Abstract
Twenty-one small Gram-negative motile coccobacilli were isolated from 15 systemically diseased African bullfrogs (Pyxicephalus edulis), and were initially identified as Ochrobactrum anthropi by standard microbiological identification systems. Phylogenetic reconstructions using combined molecular analyses and comparative whole genome analysis of the most diverse of the bullfrog strains verified affiliation with the genus Brucella and placed the isolates in a cluster containing B. inopinata and the other non-classical Brucella species but also revealed significant genetic differences within the group. Four representative but molecularly and phenotypically diverse strains were used for in vitro and in vivo infection experiments. All readily multiplied in macrophage-like murine J774-cells, and their overall intramacrophagic growth rate was comparable to that of B. inopinata BO1 and slightly higher than that of B. microti CCM 4915. In the BALB/c murine model of infection these strains replicated in both spleen and liver, but were less efficient than B. suis 1330. Some strains survived in the mammalian host for up to 12 weeks. The heterogeneity of these novel strains hampers a single species description but their phenotypic and genetic features suggest that they represent an evolutionary link between a soil-associated ancestor and the mammalian host-adapted pathogenic Brucella species.
Similar content being viewed by others
Introduction
The genus Brucella, established in 1920 by Meyer and Shaw, currently comprises twelve species1 (http://www.bacterio.cict.fr/b/brucella.html) that are historically differentiated by host tropism, pathogenicity and phenotypic traits. A decade ago the genus consisted of six “classical” Brucella species (B. melitensis, B. abortus, B. suis, B. canis, B. ovis, and B. neotomae), which are also referred to as the “core” Brucella. Three of these species (B. melitensis, B. abortus and B. suis) are important zoonotic pathogens that infect humans and a variety of other mammals, causing a disease called brucellosis. This is considered to be one of the most important zoonoses worldwide, with 500,000 human cases reported each year2. Due to its low infectious dose and ease of transmission as an aerosol, Brucella was one of the first microorganisms weaponized by the U.S. military in 1950s3 and is listed as a category B bioterrorism agent by the U.S. Centers for Disease Control and Prevention (CDC) and the National Institute of Allergy and Infectious Diseases (NIAID)4.
The group of classical Brucella species was extended in 2007 to include B. ceti and B. pinnipedialis, isolated from marine mammals5. The first phenotypically atypical species, B. microti, was initially isolated from common voles6,7 and then from soil8. B. microti is different from the classical species in that it is fast-growing and metabolically very active, resembling Ochrobactrum, the closest phylogenetic neighbour of Brucella7. Although metabolically different, the genome sequence of B. microti (strain CCM 4915T) is nearly identical to B. suis 13309, making it appear that its phenotypic differences may reflect differential gene regulation10. Subsequent whole genome analyses, however, indicate that, while B. microti is closely related to the other core Brucella, it represents a distinct basal lineage11. Most recently, B. papionis, isolated from baboons, phylogenetically clustering with the core Brucella spp.12,13, and B. vulpis from red foxes, forming a long-branched sister clade to the classical species1,14, were described.
Additional atypical Brucella isolates have been described. The first, B. inopinata (strain BO1), was isolated from a breast implant wound infection15,16. This strain showed lower sequence similarities in most housekeeping genes15 and differed at 5 nucleotide positions in the 16S rRNA sequence from the one sequence shared by all core Brucella16. Metabolically, B. inopinata (BO1) is as active as B. microti7,16. This finding was followed by a second human isolate (BO2) from a patient with chronic destructive pneumonia that revealed an identical 16S rRNA gene sequence to B. inopinata (BO1)17. In addition, the phylogenetic comparison of novel atypical strains isolated from rodents in Australia showed that these, along with the BO2 strain and B. inopinata (BO1), are united in a clade that is separate from the core Brucella spp.18.
In the past few years, more and more Brucella isolates have been reported from cold-blooded hosts, i.e. African bullfrogs (Pyxicephalus edulis)19, big-eyed tree frogs (Leptopelis vermiculatus)20, White’s tree frog21, and Pacman frog (Ceratophrys ornata)22. At present, the pathogenicity of these newly described species and atypical strains for humans is unknown but warrants investigation. Human pathogenicity cannot be excluded because human infections with B. inopinata (BO1) and strain BO2 described as B. inopinata-like have been reported15,17.
The present work is a comprehensive molecular and phenotypic study of a large collection of isolates from African bullfrogs building on a previous preliminary description of two of these strains19. We conducted an in-depth genome analysis and also evaluated the virulence of the potentially new Brucella species for mammalian hosts both in in vitro and in vivo models of infection.
Results
Historically, CO2 requirement, H2S production, urea hydrolysis, agglutination with monospecific sera, dye sensitivity, and phage lysis are determined for the identification and sub-differentiation of Brucella spp. To gain deeper insight into the composition of this novel group of amphibian Brucella sp., all currently available bullfrog isolates have been characterized using a comprehensive combination of molecular and classical microbiological approaches10.
Analysis of phenotypic characteristics
Primary isolation from various tissues of dead or moribund African bullfrogs revealed Gram-negative coccoid rods that were identified as O. anthropi by API®-20NE and Vitek2® Compact (bioMérieux, Marcy-l’Étoile, France)19. The bacterial isolates showed nonfastidious rapid growth on trypticase soy agar (TSA), sheep blood agar, and standard nutrient agar at 25–42 °C and also grew on MacConkey agar. On Columbia agar, colonies did not display haemolysis. All 21 strains could be easily cultured on Brucella agar at 37 °C without supplementary CO2. After 24 h, typical raised, convex and circular colonies, 1–2 mm in diameter, were visible. Seventeen of these strains produced translucent to whitish colonies, but four strains (09RB8913, 09RB8914, 09RB8915, and 09RB8918) developed colonies with a markedly brownish pigmentation.
All strains were both catalase- and oxidase-positive, raising suspicion of Brucella. They produced H2S, and urea was hydrolyzed within 10 to 45 min by all but two of the strains. Strains 10RB9215 and 10RB9213 had a prolonged hydrolysis time of 80 and 90 min, respectively, revealing reduced urease activity (Table 1). All isolates were able to grow in the presence of thionine at dilutions of 1/25,000, 1/50,000, and 1/100,000 and in the presence of basic fuchsin at dilutions of 1/50,000 and 1/100,000.
Spontaneous agglutination could not be induced by trypaflavine, and ruby coloured colonies were not visible after staining with crystal violet. In addition, none of the strains agglutinated with monospecific anti-R (rough) serum. All the bullfrog isolates presented a smooth lipopolysaccharide phenotype, but none agglutinated with either anti-M or anti-A monospecific sera. In three of the strains (10RB9215, 10RB9211 and 10RB9212) a slight agglutination was seen using high concentrations (1:10 or 1:20) of anti-A sera.
The bullfrog isolates were not lysed by the bacteriophages F1, F25, Tb, BK2, Iz, Wb, Fi, and R/C, neither at the routine test dilution (RTD) nor at 104x RTD (Table 1).
The Micronaut® BfR Brucella assay (Merlin Diagnostika) was used for biochemical profiling23 and the bullfrog strains generally displayed high metabolic activity, comparable to B. inopinata (BO1) and B. microti, but in contrast to the relatively low activity of classical Brucella species (Supplementary Table S1). A number of reactions considered to be typical for brucellae (except for B. papionis), i.e. H-hydroxyproline-βNA (HP: +), Glu(pNA)-OH (ENAOH: −), Pyr-pNA (PYRNA: −), suggest an allocation of the bullfrog strains to the genus Brucella. Hierarchical cluster analysis performed by the Ward’s linkage algorithm positioned the amphibian Brucella sp. strains between B. inopinata (BO1) and B. microti, close to B. suis biovars 1–4 and B. canis (Supplementary Fig. S1). The metabolic activity varied within the bullfrog isolates group (Supplementary Table S1).
Semi-solid agar testing for bacterial swarming showed that the African bullfrog strains are motile (Supplementary Fig. S2A–D), in strong contrast to the classical brucellae like B. melitensis 16 M (Supplementary Fig. S2E). Some of the bullfrog isolates migrated farther through the agar than O. anthropi LMG 3331 (Supplementary Fig. S2F), others showed a similar migration pattern, e.g. 09RB8471. Transmission electron micrographs (40,000x) showed bacterial cells (strain 10RB9206) individually arranged or in irregular clusters, exhibiting a mean average diameter of 0.5 μm and a length of 1 μm (Fig. 1A). Some produced a polar sheathed flagellum (Fig. 1B,C). The expression of the flagellum in vitro increased from 12 to 84 h of culture. In addition, individual bacteria presented pili-like structures (Fig. 1D).
Molecular analyses
In order to confirm the assignment of the African bullfrog strains to the genus and to refine their position within the known Brucella species a series of molecular typing methods with increasing resolution were applied.
Bcsp31, IS711, and Bruce-ladder multiplex PCR
The presence of the Brucella-specific IS711 element and amplification of bcsp31 using Brucella specific primers in all strains demonstrated their affiliation with the genus Brucella. Strains were identical in the Bruce-ladder PCR, a multiplex assay for the differentiation of Brucella species, displaying a previously known banding pattern with amplicon sizes of 152, 272, 450, 587, and 794 bp. IS711 was also detected by Southern blot analysis. The IS711 copy number varied from one copy to more than 14 copies depending on the genotype determined with other molecular analyses (data not shown).
16S rRNA (rrs) and recA genes
Comparative sequence analysis of all 21 bullfrog isolates confirmed the two types of 16S rRNA gene sequences previously described19 (Table 2). Type A was identical with the sequence of B. inopinata whereas type B contained a 44 nt insertion apparently derived from Ochrobactrum species. A recA-gene based phylogenetic reconstruction revealed three main clusters and two strains with a unique sequence type each (Table 2).
MLSA and MLVA
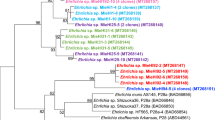
Multilocus sequence analysis (MLSA) showed the amphibian isolates to be different from all previously described Brucella spp. and comprise five distinct lineages with two singleton isolates and three larger clusters labelled A through E (Table 2). Cluster E consists of two subtypes with a different sequence at only a single locus. With the exception of the thyA allele 1, seen in seven isolates representing genotypes B and C, all alleles were novel, having not been previously reported in any Brucella examined to date. Distinct novel alleles were also found in each of the five distinct lineages. The clustering of the African bullfrog strains is entirely consistent with other molecular typing approaches (Table 2). Phylogenetic placement relative to other isolates is shown in Fig. 2 and clearly illustrates the five distinct lineages comprising clusters of six identical isolates, four identical isolates and nine isolates varying only at a single locus and two singleton isolates (10RB9215 and 10RB9213). This analysis clearly shows that the isolates could all justifiably be characterized as members of the atypical Brucella sharing a similar relationship to “core Brucella” as do B. inopinata (BO1) and B. inopinata-like isolates (strain BO2, Australian rodent strains). A recently described amphibian isolate from the UK21 was included in the analysis and found to be related to, but distinct from, the isolates described here, being most closely related to genotype D isolates.
Phylogenetic relationship of amphibian isolates with other Brucella species based on eight-locus MLSA. Numbers at nodes correspond to proportions of 500 resamplings that support the topology shown with only values >80% indicated. The bar indicates the number of substitutions per nucleotide position. UK8/14 represents a previously described amphibian isolate from the United Kingdom21.
In the comparative multilocus variable-number tandem repeat (VNTR) analysis (MLVA), the African bullfrog strains grouped together with B. inopinata (BO1), forming a separate cluster within the genus Brucella (Supplementary Fig. S3). Brucella neotomae, B. microti, B. suis bv 5, and the marine mammal brucellae, B. ceti and B. pinnipedialis, were most closely related (Supplementary Fig. S4). By use of panel 1 VNTRs, the 21 bullfrog isolates clustered into five different genotypes (MLVA-8 genotypes 101–105 [ http://microbesgenotyping.i2bc.paris-saclay.fr/]) that have not yet been previously identified in more than 3,500 other Brucella strains. On the basis of the MLVA-11 data set, seven individual genotypes could be identified, numbered 196 to 202.
Phylogenetic and comparative analyses
Phylogeny
Whole genome sequencing of nine isolates, at least two each from the three major clades and the two strains found in separate lineages, confirmed their affiliation with the genus Brucella (Fig. 3). Phylogenetically, the African bullfrog strains are basal to the classical Brucella species and are intermixed in the tree with isolates from Australian rodents and the unusual BO1 and BO2 isolates associated with human clinical disease but of unknown transmission route to humans. Four distinct clades of bullfrog isolates are apparent but limited bootstrap support for these basal groups and a high level of homoplasy (homoplasy index = 0.5091) that is substantially increased by inclusion of these lineages in phylogenies suggest substantial uncertainty for the topology of the amphibian and related Brucella strains.
The bullfrog isolates revealed a number of genomic regions that are not seen in any of the known Brucella genomes. These regions are either shared across all of the African bullfrog strains, shared across some of the strains, or found in a specific genome (Supplementary Table S2).
Since our phenotypic investigations revealed that none of the strains agglutinated with monospecific anti-Brucella sera and testing for bacterial swarming showed that they were motile, we focused on the wbk region involved in O-antigen synthesis and the flagellar genes.
LPS and O-antigen comparison
SDS-PAGE and silver stain analyses revealed that all bullfrog isolates produce S-LPS, but with a profile clearly different from A- or M-dominant S-LPS of classical Brucella species (Fig. 4A). Five distinct S-LPS profiles were found correlating with the genotypes assessed by molecular analyses. None of these five unique profiles corresponded to the atypical S-LPS profile of BO2. Furthermore, none of the bullfrog isolates had a positive ELISA when mAbs against the different classical Brucella O-chain epitopes were used (Fig. 4B), which is similar to BO2. There was a weak agglutination with anti-A polyclonal serum, indicating that the binding may be nonspecific. Nevertheless, all isolates reacted with anti-R-LPS mAb A68/10A6/B11 in Western blot revealing both the R-LPS moiety and S-LPS part as in previous studies24,25 (Fig. 4B), suggesting that the core-lipid A of the amphibian Brucella sp. strains is structurally related to that of classical Brucella species.
Silver staining (A) and Western blot (B) profiles with mAb A68/10A06/B11 (anti-R-LPS) after SDS-PAGE of proteinase K-digested S-LPS preparations of B. melitensis 16 M (M-dominant reference strain) (lanes 1), B. suis 1330 (A-dominant reference strain) (lanes 2), B. microti CCM 4915 (lanes 3), Brucella sp. strain 83/13 (wild rodent isolate from Australia) (lanes 4), B. inopinata BO1 (lanes 5), Brucella sp. strain BO2 (lanes 6), African bullfrog strains 09RB8910 (lanes 7), 10RB9207 (lanes 8), 10RB9215 (lanes 9), 10RB9213 (lanes 10), 09RB8915 (lanes 11).
As each bullfrog strain had a unique S-LPS profile, we specifically searched for the nineteen genes in the wbk region that are essential for LPS synthesis26,27, the novel genes (rmlACBD) found in the BO228 and B13-009522 isolates, as well as the flanking areas of those regions. All Brucella genomes share the same 5′ flanking region (Fig. 5), and each of the genomes described here has a unique organization beyond the conserved 5′ end of wbkF, although 10RB9215 is nearly identical to the previously described B13-0095 genome (Fig. 5 and Supplementary Table S3). None of the frog isolates had a profile similar to that seen in the classical Brucella spp. and B. inopinata (BO1) genomes or to the novel configuration that strain BO2 has.
Comparison of the wbk region, involved in the synthesis of LPS in the classical Brucella strains, across B. microti, B. inopinata (BO1), the BO2 strain, B13-0095 isolated from a Pacman frog and five African bullfrog strains. Genes that are shared across different species have similar color and border patterns, and are also indicated by background shading. Genes with sequence similarity above 91% have solid borders and shading, but those with similarity between 70–90% are indicated by dashed borders. Coding sequences appear as arrows, mobile element proteins as grey triangles and tRNAs as solid boxes. Genes united on the same contig or chromosome are found within the same rectangle. The classically known wbk region can be seen in B. microti and B. inopinata. All genes, their product description, the assigned protein family, and the gene order in the newly annotated genomes are identified by a peg identifier (Supplementary Table S3).
The rmlACBD region is shared across two of the bullfrog strains (10RB9213 and 10R9215) as well as the previously described BO2 and B13-0095. Strains 10RB9215 and B13-0095 also share four genes upstream of this region. Two of these (rfbD and tagH) have been previously noted in the B13-0095 and BO222 genomes and are also found in strain 09RB8910. All proteins were the best bidirectional BLASTP hits to each other, but while the sequence similarity was greater than 99%, these two genes have somewhat reduced sequence similarity in BO2, 10RB9213 and 09RB8910. Another gene in this area, annotated as a glycosyl transferase, is also shared across these same genomes except for 09RB8910 (Fig. 5).
The wbk region is strikingly similar between B. microti and B. inopinata (BO1), as well as highly similar in strains 10RB9215 and B13-0095. In contrast, the bullfrog isolates and BO2 have unique genes in this region (Fig. 5 and Supplementary Table S3).
Two other genes, wboA and wboB, known to be important in LPS synthesis, are located outside of the wbk region. The wboAB genes27 are missing in all bullfrog strains, strain B13-0095, and strain BO2, but are present in B. inopinata (BO1) and all of the classical species.
Flagellar genes
An initial examination of the genes from the African bullfrog strains, the BO1 and BO2 strains, and B. microti CCM 4915 revealed an interesting pattern of pseudogenization, loss, or in-frame deletions of many of the flagellar genes. The examination was expanded to look across the major species and strains of Brucella as previously described11. A summary of the presence, absence, or sequence disparities among the known flagellar genes29,30,31 is provided (Supplementary Table S4). Figure 6 shows the genes involved in the construction and regulation of the flagellum, with red lettering indicating those few genes that appear to be functional in all of the compared strains.
Regulators of flagellar gene expression and proteins participating in assembly of the flagellum. Those genes that appear to be functional across all of the compared strains (see Supplementary Table S4) are marked in red.
Most of the classical Brucella examined had pseudogenized at least one, if not many of the flagellar genes. Brucella microti was the only exception, having no pseudogenes, but it did have two genes with internal deletions (fliG and flgI) and turned out to be immotile. There is no pattern of pseudogenization in the other species of the classical clade, but instead loss of function of different flagella genes appears in each species. Within the species, some strains revealed pseudogenes that appear to be functional in their close relatives (Supplementary Table S4). Fewer pseudogenes are seen in strain BO2 and in the isolates from Australian rodents, and all genes in strain BO1 appear to be fully functional.
The flagella genes from the five African bullfrog strains we examined appear to be mutation-free. The only exceptions are in 10RB9213′s motE that has an internal deletion of 5aa, and 09RB8910′s flgI gene that has an upstream mutation which shortens it to 409aa, a mutation shared with the genomes in the ancestral clade (Supplementary Table S4). As all bullfrog strains are motile, these few mutations apparently do not influence motility.
Infection experiments
The pathogenic potential of Brucella spp. correlates with their capacity to replicate in host macrophage cells and in target organs. The fate of the amphibian Brucella strains in mammalian macrophages and in the murine model of infection is of particular interest because African bullfrogs have been described as the first cold-blooded host of Brucella.
J774 macrophage infection
Brucellae isolated from amphibians readily multiplied in J774 murine macrophage-like cells (Fig. 7A). However, there were significant differences between the ability of our amphibian strains and classical B. suis 1330 to replicate within mammalian macrophages. At 24 h post-infection, the number of intracellular bacteria was ~2 to 4 logs higher than that of B. suis 1330 (Student’s t-test, p ≤ 0.01). The observed reduction in viable intracellular counts for at least one out of four bullfrog strains (09RB8471) at 30 h post-infection may be explained by very rapid growth, resulting in macrophage lysis and killing of released bacteria in the gentamicin-containing medium. In contrast, strain 09RB8910 reached higher bacterial loads, maybe because the infected macrophages were more efficiently protected from apoptosis32. Over a period of 24 h, the overall intramacrophagic growth rate of these strains was comparable to that of B. inopinata (BO1), and slightly higher than that of B. microti CCM 4915 (Fig. 7B).
Intracellular multiplication of (A) the African bullfrog strains 09RB8471 (●), 09RB8910 (▲), 09RB8913 (▼) and 10RB9213 (■) in comparison with (B) B. microti CCM 4915 (●), B. inopinata BO1 (▼), and B. suis 1330 (▲) in murine J774 macrophage-like cells. All experiments were performed three times in triplicate each, and results of one typical experiment are shown, presented as the means ± standard deviation.
BALB/c mice infection
The ability of the bullfrog strains to replicate in murine macrophages prompted us to analyse the susceptibility of BALB/c mice. In a preliminary experiment, 105 CFU of strains 09RB8471 and 09RB8910 were intraperitoneally injected. This dose is widely used in standard murine infections with B. suis 1330 but is known to be lethal for BALB/c mice which are infected with B. microti CCM 4915, B. inopinata BO1, and the Australian rodent strain 83-21033,34. However, our amphibian isolates were not lethal for BALB/c mice using this specific dose.
The pathogenic potential of the Brucella sp. strains isolated from African bullfrogs was also assessed by following up splenic and hepatic colonization (Fig. 8A,C) as well as spleen and liver weights (Fig. 8B,D) from day 3 to 84 post infection with 104 CFU. The bacterial replication rates in spleen and liver peaked between the third and seventh day after infection (Fig. 8A,C), similar to other Brucella species35 including B. suis 1330. The bullfrog strains were eliminated much more rapidly from spleen and liver than B. suis 1330 but still persisted 84 days post infection (except for 09RB8910 in the liver, which was totally cleared by this time point). Remarkably, at the end of the experiment (84 d post infection) strain 09RB8913 showed persistence in the spleen comparable to B. suis 1330.
Bacterial counts and organ weights of spleen and liver. Growth curves of Brucella sp. strains 09RB8471 (○), 09RB8910 (△), 09RB8913 (▽), 10RB9213 (□), and B. suis 1330 (●) in spleens (A) and livers (C) of BALB/c mice after intraperitoneal inoculation of 104 colony-forming units (CFU) of the bacteria. For each infection experiment, five mice were killed per time point after 3, 5, 7, 14, 28, 56, and 84 days and the number of viable bacteria was counted. Spleen (B) and liver weights (D) were also determined. Data are presented as mean values ± standard deviation.
In the course of infection, the colonization rate of the target organs was significantly different between BALB/c mice infected with B. suis 1330 and those infected with the bullfrog strains, except for 10RB9213 in the spleen three and five days post infection, and for 10RB9213 and 09RB8913 in the liver 12 weeks post infection.
Neither spleen nor liver weight significantly increased due to the infection with the amphibian Brucella sp. strains indicating a very mild inflammatory reaction.
Discussion
The bullfrog strains genetically group together, although showing considerable heterogeneity, and are basal to the core Brucella species (Figs 2 and 3). This amphibian group is distantly related to Ochrobactrum but nested within existing atypical Brucella species, including isolates from Australian rodents, B. inopinata BO1, and Brucella sp. strain BO2. Although it is likely that more Brucella species from the classic clade – species that appear to be host associated – will be discovered, new species within these new basal lineages and their genetic diversity challenge our concepts of what defines Brucella.
Greater genetic diversity exists among the relatively few members of the basal Brucella clade than in the hundreds of strains in the classical clade. This diversity may come from the ability of these basal species to exchange DNA with each other and with other microbes in the environment using horizontal gene transfer. A similar situation is seen in the Mycobacterium complex where M. canettii, an opportunistic pathogen for humans, undergoes horizontal exchanges with other mycobacteria, while M. tuberculosis, the host adapted obligatory pathogen, is strictly clonal36.
Our analysis expands the previously described ancestral clade of Brucella11 which included the isolates from Australian rodents (83/13 and NF 2653) and two novel isolates from human infections (B. inopinata BO116 and Brucella sp. BO217). Figure 3 shows that the basal clade of atypical strains can be divided into two subclades; the first contains the Australian rodent isolates and five of the bullfrog strains (10RB9215, 10RB9213, 10RB9214, 10RB9212, 09RB8910). The other major subclade contains B. inopinata (BO1), BO2, and the Pacman frog strain B13-0095, as well as four of our bullfrog strains 09RB8918, 09RB8913, 10RB9210, 09RB8471. As part of these two subclades, our bullfrog strains thus span much of the known diversity within the atypical Brucella species. Interestingly, B. vulpis is related but is basal to all of these other atypical species. Combined, these results suggest substantial undiscovered diversity in the genus.
Comparative whole genome analysis revealed a set of genes present on the chromosomes of the African bullfrog strains that are not found in other Brucella species but found in soil bacteria such as Rhizobium and Agrobacterium (data not shown). In contrast to their soil- and plant-associated relatives, which harbour virulence factors such as plasmids and temperate phages necessary to survive in fast changing harsh environmental conditions, having these type of genes seems to be superfluous for an organism adapted to intracellular survival37. Recently, we described a temperate phage residing in the human isolate B. inopinata that was very similar to a temperate phage of O. anthropi and to several prophages identified in rhizobiales38. These findings and the detection of pili-like structures in the Brucella sp. isolated from African bullfrogs may hint that horizontal gene transfer does occur between Brucella and other bacteria in the same niche, such as soil associated bacteria.
This notion that the basal species more readily exchange DNA with each other and other microbes is strengthened by the great diversity of new genomic regions found in the five strains more deeply analyzed, and is also apparent in the heterogeneity of the wbk region (Fig. 5 and Supplementary Table S3). Across the classical clade, the composition and order of wbk is conserved, with the only mutations seen in some of the genes from those species known to have a rough phenotype. When BO2 was first described, phenotypic analysis and sequencing showed that it had a different structure in its wbk region that included four new genes predicted to be involved in constructing a rhamnose-based O-antigen, making it unique at that time among Brucella28 but well described in other bacteria39,40. These genes, rmlACBD, encode a rhamnose-based O-antigen28 that is different from the N-formyl-perosamine-based O-antigen produced by classic Brucella41. As our knowledge of Brucella expands with the amphibian isolates, our understanding of the wbk region is rapidly evolving and suggests that this is an active region of change within these genomes. Strains 10RB9213 and 10RB9215 both have those same rml genes, but have radically different genes in the upstream flanking region. In fact, each of the amphibian representatives from this study has a unique set of genes in the wbk region (Fig. 5 and Supplementary Table S3), although they share (for the most part) the flanking regions at either end. They each have a variety of genes that are unique not only among strains that have been described previously, but also unique compared to each other. The sole exception is the African bullfrog strain 10RB9215 and the Pacman frog isolate B13-0095, which are almost identical in this region. It appears that horizontal transfer is a major diversifying force in this area, but it is impossible to say just what gene structures the ancestor to all the Brucella originally had between the conserved flanking regions. It is also impossible to discern what type of O-antigen that ancestor possessed. Comparison to Ochrobactrum genomes show that they have the rmlACBD operon, too, but here the same flanking regions are widely dispersed, with the 5′ and 3′ ends on separate chromosomes (as per O. anthropi ATCC 49188, data not shown).
The flagellar system, which is phylogenetically related to the type III secretion system and as such may be involved in the secretion of virulence factors, appears to be essential for the infectious cycle and persistence of Brucella in mammalian hosts. Brucella melitensis has been shown to produce a sheathed flagellum during the early stages of exponential growth29,42, and while B. melitensis with mutated flagellar genes were not attenuated in cellular models of infection, BALB/c mice were able to clear them within 12 weeks of infection29. When comparisons are made across all Brucella and the sequences of the genes that produce the flagellum, an extensive pattern of pseudogenization is apparent (Fig. 6 and Supplementary Table S4), suggesting that most, if not all of the known classical species are not motile. In contrast, the African bullfrog strains have a flagellum (Fig. 1) and are motile (Supplementary Fig. S2). BO2 also has a full component of functional genes, and motility has recently been demonstrated in this strain22. One could hypothesize that the strains in the classical clade, with the possible exception of B. microti, accumulated mutations in their flagellar genes in a random manner because they no longer require mobility. Their predominantly intracellular lifestyle may not require the presence of a flagellum.
Although primarily isolated from a cold-blooded host, the African bullfrog strains revealed long-term survival in mammals comparable to the classic Brucella species in BALB/c mice (up to 84 d after inoculation). In contrast to the hepatosplenomegaly observed after B. suis infection, intraperitoneal infection of BALB/c mice with the African bullfrog strains proceeded in the form of asymptomatic persistence. This might be explained by differences in the expression of flagellin, which is known to play a key role in the immunological stand-off between Brucella and its host43. Both the downregulation of flagellin expression as well as the changes in the amino acid sequence of flagellin may help to avoid recognition by the innate immune system of the host43. Whether the variable flagellar genes of the amphibian Brucella sp. strains may contribute to the stealthy strategy of this pathogen remains unknown. Interestingly, motility of the flagellum necessary to cope with environmental stress and the flagellum as a virulence factor for persistent infection in a mammalian host do exist in parallel in the bullfrog isolates.
A major aspect of Brucella virulence is the capacity of the bacteria to replicate inside macrophages and their ability to escape the host immune system. We tested the pathogenic potential of the novel amphibian isolates in both in vitro and in vivo infection models. The African bullfrog strains showed an enhanced capacity to replicate in macrophages at a level comparable to B. microti, B. inopinata, and the Australian rodent isolates33,34 and inconsistent with classical B. suis. The high replication rate in phagocytic cells correlates well with the fast growth of the bullfrog strains in culture medium. Specific metabolic pathways might be responsible for the faster growth and metabolism of the bullfrog isolates differed essentially from other brucellae (Supplementary Table S1 and Supplementary Fig. S1). However, despite the higher replication rate of these atypical Brucella strains in vitro, they apparently do not cause death in the murine model of infection. New virulence factors of the amphibian Brucella sp. strains such as the functional flagellum or a novel lipopolysaccharide might be responsible for their long-term persistence in mammals but further experiments are needed to address this hypothesis.
Despite their close relationship to the atypical fast-growing brucellae, the African bullfrog strains revealed long-term survival in mammalian hosts comparable to strains from the classical clade. Although the pathogenicity of brucellae in vivo usually correlates with their ability to replicate in macrophages, the bullfrog strains neither killed BALB/c mice with a standard infectious dose of 105 bacteria such as other atypical brucellae nor caused a systemic inflammatory response associated with an increase in liver and spleen weights as seen in classical brucellae. Hence, according to our experimental data and considering the potential misidentification as Ochrobactrum sp. as well as the difficulties in serological tests due to modifications of the LPS, the potential infections of livestock and humans by the amphibian Brucella sp. cannot be easily assessed.
Concluding Remarks
In the last decade, the emergence of several new Brucella species as well as atypical Brucella strains have enhanced our understanding of the evolution of the genus from a soil-associated motile bacterium to a host adapted clonal pathogen. Our results greatly extend understanding of the genomic and phenotypic diversity of atypical Brucella. The amphibian strains that are described here form a remarkable group of heterogeneous brucellae, characterized by some unique features. First, Brucella-like organisms have been described from African bullfrogs and other frogs, but this is the first comprehensive description of variable Brucella genomes from a single cold-blooded host. Interestingly, the Brucella sp. strains isolated from African bullfrogs revealed versatile adaptability both to cold-blooded animals and endotherms. Second, while a single amphibian strain was recently shown to be motile, our studies confirm that motility is a common feature of diverse organisms that are progenitors to the classical zoonotic Brucella. Third, the bullfrog strains show long-term persistence in BALB/c mice without causing disease. The zoonotic potential and pathogenicity of these Brucella sp. strains must be assessed carefully because of their close phylogenetic relationship to the human opportunistic pathogens B. inopinata (BO1) and Brucella sp. strain BO2.
Our data support the hypothesis that the classical core Brucella members have emerged from soil bacteria that are characterized by motility and a broad metabolic activity. Brucella evolved to become a highly virulent but host specific clonal pathogen by genome reduction and adaptation. However, the apparent competence of the amphibian strains to exchange DNA may allow Brucella to adapt to changing environments quickly and give it a broad range of hosts.
Methods
Bacterial strains
A total of 21 strains (Table 1) isolated from tissue samples of various moribund or dead African bullfrogs kept in a quarantine unit of the Frankfurt Zoo (Germany) were analyzed44. The animals had been originally imported from Tanzania.
The bacteria were grown on Brucella agar and Columbia agar for 48 h at 37 °C both with and without 10% CO2. Molecular analyses (16S rRNA [rrs], recA, multilocus sequence typing [MLST], and multiple-locus variable-number tandem repeat analysis [MLVA]) as well as phenotypic characterization (biochemical profiling, agglutination, phage lysis) were performed as described previously for B. microti isolates10. Whole genome sequencing was conducted on nine representative strains.
Phenotyping
Classical microbiological methods were employed, that determine CO2 requirement, H2S production, urea hydrolysis, agglutination with monospecific sera (anti-A, anti-M, and anti-R), dye sensitivity (basic fuchsin and thionine), and phage lysis (F1, F25, Tb, BK2, Iz, Wb, R/C) for the identification and sub-differentiation of the African bullfrog strains45.
Since members of the atypical group of novel emerging Brucella species display lipopolysaccharide (LPS) heterogeneity25,28, structural differences in LPS were investigated using monoclonal antibodies (mAbs) specific for O-polysaccharide (O-PS) epitopes: 2E11 (IgG3; M epitope), 12G12 (IgG1; C [A = M] epitope), 12B12 (IgG3; C [M > A] epitope), 18H08 (IgA; C/Y [A = M] epitope), 05D4 (IgG1; C/Y [A > M] epitope). An ELISA was set up with hybridoma supernatants including mAbs and whole-cell bacterial antigens. SDS-PAGE of proteinase K-digested S-LPS preparations followed either by silver staining or Western blotting was performed as described previously25.
Metabolic activity was evaluated using a commercial biotyping system (Micronaut; Merlin Diagnostika, Bornheim-Hersel, Germany) as described previously23. The 96-well Micronaut® BfR Brucella assay tests for 29 aminopeptidases, two phosphatases, four glucosidases, one esterase, and the metabolism of eleven monosaccharides, three disaccharides, seven sugar derivates, 15 amino acids, eleven organic acids, one salt, one amino acid derivate, one peptide, one base, and six classical reactions (nitrite and nitrate reduction, pyrazinamidase, Voges-Proskauer medium, urease, and H2S production). Biotyping experiments were carried out in triplicates. Hierarchical cluster analysis was conducted by Ward’s linkage algorithm using binary coded data based on empirically set cutoffs (Bionumerics v. 7.5, Applied Maths, Sint-Martens-Latem, Belgium). All characters were considered equal within the data set.
Motility of the bacteria was tested using a 0.3% semisolid agar including TTC (2,3,5 triphenyltetrazolium chloride) to visualize dynamics of bacterial swarming. Exemplarily, Brucella sp. strain 10RB9206 was harvested after 12, 24, 36, 48, 60, 72, and 84 h of culture to analyze the ultrastructure of a prospective flagellum using transmission electron microscopy (JEM-1010 electron microscope). Bacterial samples were prepared for microscopic examination by negative staining with 1% solution of uranyl acetate dissolved in distilled water (pH 4.2 to 4.5).
DNA preparations
A single colony of each strain was transferred from the agar plate to 200 μl 5x lysis buffer D (PCR Optimizer kit; Invitrogen, De Schelp, The Netherlands) diluted 1:5 in distilled water, supplemented with 0.5% Tween 20 (ICI America Inc., Merck, Hohenbrunn, Germany) and 2 mg/ml proteinase K (Roche Diagnostics, Mannheim, Germany). The bacterial samples were incubated at 56 °C for 1 h and inactivated at 95 °C for 10 min, before crude DNA samples were purified using the QIAamp DNA Mini kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions.
High quality genomic DNA (gDNA) was prepared for whole genome sequencing by using a Qiagen genomic extraction kit and Qiagen Genomic-tip 100/G (Qiagen, Hilden, Germany) according to manufacturer’s recommendations. Bacterial lysis was optimized by three freeze/thaw cycles in a lysozyme/lysostaphin buffer.
IS711, bcsp31, and Bruce-ladder multiplex PCR
The presence of both the Brucella-specific bscp31 gene and insertion element IS711 was determined19 and the Bruce-ladder multiplex PCR was used in its modified version46. Presence of IS711 and its copy number were also assessed by Southern blot analysis as described previously47.
Analysis of 16S rRNA and recA genes
16S rRNA (rrs) and recA (recombinase A) genes were amplified and sequenced as described previously48,49. Almost the entire rrs sequence was amplified using universal primers, i.e. 27f and 1492r (all primer sequences can be retrieved from Al Dahouk et al.)10. The primer pair recA-BrucOchro-f and recA-BrucOchro-r generated a fragment including the whole recA gene (1,086 bp). 15 pmol of each primer were added to 50 μl Ready-To-Go master mix (Eppendorf GmbH, Hamburg, Germany) and PCRs were carried out in a Perkin-Elmer GeneAmp 2400 thermal cycler (Perkin-Elmer, Applied Biosystems, Foster City, CA, USA). A total of 30 cycles were performed: 30 s of denaturation at 94 °C, 30 s of annealing at 58 °C (rrs) or 65 °C (recA), and elongation at 72 °C for 90 s (rrs) or 60 s (recA). The run was completed with a final elongation step of 7 min at 72 °C. PCR products were analyzed by agarose gel electrophoresis (1% [wt/vol] in Tris-Acetate-EDTA [TAE] buffer). The purified recA and rrs fragments were sequenced with an ABI Prism 3100 genetic analyzer (Applied Biosystems, Foster City, CA, USA) using recA-BrucOchro-f/recA-BrucOchro-r primers and an internal primer set consisting of 341fw, 518r, and 926 f, respectively. ClustalW, version 1.8, was used (http://clustalw.genome.jp/) to generate multiple sequences alignments.
MLSA and MLVA
Multilocus sequence analysis was performed as described previously50 by determining DNA sequences at nine independent genetic loci. In order to establish the relationships of the amphibian isolates, these sequence data were compared with equivalent data from the 27 sequence types (STs) originally described, representing all Brucella species and biovars recognized at the time as well as more recently emerged Brucella spp. including the type strains of B. microti, B. papionis, B. inopinata, an organism described as B. inopinata-like (strain BO2)17 and a representative of a group of atypical isolates from Australian rodents18. Sequences for eight of the nine loci examined (excluding one not present in the Ochrobactrum outgroup) were concatenated as described previously19,21. Alignment and phylogenetic analysis were performed within the MEGA 5.2 package using the Jukes-Cantor distance and the neighbor joining approach.
Multiple-locus variable-number tandem repeat (VNTR) analysis (MLVA-16) was carried out as described by Le Flèche and colleagues51 and modified by Al Dahouk and colleagues52 using eight minisatellite markers (panel 1: bruce06, bruce08, bruce11, bruce12, bruce42, bruce43, bruce45, bruce55) and eight microsatellite markers (panel 2A: bruce18, bruce19, bruce21; panel 2B: bruce04, bruce07, bruce09, bruce16, bruce30). Alleles were called according to version 3.6 of the Brucella table for allele assignment (MLVA allele-coding convention to convert allele size (bp) into number of repeats (u)).
Phylogenetic analysis
Comparative genomic analyses were conducted following previously described methodology53,54 to discover single nucleotide polymorphisms (SNPs). Representative strains clustering in different groups were selected according to molecular typing data. Genome sequencing was carried out by the sequencing services provider GATC Biotech, Konstanz, Germany using either MiSeq (Illumina Inc., San Diego, CA, USA) or PacBio (Pacific Biosciences, Menlo Park, CA, USA). De novo assembly of the Illumina and PacBio sequencing reads was done with CLC Genomics Workbench 9.0 (CLC bio, Aarhus, Denmark) and SMRT Analysis Software v. 2.3 (Pacific Biosciences), respectively. Illumina reads were assembled using standard parameters. For the assembly of PacBio reads, the HGAP3 algorithm with a read length minimum of 2,500 bp was used. SNP analysis was conducted with the NASP v. 1.0.0 pipeline (https://github.com/TGenNorth/NASP) using default settings for the programs implemented in the pipeline. Briefly, assemblies of the amphibian Brucella sp. in FASTA format were aligned to the B. abortus 2308 reference genome (GenBank accession numbers NC_007618, NC_007624) and analyzed for SNPs with MUMmer 3.2355; SNP discovery using sequence reads gave nearly identical results (data not shown). Reads were aligned to the reference with BWA v. 0.3.756, and SNPs discovered in the alignments with the Genome Analysis Toolkit v. 2.5 Unified Genotyper57. We required SNP loci had have a base call in all samples, i.e., no missing data. SNP loci in duplicated regions, determined by an alignment of the reference to itself with MUMmer, were excluded from analyses. Phylogenetic trees were built using maximum parsimony in PAUP* with 100 bootstrap replicates to indicate the amount of support for various branches58. Trees were visualized with FigTree v. 1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/). Eight of the classical Brucella species were included in the tree for phylogenetic context and to contrast the amounts of genetic diversity among the various clades. Ochrobactrum anthropi ATCC 49188T (GenBank accession numbers CP000758-CP000763) was used as an outgroup.
Comparative analysis
The comparative gene-based analysis included the genomes of strains 10RB9213, 10RB9215, 09RB8910, 09RB8913, and 09RB8471. An initial examination using 16S rRNA revealed that some bullfrog isolates were closely related to B. inopinata BO119, so these genomes were compared to B. inopinata BO1. Brucella sp. BO2 and B. microti CCM 4915T were also chosen for the initial comparison. All genomes were annotated with RASTtk to provide consistency across the results. A list of all genomes used, as well as their annotation statistics, is provided (Table 3).
Several methodologies were used to compare the genomes, all of which are available in PATRIC53. The Protein Family Sorter54 was used to look for unique regions in individual genomes. Regions determined to be unique to each bullfrog strain, or shared across them, were verified by BLAST against the representative genome database at NCBI. In addition, these unique regions were examined by BLAST against both the complete plasmid and complete bacteriophage databases.
A detailed comparison of specific genomes (10RB9215, B13-0095, 10RB9213, BO2, 09RB8910, 09RB8913, 09RB8471, B. microti CCM 4917 and B. inopinata BO1) was conducted to analyze the wbk region. Contigs from each of these genomes were re-annotated using the current version of RASTtk59 available in the PATRIC bioinformatics resource53,60. Annotation in PATRIC now assigns protein families that are scoped at the genus level, and these PLFams were compared for this analysis61. The annotated genomes were first compared in a bi-directional, best BLASTP hit analysis using PATRIC’s Proteome Comparison tool60. The wbk region was selected from genome, with special attention paid to the flanking genes and the contigs they were isolated on, as well as the strength of the BLASTP hit, which included the % Sequence Identity, the % Sequence Coverage, and the directionality (uni or bi) of the hits. Only bidirectional hits were used. The PLFams were noted for all of the genes that matched these criteria. If there was a discrepancy between the PLFam assigned to each gene, a multiple sequence alignment was generated in PATRIC and examined. Following this examination, it was determined that genes with a Sequence Identity greater than 70% would be considered homologs.
A comparison of differences in metabolic pathways among the strains was made using the Comparative Pathway Tool53,54. When gene-specific differences were identified as unique to a particular genome, we looked more broadly across the different species of Brucella to examine consistency of results. We examined all available genomes at the species level to see if those differences were confined to the particular genome, which could indicate sequencing error, or were shared across most of the genomes available for the species.
Macrophage infection model
The replication of brucellae within macrophages is a prerequisite for virulence of the pathogen in a given host species. Using macrophage-like murine J774-cells, we therefore compared the behaviour of Brucella strains isolated from bullfrogs to that of the pathogenic B. suis strain 1330 and of the two fast-growing species B. microti (strain CCM 4915) and B. inopinata (strain BO1). For macrophage and murine infections, all strains have been cultivated on Tryptic Soy Broth (Biokar Diagnostics, Allonne, France) for 48–72 h.
Based on molecular and biotyping data, 4 of the most diverse of the 21 bullfrog isolates were selected for in vitro infection experiments; namely strains 09RB8471, 09RB8910, 09RB8913, and 10RB9213. Infection experiments were performed as described previously33. Briefly, adherent cells were resuspended at 2.5 × 105 cells/ml in RPMI 1640 cell culture medium supplemented with 10% foetal calf serum (FCS), and incubated for 24 h at 37 °C with 5% CO2 prior to infection. The cells were infected at a multiplicity of infection (MOI) of 20 with early-stationary phase bacteria cultured in TS medium. After 30 min, cells were washed twice with phosphate-buffered saline (PBS) and incubated in RPMI 1640/10% FCS with gentamicin (30 μg/ml) for at least 1 h to kill non-phagocytosed bacteria. Ninety minutes, 4, 24, and 30 h post-infection, cells were washed twice with PBS and lysed in 0.2% Triton X-100. CFUs were determined by plating serial dilutions on TS agar, followed by incubation at 37 °C for 2–3 days. All experiments were performed twice in triplicate.
Murine infection model
The mouse experiments were approved by the ethical review committee of the Centro de Investigación y Tecnología Agroalimentaria, Unidad de Sanidad Animal, Zaragoza, Spain (approval no. I95/2010–1 and I111/2010-1). Well-established experimentation guidelines were followed33,34. Eight weeks old, female BALB/c mice were provided by Charles River Laboratories (Chatillon-sur-Chalaronne, France). The four bullfrog isolates chosen for the macrophage infection experiments and B. suis 1330 as a control were intraperitoneally injected into the mice using 104 CFU in a single dose. Five animals per group were euthanized by CO2 asphyxiation after 3, 5, 7, 14, 28, 56, and 84 days. Spleens and livers were aseptically removed, weighed, homogenized, serially diluted in PBS, and plated onto Blood Agar Base (BAB) plates to count bacteria. Student’s t-test was applied to test for significant differences between the groups in the course of time, with p values ≤ 0.05 considered significant.
Additional Information
How to cite this article: Al Dahouk, S. et al. Brucella spp. of amphibians comprise genomically diverse motile strains competent for replication in macrophages and survival in mammalian hosts. Sci. Rep. 7, 44420; doi: 10.1038/srep44420 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Scholz, H. C. et al. Brucella vulpis sp. nov., isolated from mandibular lymph nodes of red foxes (Vulpes vulpes). Int. J. Syst. Evol. Microbiol. 66, 2090–2098 (2016).
Pappas, G., Papadimitriou, P., Akritidis, N., Christou, L. & Tsianos, E. V. The new global map of human brucellosis. Lancet Infect. Dis. 6, 91–99 (2006).
Christopher, G. W., Agan, M. B., Cieslak, T. J. & Olson, P. E. History of U.S. military contributions to the study of bacterial zoonoses. Mil. Med. 170, 39–48 (2005).
Pappas, G., Panagopoulou, P., Christou, L. & Akritidis, N. Brucella as a biological weapon. Cell. Mol. Life Sci. 63, 2229–2236 (2006).
Foster, G., Osterman, B. S., Godfroid, J., Jacques, I. & Cloeckaert, A. Brucella ceti sp. nov. and Brucella pinnipedialis sp. nov. for Brucella strains with cetaceans and seals as their preferred hosts. Int. J. Syst. Evol. Microbiol. 57, 2688–2693 (2007).
Hubálek, Z. et al. Brucellosis of the common vole (Microtus arvalis). Vector Borne Zoonotic Dis. 7, 679–687 (2007).
Scholz, H. C. et al. Brucella microti sp. nov., isolated from the common vole Microtus arvalis . Int. J. Syst. Evol. Microbiol. 58, 375–382 (2008).
Scholz, H. C. et al. Isolation of Brucella microti from soil. Emerg. Infect. Dis. 14, 1316–1317 (2008).
Audic, S., Lescot, M., Claverie, J. M. & Scholz, H. C. Brucella microti: the genome sequence of an emerging pathogen. BMC Genomics 10, 352 (2009).
Al Dahouk, S. et al. Intraspecies biodiversity of the genetically homologous species Brucella microti . Appl. Environ. Microbiol. 78, 1534–1543 (2012).
Wattam, A. R. et al. Comparative phylogenomics and evolution of the Brucellae reveal a path to virulence. J. Bacteriol. 196, 920–930 (2014).
Schlabritz-Loutsevitch, N. E. et al. A novel Brucella isolate in association with two cases of stillbirth in non-human primates – first report. J. Med. Primatol. 38, 70–73 (2009).
Whatmore, A. M. et al. Brucella papionis sp. nov., isolated from baboons (Papio spp.). Int. J. Syst. Evol. Microbiol. 64, 4120–4128 (2014).
Hofer, E. et al. A potential novel Brucella species isolated from mandibular lymph nodes of red foxes in Austria. Vet. Microbiol. 155, 93–99 (2012).
De, B. K. et al. Novel Brucella strain (BO1) associated with a prosthetic breast implant infection. J. Clin. Microbiol. 46, 43–49 (2008).
Scholz, H. C. et al. Brucella inopinata sp. nov., isolated from a breast implant infection. Int. J. Syst. Evol. Microbiol. 60, 801–808 (2010).
Tiller, R. V. et al. Identification of an unusual Brucella strain (BO2) from a lung biopsy in a 52 year-old patient with chronic destructive pneumonia. BMC Microbiol. 10, 23 (2010).
Tiller, R. V. et al. Characterization of novel Brucella strains originating from wild native rodent species in North Queensland, Australia. Appl. Environ. Microbiol. 76, 5837–5845 (2010).
Eisenberg, T. et al. Isolation of potentially novel Brucella spp. from frogs. Appl. Environ. Microbiol. 78, 3753–3755 (2012).
Fischer, D. et al. Abscesses associated with a Brucella inopinata-like bacterium in a big-eyed tree frog (Leptopelis vermiculatus). J. Zoo Wildl. Med. 43, 625–628 (2012).
Whatmore, A. M. et al. Isolation of Brucella from a White’s tree frog (Litoria caerulea). JMM Case Rep. 2, doi: 10.1099/jmmcr.0.000017 (2015).
Soler-Lloréns, P. F. et al. A Brucella spp. isolate from a Pacman frog (Ceratophrys ornata) reveals characteristics departing from classical brucellae. Front. Cell. Infect. Microbiol. 6, 116 (2016).
Al Dahouk, S. et al. Differential phenotyping of Brucella species using a newly developed semi-automated metabolic system. BMC Microbiol. 10, 269 (2010).
Cloeckaert, A., Jacques, I., Bowden, R. A., Dubray, G. & Limet, J. N. Monoclonal antibodies to Brucella rough lipopolysaccharide: characterization and evaluation of their protective effect against B. abortus . Res. Microbiol. 144, 475–484 (1993).
Zygmunt, M. S., Jacques, I., Bernardet, N. & Cloeckaert, A. Lipopolysaccharide heterogeneity in the atypical group of novel emerging Brucella species. Clin. Vaccine Immunol. 19, 1370–1373 (2012).
Godfroid, F. et al. Genetic organisation of the lipopolysaccharide O-antigen biosynthesis region of Brucella melitensis 16M (wbk). Res. Microbiol. 151, 655–668 (2000).
González, D. et al. Brucellosis vaccines: assessment of Brucella melitensis lipopolysaccharide rough mutants defective in core and O-polysaccharide synthesis and export. PLoS One 3, e2760 (2008).
Wattam, A. R. et al. Comparative genomics of early-diverging Brucella strains reveals a novel lipopolysaccharide biosynthesis pathway. MBio. 3, e00246–12 (2012).
Fretin, D. et al. The sheathed flagellum of Brucella melitensis is involved in persistence in a murine model of infection. Cell. Microbiol. 7, 687–698 (2005).
Léonard, S. et al. FtcR is a new master regulator of the flagellar system of Brucella melitensis 16M with homologs in Rhizobiaceae. J. Bacteriol. 189, 131–141 (2007).
Terashima, H., Kojima, S. & Homma, M. Flagellar motility in bacteria structure and function of flagellar motor. Int. Rev. Cell Mol. Biol. 270, 39–85 (2008).
Gross, A., Terraza, A., Quahrani-Bettache, S., Liautard, J. P. & Dornand, J. In vitro Brucella suis infection prevents the programmed cell death of human monocytic cells. Infect Immun. 68, 342–351 (2000).
Jiménez de Bagüés, M. P. et al. The new species Brucella microti replicates in macrophages and causes death in murine models of infection. J. Infect. Dis. 202, 3–10 (2010).
Jiménez de Bagüés, M. P., Iturralde, M., Arias, M. A., Pardo, J., Cloeckaert, A. & Zygmunt, M. S. The new strains Brucella inopinata BO1 and Brucella species 83-210 behave biologically like classic infectious Brucella species and cause death in murine models of infection. J. Infect. Dis. 210, 467–472 (2014).
Grilló, M. J., Blasco, J. M., Gorvel, J. P., Moriyón, I. & Moreno, E. What have we learned from brucellosis in the mouse model? Vet. Res. 43, 29 (2012).
Blouin, Y. et al. Progenitor “Mycobacterium canettii” clone responsible for lymph node tuberculosis epidemic, Djibouti. Emerg. Infect. Dis. 20, 21–28 (2014).
Moreno, E. & Moriyón, I. Brucella melitensis: a nasty bug with hidden credentials for virulence. Proc. Natl. Acad. Sci. USA 99, 1–3 (2002).
Hammerl, J. A., Göllner, C., Al Dahouk, S., Nöckler, K., Reetz, J. & Hertwig, S. Analysis of the first temperate broad host range brucellaphage (BiPBO1) isolated from B. inopinata . Front. Microbiol. 7, 24 (2016).
Giraud, M. F. & Naismith, J. H. The rhamnose pathway. Curr. Opin. Struct. Biol. 10, 687–696 (2000).
Reeves, P. Evolution of Salmonella O-antigen variation by interspecific gene transfer on a large scale. Trends Genet. 9, 17–22 (1993).
Iriarte, M. et al. Brucella lipopolysaccharide: structure biosynthesis and genetics In Brucella: Molecular and Cellular Biology (eds López-Goñi, I. & Moriyón, I. ) 152–183 (Horizon Biosciences, Norfolk, UK, 2004).
Ferooz, J. & Letesson, J. J. Morphological analysis of the sheathed flagellum of Brucella melitensis . BMC Res. Notes 3, 333 (2010).
Terwagne, M. et al. Innate immune recognition of flagellin limits systemic persistence of Brucella. Cell. Microbiol . 15, 942–960 (2013).
Mühldorfer, K. et al. The role of “atypical” Brucella in amphibians: are we facing novel emerging pathogens? J Appl Microbiol. 122, 40–53 (2017).
Alton, G. G., Jones, L. M., Angus, R. D. & Verger, J. M. Techniques for the brucellosis laboratory (Institut National de la Recherche Agronomique, Paris, France, 1988).
Mayer-Scholl, A., Draeger, A., Göllner, C., Scholz, H. C. & Nöckler, K. Advancement of a multiplex PCR for the differentiation of all currently described Brucella species. J. Microbiol. Methods 80, 112–114 (2010).
Cloeckaert, A., Bernardet, N., Koylass, M. S., Whatmore, A. M. & Zygmunt, M. S. Novel IS711 chromosomal location useful for identification of marine mammal Brucella genotype ST27, which is associated with zoonotic infection. J. Clin. Microbiol. 49, 3954–3959 (2011).
Scholz, H. C. et al. Genotyping of Ochrobactrum anthropi by recA-based comparative sequence, PCR-RFLP, and 16S rRNA gene analysis. FEMS Microbiol. Lett. 257, 7–16 (2006).
Scholz, H. C. et al. Genetic diversity and phylogenetic relationships of bacteria belonging to the Ochrobactrum-Brucella group by recA and 16S rRNA gene-based comparative sequence analysis. Syst. Appl. Microbiol. 31, 1–16 (2008).
Whatmore, A. M., Perrett, L. L. & MacMillan, A. P. Characterisation of the genetic diversity of Brucella by multilocus sequencing. BMC Microbiol. 7, 34 (2007).
Le Flèche, P. et al. Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC Microbiol. 6, 9 (2006).
Al Dahouk, S. et al. Evaluation of Brucella MLVA typing for human brucellosis. J. Microbiol. Methods 69, 137–145 (2007).
Wattam, A. R. et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 42 (Database issue), D581–91 (2014).
Wattam, A. R., Gabbard, J. L., Shukla, M. & Sobral, B. W. Comparative genomic analysis at the PATRIC, a bioinformatic resource center. Methods Mol. Biol. 1197, 287–308 (2014).
Kurtz, S. et al. Versatile and open software for comparing large genomes. Genome Biol. 5, R12 (2004).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler Transform. Bioinformatics 26, 589–595 (2010).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research 20, 1297–1303 (2010).
Swofford, D. L. PAUP*. Phylogenetic analysis using parsimony (* and other methods), version 4.0. (Sinauer Associates, Sunderland, MA, USA, 2002).
Bretti, T. et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. 5, 8365 (2015).
Wattam, A. R. et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 45, D535–D542 (2017).
Davis, J. J. et al. PATtyFams: Protein families for the microbial genomes in the PATRIC database. Front Microbiol. 7, 118 (2016).
Acknowledgements
We are grateful to Nelly Bernardet, Cornelia Göllner, Anna-Louisa Hauffe, Robert Schneider, Astrid Thomas, and Phillip Vette for excellent technical assistance and Miguel Angel Navarro for animal care. We also thank Philippe Le Flèche for his assistance in MLVA typing, Mark S. Koylass and Jakub Muchowski for MLST analysis, Charles H.D. Williamson for genome assembling, and Jochen Reetz for electron microscopy. Thanks to Risa Y. Dickerman for her help in creating the flagellum figure. A.R. Wattam is supported in part by federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract no. HHSN272201400027C. The work of M.P. Jiménez de Bagüés was supported by the Spanish grant INIA-RTA2013-00065-C02-01. The work of S. Köhler and A. Occialini was funded by the Federal Institute for Risk Assessment, Germany (BfR grant no. 1329-485). Brucella research work at APHA is supported by the Department of Environment, Food and Rural Affairs (projects SE0314 and SE0316), at the German Federal Institute for Risk Assessment by the Federal Ministry of Food and Agriculture (BfR project no. 47-003, 1322-503, and 1322-619), and at the Hessian State Laboratory by the Hessian Ministry of Environment, Climate Change, Agriculture and Consumer Protection. The development of genotyping methods for strain identification of dangerous pathogens as part of microbial forensics is supported by the French DGA (Direction Générale de l’Armement) via the MicroType project ANR-14-ASMA-0002-02. The Bundeswehr Institute of Microbiology is a member of the European Biodefense Laboratory Network (EBLN) supported by the European Defence Agency (EDA).
Author information
Authors and Affiliations
Contributions
S.A.D. and H.C.S. designed the study. T.E. and F.M. isolated the novel Brucella strains from African bullfrogs and did initial phenotyping work. S.A.D. and J.A.H. were responsible for extensive phenotyping of the bullfrog strains. G.V., A.M.W. and H.C.S. oversaw molecular typing work. Phylogenetic genome analysis was executed by K.P.D. and J.T.F.; A.C. and M.S.Z. characterized the novel LPS of the bacteria. J774 macrophage infection and BALB/c mice infection experiments were conducted by S.K., A.O. and M.P.J.B., respectively. A.R.W. did the extensive bioinformatics analysis. S.A.D., A.R.W. and H.C.S. drafted the manuscript and all authors approved its final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Al Dahouk, S., Köhler, S., Occhialini, A. et al. Brucella spp. of amphibians comprise genomically diverse motile strains competent for replication in macrophages and survival in mammalian hosts. Sci Rep 7, 44420 (2017). https://doi.org/10.1038/srep44420
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep44420
This article is cited by
-
A molecular and bacteriological survey of Brucella spp. in wild venomous snake in Iran
European Journal of Wildlife Research (2024)
-
Oral vaccination with novel Lactococcus lactis mucosal live vector-secreting Brucella lumazine synthase (BLS) protein induces humoral and cellular immune protection against Brucella abortus
Archives of Microbiology (2023)
-
Detection of Brucella spp. in raw milk from various livestock species raised under pastoral production systems in Isiolo and Marsabit Counties, northern Kenya
Tropical Animal Health and Production (2020)
-
Expanding the host range: infection of a reptilian host (Furcifer pardalis) by an atypical Brucella strain
Antonie van Leeuwenhoek (2020)
-
The CO2-dependence of Brucella ovis and Brucella abortus biovars is caused by defective carbonic anhydrases
Veterinary Research (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.