g067870, Solyc01g067880) genes were designed (Table S1) and used in PCR experiment to demonstrate that a 38.6 kb-deletion is on wls-2297 mutant plants. Amplification was performed using in a volume of 30 μl using 10 ng of total DNA, 50 ng of each primer, 0.25 mM dNTPs, 2.5 mM MgCl2, and 1 U BIOTAQTM DNA Polymerase (Bioline) in 1X Taq buffer. DNA was amplified under the following thermal cycling conditions: 94 °C for 5 min, followed by 35 cycles at 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 2 min, and a final extension of 5 min at 72 °C. PCR products were analysed in 1% agarose gels in 1X SB buffer (200 mM NaOH, 750 mM boric acid, pH 8.3) and visualized with ethidium bromide. In addition, to amplify the genomic region located between the T-DNA flanking regions a Long-PCR assay was carried out using Elongase® Enzyme Mix (Invitrogen) following the instructions given by the manufacturer. The primers used in this experiment were Genotyping-F1 and Genotpyping-R2 (Table S1). Genomic DNA from wild type (cv. Moneymaker) and the wls-2297 mutant plants was used as template.
RNA preparations and gene expression analysis
Total RNA was purified from using Trizol reagent (Invitrogen) following the manufactures instructions. Contamination of genomic DNA was removed by using the DNA-freeTM kit (Ambion), and cDNA was synthetized from the DNA-free total RNA by means of a M-MuLV reverse transcriptase (Invitrogen) using as primers a mixture of random hexamer and 18-mer oligo(dT).
Expression analysis were performed by real-time PCR assays using the SYBR Green PCR Master Mix kit in a 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) and a pair of specific primer pairs for each gene analysed (Table S1). Data obtained were collected and analysed using the System Sequence Detection Software v1.2. Results were calculated using ΔΔCt method by comparison to a data point from the wild type samples and are expressed in arbitrary units. The housekeeping UBIQUITINE3 gene was used as an internal reference in the gene expression analyses performed. Absence of genomic DNA contaminating in the RNA sample analysed by RT-PCR was demonstrated as previously described55. Mean comparison (Fisher’s Least Significant Difference test, LSD) was used to determine significant differences among gene expression levels. Analyses were performed using the Statgraphics Centurion XVI software package and data presented as means ± standard error.
Generation of DXS transgenic tomato plants
The DXS1 (Solyc01g067890) complete open reading frame (ORF) was amplified from tomato cv. Moneymaker cDNA using a pair of specific primers 35SDXS1F and 35SDXSR to introduce a BamHI restriction site 127 bp upstream the initiation codon and a KpnI restriction sites downstream the stop codon (Table S1). The PCR product was cloned into a p-GEM®-T-Easy plasmid and sequenced in order to confirm the absence of mutations in the amplicon. The selected plasmid was digested with BamHI and KpnI, and the DXS1 cDNA was subcloned under the control of the constitutive CaMV promoter together a plant kanamycin resistant selection marker into the binary vector pROKII to generate an overexpression gene construct used to transform plants of the cv. Moneymaker. The same DXS1 cDNA was subcloned under the control of the constitutive CaMV promoter together with a plant hygromycin resistant selection marker into the binary vector pCAMBIA1302 to generate an overexpression gene construct used to complement the wls-2297 mutant seedlings.
In order to silencing the DXS1 gene, an interference RNA (RNAi) approach was followed. A 330 bp specific fragment of the DXS1 cDNA was amplified, using primers RNAiDXS1F, to introduce a XbaI and a XhoI restriction sites, and RNAiDXS1R, to introduce a ClaI and a KpnI restriction sites (Fig. S2a; Table S1), and cloned into p-GEM®-T-Easy. The insert of RNAiDXS1 was liberated from the p-GEM®-T-Easy twice, firstly by XhoI and KpnI digestion and secondly by XbaI and ClaI digestion. Both inserts liberated were cloned as inverted repeats into vector pKannibal. The resulting plasmid was digested with NotI and fragment isolated was cloned into the binary vector pART27 to express inverted repeat sequences of DXS1 separated by intronic sequences under the control of the constitutive promoter 35S. In order to generate RNAi lines to inhibit the three POX genes (Solyc01g067860, Solyc01g067870, Solyc01g067880) simultaneously, a pair of primers annealing in a conserved domain for the three POX, RNAiPOXsF and RNAiPOXsR (Fig. S2b; Table S1), were used to generate the silencing construct obtained as described for the DXS1 gene.
Overexpression and silencing binary vectors were electroporated into Agrobacterium tumefaciens and Agrobacterium-mediated transformation of Moneymaker tomato cultivar cotyledons was performed following the protocols previously described56. The ploidy level in transgenic plants was evaluated by flow cytometry according to the protocol described by Atarés et al.57 and diploid transgenic lines were selected for further phenotypic and expression analyses.
Additional Information
How to cite this article: García-Alcázar, M. et al. Albino T-DNA tomato mutant reveals a key function of 1-deoxy-D-xylulose-5-phosphate synthase (DXS1) in plant development and survival. Sci. Rep. 7, 45333; doi: 10.1038/srep45333 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Lambers, H., Chapin, F. S. 3rd & Pons, T. L. Plant Physiologycal Ecology (eds Lambers, H., Chapin, F. S. 3rd & Pons, T. L. ) 11–12 (Springer Science + Business Media, 2008).
Broun, P. & Somerville, C. Progress in plant metabolic engineering. Proc. Natl. Acad. Sci. USA 98, 8925–8927 (2001).
McGarvey, D. & Croteau, R. Terpenoid metabolism, Plant Cell 7, 1015–1026 (1995).
Rodríguez-Concepción, M. & Boronat, A. Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol. 130, 1079–1089 (2002).
Bach, T. J & Rohmer, M. Isoprenoid Synthesis in Plants and Microorganisms: New Concepts and Experimental Approaches (ed. Bach, T.J . & Rohmer, M. ) (Springer Science & Business Media, 2013).
Gutensohn, M. & Dudareva, N. Involvement of compartmentalization in monoterpene and sesquiterpene biosynthesis in plants. In Isoprenoid Synthesis in Plants and Microorganisms: New Concepts and Experimental Approaches ( Bach, T. J. & Rohmer, M. eds) 155–168 (Springer Science + Buiness Media, 2013).
Vranova, E., Coman, D. & Gruissem, W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 64, 665–700 (2013).
Spurgeon, S. L & Porter J. W. Biosynthesis of carotenoids. In: Biosynthesis of isoprenoid compounds, vol2 ( Porter J. W ., Spurgeon S. L. eds) 1–122 (John Wiley & Sons, 1981).
Goldstein J. L. & Brown M. S. Regulation of the mevalonate pathway. Nature 343, 425–430 (1990).
Rohmer, M. Isoprenoid biosynthesis via the mevalonate-independent route, a novel target for antibacterial drugs? Prog. Drug Res. 50, 135–154 (1998).
Rohmer, M. The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat. Prod. Rep. 16, 565–574 (1999).
Lichtenthaler H. K. The 1-deoxy-D-xylulose-5-phospate pathway of isoprenoid biosynthesis in plants. Annu. Rev. Plant Physiol. Plant. Mol. Biol. 50, 47–65 (1999).
Arigoni, D. et al. Terpenoid biosynthesis from 1-deoxy-D-xylulose in higher plants by intramolecular skeletal rearrangement. Proc. Natl. Acad. Sci. USA 94, 10600–10605 (1997).
Eisenreich, W. et al. The deoxyxylulose phosphate pathway of terpenoid biosynthesis in plants and microorganisms. Chem. Biol. 5, R221–R233 (1998).
Sprenger, G. A. et al. Identification of a thiamin-dependent synthase in Escherichia coli required for the formation of the 1-deoxy-D-xylulose 5-phosphate precursor to isoprenoids, thiamin, and pyridoxol. Proc. Natl. Acad. Sci. USA 94, 12857–12862 (1997).
Lange, B. M., Wildung, M. R., McCaskill, D. & Croteau, R. A family of transketolases that directs isoprenoid biosynthesis via a mevalonate-independent pathway. Proc. Natl. Acad. Sci. USA 95, 2100–2104 (1998).
Bouvier, F., d’Harlingue, A., Suire, C., Backhaus, R. A. & Camara, B. Dedicated roles of plastid transketolases during the early onset of isoprenoid biogenesis in pepper fruits1. Plant Physiol. 117, 1423–1431 (1998).
Estévez, J. M. et al. Analysis of the expression of CLA1, a gene that encodes the 1-deoxyxylulose 5-phosphate synthase of the 2-C-methyl-D-erythritol-4-phosphate pathway in Arabidopsis. Plant Physiol. 124, 95–104 (2000).
Lois, L. M., Rodríguez-Concepción, M., Gallego, F., Campos, N. & Boronat, A. Carotenoid biosynthesis during tomato fruit development: regulatory role of 1-deoxy-D-xylulose 5-phosphate synthase. Plant J. 22, 503–513 (2000).
Córdoba, E. et al. Functional characterization of the three genes encoding 1-deoxy-D-xylulose 5-phosphate synthase in maize. J. Exp. Bot. 62, 2023–2038 (2011).
Kim, Y. B. et al. Regulation of resin acid synthesis in Pinus densiflora by differential transcription of genes encoding multiple 1-deoxy-D-xylulose 5-phosphate synthase and 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase genes. Tree Physiol. 29, 737–749 (2009).
Walter, M. H., Hans, J. & Strack, D. Two distantly related genes encoding 1-deoxy-d-xylulose 5-phosphate synthases: differential regulation in shoots and apocarotenoid-accumulating mycorrhizal roots. Plant J. 31, 243–254 (2002).
Floss, D. S. et al. Knock-down of the MEP pathway isogene 1-deoxy-D-xylulose 5-phosphate synthase 2 inhibits formation of arbuscular mycorrhiza-induced apocarotenoids, and abolishes normal expression of mycorrhiza-specific plant marker genes. Plant J. 56, 86–100 (2008).
Iriti, M. & Faoro. F. Bioactivity of grape chemicals for human health. Nat. Prod. Commun. 4, 611–634 (2009).
Islam, M. A. et al. Dietary Phytochemicals: Natural swords combating inflammation and oxidation-mediated degenerative diseases. Oxid. Med. Cell Longev. 2016, 5137431, doi: 10.1155/2016/5137431 (2016).
Estévez, J. M., Cantero, A., Reindl, A., Reichler, S. & León, P. 1-Deoxy-D-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J. Biol. Chem. 276, 22901–22909 (2001).
Carretero-Paulet, L. et al. Enhanced flux through the methylerythritol 4-phosphate pathway in Arabidopsis plants overexpressing deoxyxylulose 5-phosphate reductoisomerase. Plant Mol. Biol. 62, 683–695 (2006).
Enfissi, E. M. et al. Metabolic engineering of the mevalonate and non-mevalonate isopentenyl diphosphate-forming pathways for the production of health-promoting isoprenoids in tomato. Plant Biotechnol. J. 3, 17–27 (2005).
Zhang, M., Li, K., Zhang, C., Gai, J. & Yu, D. Identification and characterization of class 1 DXS gene encoding 1-deoxy-D-xylulose-5-phosphate synthase, the first committed enzyme of the MEP pathway from soybean. Mol. Biol. Rep. 36, 879–887 (2009).
Araki, N., Kusumi, K., Masamoto, K. & Iba, K. Temperature-sensitive Arabidopsis mutant defective in 1-deoxy-D-xylulose 5-phosphate synthase within the plastid non-mevalonate pathway of isoprenoid biosynthesis. Physiol. Plant 108, 19–24 (2000).
Mandel, M. A., Feldmann, K. A., Herrera-Estrella, L., Rocha-Sosa, M. & León, P. CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. Plant J. 9, 649–668 (1996).
Carretero-Paulet, L. et al. Functional and evolutionary analysis of DXL1, a non-essential gene encoding a 1-deoxy-D-xylulose 5-phosphate synthase like protein in Arabidopsis thaliana . Gene 524, 40–53 (2013).
Besser, K. et al. Divergent regulation of terpenoid metabolism in the trichomes of wild and cultivated tomato species. Plant Physiol. 149, 499–514 (2009).
Hofberger, J. A. et al. Large-Scale Evolutionary analysis of genes and supergene clusters from terpenoid modular pathways provides insights into metabolic diversification in flowering plants. PLoS One 10, e0128808, doi: 10.1371/journal.pone.0128808 (2015).
Vaucheret, H. et al. Transgene-induced gene silencing in plants. Plant J. 16, 651–659 (1998).
Cordoba, E., Salmi, M. & León, P. Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. J. Exp. Bot. 60, 2933–2943 (2009).
Ruiz-Sola, M. A. & Rodríguez-Concepción, M. Carotenoid biosynthesis in Arabidopsis: a colorful pathway. Arabidopsis Book 10, e0158, 10.1199/tab.0158 (2012).
Sallaud, C. et al. High throughput T-DNA insertion mutagenesis in rice: a first step towards in silico reverse genetics. Plant J. 39, 450–464 (2004).
Alonso, J. M. et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana . Science 301, 653–657 (2003).
Menda, N., Semel, Y., Peled, D., Eshed, Y. & Zamir, D. In silico screening of a saturated mutation library of tomato. Plant J. 38, 861–872 (2004).
Muñoz-Bertomeu, J., Arrillaga, I., Ros, R. & Segura, J. Up-regulation of 1-deoxy-D-xylulose-5-phosphate synthase enhances production of essential oils in transgenic spike lavender. Plant Physiol. 142, 890–900 (2006).
Guevara-García, A. et al. Characterization of the Arabidopsis clb6 mutant illustrates the importance of posttranscriptional regulation of the methyl-D-erythritol 4-phosphate pathway. Plant Cell 17, 628–643 (2005).
McHale, N. A., Hanson, K. R. & Zelitch, I. A Nuclear mutation in Nicotiana sylvestris causing a thiamine-reversible defect in synthesis of chloroplast pigments. Plant Physiol. 88, 930–935 (1988).
Xiang, S., Usunow, G., Lange, G., Busch, M. & Tong, L. Crystal structure of 1-deoxy-D-xylulose 5-phosphate synthase, a crucial enzyme for isoprenoids biosynthesis. J. Biol. Chem. 282, 2676–2682 (2007).
Paetzold, H. et al. The isogene 1-deoxy-D-xylulose 5-phosphate synthase 2 controls isoprenoid profiles, precursor pathway allocation, and density of tomato trichomes. Mol. Plant. 3, 904–916 (2010).
Lichtenthaler, H. K., Rohmer, M. & Schwender, J. Two independent biochemical pathways for isopentenyl diphosphate and isoprenoid biosynthesis in higher plants. Physiol. Plant. 101, 643–652 (1997).
Hemmerlin, A. et al. Cross-talk between the cytosolic mevalonate and the plastidial methylerythritol phosphate pathways in tobacco bright yellow-2 cells. J. Biol. Chem. 278, 26666–26676 (2003).
Laule, O. et al. Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana . Proc. Natl. Acad. Sci. USA 100, 6866–6871 (2003).
Pankratov, I. et al. Fruit carotenoid-deficient mutants in tomato reveal a function of the plastidial isopentenyl diphosphate isomerase (IDI1) in carotenoid biosynthesis. Plant J. 88, 82–94 (2016).
Rodríguez-Concepción, M. et al. 1-Deoxy-D-xylulose 5-phosphate reductoisomerase and plastid isoprenoid biosynthesis during tomato fruit ripening. Plant J. 27, 213–222 (2001).
Campisi, L. et al. Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J. 17, 699–707 (1999).
Murashige, T. & Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plantarum 15, 473–497 (1962).
Lichtenthaler, H. K. & Buschmann, C. Chlorophylls and Carotenoids: Measurement and characterization by UV-VIS Spectroscopy. In Current protocols in food analytical chemistry (ed. Wrolstad, R. E. et al.) F4.3.1–F4.3.8 (John Wiley & Sons, 2001).
Loh, E. Anchored PCR: amplification with single-sided specificity. Methods 2, 11–19 (1991).
Giménez, E. et al. Functional analysis of the Arlequin mutant corroborates the essential role of the ARLEQUIN/TAGL1 gene during reproductive development of tomato. PLoS One 5, 14427, 10.1371/journal.pone.0014427 (2010).
Ellul, P. et al. The ploidy level of transgenic plants in Agrobacterium-mediated transformation of tomato cotyledons (Lycopersicon esculentum L. Mill.) is genotype and procedure dependent. Theor. Appl. Genet. 106, 231–238 (2003).
Atarés, A. et al. An insertional mutagenesis programme with an enhancer trap for the identification and tagging of genes involved in abiotic stress tolerance in the tomato wild-related species Solanum pennellii . Plant Cell Rep. 30, 1865–1879 (2011).
Acknowledgements
This work was supported by research grants from the Spanish Ministry of Economy and Competitiveness and the UE-European Regional Development Fund (AGL2015-64991-C3-1-R, and AGL2015-64991-C3-3-R), and Junta de Andalucia (P12-AGR-1482). PhD fellowship to M.G.-A. was funded by the FPU Programme of the Spanish Ministry of Science and Innovation. The authors thank research facilities provided by the Campus de Excelencia Internacional Agroalimentario (CeiA3).
Author information
Authors and Affiliations
Contributions
M.G.-A., B.P., C.C., B.G.-S., S.S. and T.A. performed field and laboratory experiments. M.G.-A., E.G., F.Y.-L., J.C. and R.L. wrote the manuscript. J.C., V.M., T.A., and R.L. conceived the research plans, supervised the study and collaborated in data analysis. All authors revised the draft manuscript and read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
García-Alcázar, M., Giménez, E., Pineda, B. et al. Albino T-DNA tomato mutant reveals a key function of 1-deoxy-D-xylulose-5-phosphate synthase (DXS1) in plant development and survival. Sci Rep 7, 45333 (2017). https://doi.org/10.1038/srep45333
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep45333
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.