Abstract
Helicobacter pylori chronically colonises half of the world’s human population and is the main cause of ulcers and gastric cancers. Its prevalence and the increase in antibiotic resistance observed recently reflect the high genetic adaptability of this pathogen. Together with high mutation rates and an efficient DNA recombination system, horizontal gene transfer through natural competence makes of H. pylori one of the most genetically diverse bacteria. We show here that transformation capacity is enhanced in strains defective for recN, extending previous work with other homologous recombination genes. However, inactivation of either mutY or polA has no effect on DNA transformation, suggesting that natural competence can be boosted in H. pylori by the persistence of DNA breaks but not by enhanced mutagenesis. The transformation efficiency of the different DNA repair impaired strains correlates with the number of transforming DNA foci formed on the cell surface and with the expression of comB8 and comB10 competence genes. Overexpression of the comB6-B10 operon is sufficient to increase the transformation capacity of a wild type strain, indicating that the ComB complex, present in the bacterial wall and essential for DNA uptake, can be a limiting factor for transformation efficiency.
Similar content being viewed by others
Introduction
Helicobacter pylori chronically colonises the stomach of approximately half of the world’s population. The infection triggers chronic inflammation, remaining in most cases asymptomatic but that can evolve into serious pathologies such as peptic ulcer or gastric cancer1,2. H. pylori infection can be eradicated by antibiotics treatment, but the last decade has seen dramatic increases in the appearance of resistant strains3. The amazing genome plasticity observed in comparative analysis of isolates could explain the high adaptability of H. pylori and its success as a pathogen4,5. Several reasons can be pointed out to explain the high genetic variability of H. pylori. This species presents one of the highest mutation frequencies known6 combined with abundant chromosomal rearrangements7. The widespread presence of strain-specific genes suggests that horizontal gene transfer (HGT) plays a prominent role in the genetic diversity of the species. First evidence for natural transformation resulting in HGT in H. pylori was reported in 19908 and further confirmed to occur in vivo during human infection5,9,10. The transformation capacity reported for different H. pylori isolates varies enormously. Transformation frequencies of up to 50% have been described under laboratory conditions11,12. This amazing transformation efficiency is likely to contribute to the increase in antibiotic resistance spreading observed the last ten years3,13.
Natural transformation has been studied for many years in several bacterial models, originally in the Gram-positive Streptococcus pneumoniae and Bacillus subtilis (for a review see ref. 14). In all the systems studied so far, including Gram-negative species, transforming double-stranded DNA (dsDNA) from the environment is imported through the bacterial membrane and processed to yield single-stranded DNA (ssDNA) before its internalisation into the cytoplasm. There, ssDNA is recognised by the Homologous Recombination (HR) machinery and integrated into the host chromosome. The loading of the RecA recombinase is facilitated by DprA, a recombination mediator protein dedicated to DNA transformation15. In the majority of bacteria, Gram-positive as well as Gram-negative, internalisation occurs through protein complexes homologue to Type 4 Pilus (T4P)14. However, in H. pylori this T4P is replaced by conjugation-type proteins with homology to Type 4 secretion system components from Agrobacterium tumefaciens. Computational and genetic analysis revealed that the membrane complex is encoded in H. pylori by two operons, comB2-B4 and comB6-B10, named after the A. tumefaciens orthologues16,17. This complex is essential for the entrance of transforming DNA (tDNA) into the periplasm. Based on the VirB/D system, the ComB7, ComB9 and ComB10 proteins are proposed to be involved in the formation of the outer membrane channel, while ComB6 and ComB8 are supposed to be anchored to the inner membrane18. The passage through the inner membrane is then carried out by the ComEC protein11,12,19. It has been proposed that DNA transport between the cell surface and the cytoplam in H. pylori occurs in two distinct steps12.
The conditions inducing natural competence for transformation, the fraction of the population affected and the growth phase in which this occurs vary from species to species. In most cases, natural competence for transformation is induced through complex regulatory networks responding to specific environmental cues (for reviews see refs 14, 20 and 21). H. pylori, on the other hand, was first presented as being constitutively competent8. Consistently, no orthologues for the competence regulatory genes studied in the other models were identified in its genome. However, it was shown that competence varies during growth in H. pylori22 and that modification of O2 and CO2 conditions can be determinant for DNA transformation efficiency in H. pylori23. More recent studies have shown that pH and oxidative stress are critical for defining H. pylori DNA uptake capacity. Indeed, DNA uptake is reversibly shut down below pH 6.524. In agreement with the reported increase in transformation capacity of strains deficient in the HR repair genes addA, addB and recO25,26, accumulation of DNA damage was proposed to boost transformation27. Interestingly, DNA damage caused by UV, fluoroquinolones or mitomycin C was shown to induce competence in the Gram-positive S. pneumoniae28,29 and in the Gram-negative Legionella pneumophila30. In the case of S. pneumoniae, the mutational burden in coding sequences caused by inactivation of a DNA repair pathway induces competence31. Because translational errors were also shown to induce competence32, the effect of mutations in coding sequences on competence was proposed to be a response to the presence of misfolded proteins.
Here, we used a genetic approach to explore the impact of DNA damage on H. pylori competence. We show that transformation capacity is induced in strains mutated in HR genes such as addA, recO or recN, but not in the polA and mutY genes involved in other DNA repair pathways. Furthermore, transformation efficiency correlates with DNA uptake capacity reflected in the formation of tDNA foci on the bacteria and, ultimately, with the comB6-B10 operon expression levels.
Results
DNA damage, mutations and bacterial transformation capacity
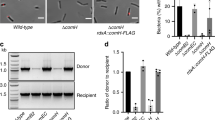
Previous reports showed that strains deficient in either RecOR or AddAB recombination mediator complexes display increased transformation frequencies25,26. Interestingly, the expression of competence genes in H. pylori, among which several comB, was found to be elevated in addA mutants as well as after treatment with ciprofloxacin, an antibiotic that inhibits several DNA processing enzymes and therefore generates DNA breaks27. These results suggested that the persistence of DNA strand breaks in the bacterial chromosome triggers an increase in transformation capacity. We confirmed the enhanced transformation frequencies of recO and addA mutants (Table 1 and Fig. 1A), albeit with different values likely due to improved bacterial culture conditions and transformation protocol with which we obtain a 10-fold increase in the yield of recombinants for a wt strain (see Methods). Because addA strains are slow growers and to avoid a possible bias in transformation efficiencies due to the heterogeneity of phase growth status characteristic of plate cultures, we repeated the experiment for wt and addA strains in the same growth phase using liquid cultures in controlled conditions. In that case we obtained a 6-fold increase in recombination frequencies for the mutant strain compared to the wt (data not shown). Consistently with the hypothesis that persistence of strand breaks can induce transformation, disruption of recN, another gene involved in strand break repair33, also resulted in a significant increase of recombinant frequencies when bacteria were transformed with genomic DNA from a streptomycin resistant strain (Table 1 and Fig. 1A). To test whether the mutation burden also affected transformation capacity, as proposed for S. pneumoniae31, we used as recipients strains defective in DNA repair pathways affecting mutation frequencies. We have previously shown that, due to its role in translesion synthesis, a deficiency in DNA Polymerase I leads to a 10-fold decrease in spontaneous mutation rates34. Despite this, the transformation capacity of a polA mutant strain was not significantly different from that of the wt (Fig. 1A and Table 1). We then tested a strain defective in MutY, a base excision repair protein involved in the avoidance of mutations induced by 8-oxoguanine35. Inactivation of mutY, while increasing mutation frequencies by 10-fold35,36,37, did not change significantly the transformation efficiency compared to that of the wt strain (Table 1 and Fig. 1A). Taken together, these results suggest that in H. pylori, DNA breaks, but not mutations, induce transformation capacity.
(A) Transformation frequencies. The bar represents the median, the boxes display the inner quartile range and the whiskers represent the minimum and maximum values. Number of experiments and p value determined with Mann-Whitney U test are indicated in Table 1. (B) Microscopy images of fixed bacteria. For each of the indicated strains, merged images are presented with red for FMX64, blue for DAPI and green for ATTO488. Scale bars correspond to 5 μm. (C) Proportion of bacteria with foci. At least 3 independent experiments were analyzed and more than 1200 bacteria were counted for each strain. The percentage of bacteria harboring foci are indicated for the different strains. The bar represents the median, the boxes display the inner quartile range and the whiskers represent the minimum and maximum values. (D) Western blot of wild-type and different mutants against ComB10 (upper line) and ComB8 (lower line).
Transformation capacity, tDNA foci formation and ComB expression level in DNA repair mutants
We next analysed whether the variations in transformation frequencies determined by the disruption of DNA repair genes were due to changes in the capacity of the bacteria to capture exogenous DNA or to process it once inside the cell. For that purpose, we used transforming DNA (tDNA) labelled with fluorescent ATTO-dUTP. ATTO-labelled tDNA forms ComB-dependent foci within the periplasm. The DNA present in the foci is eventually internalised into the cytoplasm and gives rise to recombinant bacteria11. When we analysed tDNA foci formation in the DNA strand break repair deficient strains, we observed in all cases an increase in the frequency of bacteria displaying foci (Fig. 1B and C, Table 2). These results indicate a correlation between the fraction of cells displaying tDNA foci, therefore DNA uptake, and the transformation capacity of the strain. It is worth noting that while this manuscript was in preparation a similar correlation between these two parameters was described by Kruger et al.24. Because the formation of tDNA foci requires the expression of a functional ComB complex (Tables 1 and 2 and refs 11,12) and comB genes RNA levels were found to be up-regulated in addA mutants27, we analysed the relative expression levels of ComB8 and ComB10 proteins in the different DNA repair mutant strains. As shown in Fig. 1D, the enhanced foci formation and transformation capacity of the recombination-deficient strains were associated with an increase in the expression of ComB8 and ComB10. In the case of mutY and polA strains, in which we did not observe variations in transformation frequencies nor in foci formation compared to the wild type (Table 2 and Fig. 1A and C), the levels of ComB8 and ComB10 were not significantly altered. In all the mutants tested here, the transformation efficiencies were positively correlated with the frequencies of bacteria with foci (r2 = 0.80; p = 0.0065) (Fig. S1B) and with the ComB8 expression levels (r2 = 0.65; p = 0.0299) (Fig. S1C).
Overexpression of comB6-B10 operon is sufficient to increase transformation
The results presented above suggested that the levels of ComB proteins, at least ComB8 and ComB10, could be a limiting factor for transformation in H. pylori. In order to test this hypothesis, we replaced in strain 26695 the promoter of the comB6-B10 operon by the ureA strong promoter38 (Fig. 2A). We obtained 2 independent clones that present an average of 2.6-fold induction for ComB8 expression (Fig. 2B, and Table 3). Analysis of those clones revealed significant increases in the frequencies of transformation and of foci formation (Table 3 and Fig. 2C). These results show that the level of ComB complexes present in the bacterial membrane determines the amount of tDNA taken up by the bacteria and is a limiting factor for transformation.
(A) Schematic representation of the genomic construction for over expression of comB6 operon. (B) Western blot against ComB10 or ComB8 of wild-type and two different strains constructed as indicated in A. (C) Correlation between transformation, foci formation and ComB8 expression in three independent clones overexpressing ComB8. In each experiment, all the values were reported to the value observed with the wt strain.
Transformation capacity, tDNA foci formation and ComB expression levels in different genetic backgrounds
Large variations in transformation frequencies have been reported amongst H. pylori isolates39. To determine if those differences are also a reflection of the DNA uptake capacities, we investigated the links between transformation, foci formation and ComB expression in different H. pylori genetic backgrounds. Because we monitored the replacement of a single nucleotide required for conferring streptomycin resistant, a major effect of restriction modification systems can be ruled out40. Moreover, the Mismach Repair pathway is absent in H. pylori41 and then cannot interfere in the experiment. The results presented in Table 4 and Fig. 3 show a general trend positively correlating transformation efficiencies with both, fraction of cells with foci (r2 = 0.75; p = 0.0115) and ComB8 expression (r2 = 0.59; p = 0.0426) (Fig. S1).
(A) Transformation frequencies. The bar represents the median, the boxes display the inner quartile range and the whiskers represent the minimum and maximum values. Number of experiments and p value determined with Mann-Whitney U test are indicated in Table 1. (B) Proportion of bacteria with foci. At least 3 independent experiments were analyzed and more than 1200 bacteria were counted for each strain. The percentage of bacteria harboring foci are indicated for the different strains. The bar represents the median, the boxes display the inner quartile range and the whiskers represent the minimum and maximum values.
Discussion
Several antibiotics induce genetic transformability in S. pneumoniae28. While some of them, like mitomycin C or fluoroquinones, induce DNA damage, others, like kanamycin and streptomycin, inhibit protein synthesis, suggesting that the induction of competence is a general stress response. In H. pylori there is contradictory data on the effect of antibiotics on competence. While an increase of transformation frequency was reported after ciprofloxacin exposure27, another group did not find an increase in the fraction of competent cells after treatment with either ciprofloxacin or mitomycin C24. Besides the fact that culture conditions used were different, such a discrepancy could also be explained by the fact that the end points determined were not the same, allowing the possibility that steps downstream of DNA uptake (i.e. DNA processing in the cytoplasm) are affected by the antibiotic treatments. Here, to avoid the pleiotropic effects of antibiotics, we used a genetic approach to analyse the impact of DNA damage on competence.
In a H. pylori strain deficient in AddA, a component of the nuclease/helicase recombination mediator AddAB, the expression of the genes coding for the competence T4SS is constitutively induced27. We had previously shown that inactivating genes coding for the components of the RecOR or AddAB mediator complexes, implicated in the formation of the RecA nucleofilament, leads to an increase in transformation frequencies25,26. Here, we extended these observations by showing that mutating recN, another gene involved in HR and DNA repair33,42, in strain 26695 also leads to increased transformation frequencies. As in most organisms, inactivation of HR in H. pylori sensitizes bacteria to agents inducing DNA strand breaks25,26,33,43,44,45. The increase in transformation frequencies in mutants affecting the initial steps of recombinational repair suggests that persistence of DNA strand breaks can be a cause for the enhancement of transformation capacity in H. pylori. Interestingly, disabling uvrA or uvrB genes, involved in the recognition of the lesion during nucleotide excision repair pathway, does not lead induction of transformation in H. pylori46. In S. pneumoniae it was suggested that a mutational burden itself could induce competence, possibly through the production of misfolded proteins31. Here, we show that in H. pylori the hypermutator phenotype obtained by inactivation of MutY, a protein implicated in base excision repair35, does not increase transformation frequencies (Table 1). Moreover, a polA mutant also deficient in DNA repair but displaying a hypomutator phenotype34 shows no modification of its transformation capacity. These results indicate that mutation frequency variations do not impact transformation capacity and suggest that it is the presence of persistent DNA breaks that is at the origin of the enhancement of transformability in H. pylori.
The lack of a known signalling pathway regulating competence in H. pylori raises the question of the mechanisms underlying the increased transformability observed in DNA repair deficient strains. Interestingly, the HR mutants ∆addA, ∆recO and ∆recN, that display higher transformability present higher levels of ComB8 and ComB10 proteins (Fig. 1), suggesting that the amount of the outer membrane ComB complex could be a limiting factor in the transformation process. This, together with the increased expression of comB genes transcripts in addA mutants27, indicates that the availability of the complex is likely to determine the DNA uptake capacity of the cell. Supporting this notion, overexpression of the comB6-10 operon in strain 26695 leads to an increase in tDNA uptake, as reflected by foci formation, and in transformation frequencies (Fig. 2). Overexpression of ComB10 alone was not able to boost transformation27 indicating that at least another one of the proteins coded by the operon is required. In view of the results presented here, it is puzzling to note that comB3 and comB4 are themselves constitutively induced in addA strains27. A possible explanation for these somehow contradictory results is that there is a positive feedback mechanism regulating the expression of both operons as suggested by the enhanced expression of comB9 in the comB4 merodiploid27.
While the analysis of the relation between transformation frequencies, DNA foci formation and comB expression in the seven wt isolates shows a positive correlation between those parameters in most cases (Fig. 4 and Fig. 1S), in some strains other factors seem to be affecting the relative efficiencies of transformation. We cannot rule out differences in downstream steps of the transformation process such as the handling of the DNA once inside the cytoplasm. However the comB6-10 operon overexpression experiment shows that all other factors being equal, the level of proteins coded by this operon is a limiting factor during transformation.
Taken together our results suggest a model by which the persistence of DNA breaks in the H. pylori genome induces the expression of the comB6-10 operon, to increase the levels of the outer membrane ComB competence complex. That allows an enhanced uptake of exogenous DNA into the periplasm, which is a limiting step in the transformation process. In this model, however, the link between DNA damage and the induction of ComB expression remains to be determined.
Methods
Oligonucleotides, enzymes and reagents
Oligonucleotides used in this work were from Eurogentec. Restriction endonucleases, DNA polymerases, and DNA modifying enzymes were purchased from New England Biolabs. Culture media and antibiotics were from AES Chemunex and Sigma Aldrich, respectively. Fluorescent nucleotides Atto-448-dUTP (aminoallyl-dUTP) (Jena bioscience) were purchased from Euromedex and integrated in DNA by PCR as decribed before11.
Strains and growth conditions
Except when indicated, all Helicobacter pylori strains were in 26695 background47 and are listed in Table S1. Plate cultures were grown at 37 °C in microaerophilic conditions (5% O2, 10% CO2, using the MAC-MIC system from AES Chemunex) on blood agar base medium (BAB) supplemented with 10% defibrillated horse blood (AES) and an antibiotics mix. Plates were incubated from 24 h to 5 days depending on the experiment or the mutant selected. Liquid cultures were grown at 37 °C with gentle shaking under microaerophilic conditions in brain heart infusion media (BHI) supplemented with 10% defibrillated and de-complemented foetal bovine serum (Invitrogen).
To generate the corresponding mutant derivatives, the gene of interest (see list in Table S2) cloned with its flanking regions into pJET2.1 (Fermentas) was disrupted by a non-polar cassette carrying either kanamycin- (Km), apramycin- (Apra), or chloramphenicol- (Cm) resistance genes. DNA was introduced into H. pylori strains by natural transformation and mutants were selected by growth on either 20 μg ml−1 Km, 12,5 μg ml−1 Apra or 8 μg ml−1 Cm. Allelic replacement was verified by PCR.
Construction of the strain overproducing comB6-B10 operon
The construction of the integrative plasmid was based on single strand annealing technology as described elsewhere48 where the different elements have been amplified by PCR using overlapping primers. Primers listed in Table S3 displayed an overlap sequence ranging from 15 to 21 nucleotides with a Tm ranging from 49 °C to 63 °C.
Briefly, the 380 bp of the hp0036 3′ sequence was amplified with Hp0036_F primer that overlapped with pUC19_R and with Hp0036_R primer that overlapped with HpKanR_F primer. The non-polar cassette carrying either Km resistance gene was amplified with HpKanR_F and HpKanR_R overlapping with PromUreA_F. The promUreA domain corresponding to the 325 pb sequence directly upstream of ureA gene was amplified with PromUreA_F and PromUreA_R overlapping with HpComB6_F. The hpComB6-1-133 domain was amplified with HpComB6_F and HpComB6_R overlapping with pUC19_F used to amplify pUC19. Finally, pUC19 was linearized with BamHI and used as template for PCR amplification with pUC19_F and pUC19_R.All PCR were performed with Phusion DNA polymerase. After amplification, the PCR products were treated with DpnI for parental template removal, and purified before assembly. Screening of correct plasmid was conducted by restriction and PCR and positive clones were confirmed by DNA sequencing.
The plasmid was introduced into H. pylori strains by natural transformation and mutants were selected by growth on 20 μg ml−1 Km, Allelic replacement was verified by PCR and sequenced. Overexpression of the operon was verified by Western Blot against ComB8 and ComB10.
Natural transformation assay
200 ng of genomic DNA from strain LR133 (StrR)25 was mixed with 105 cells from overnight plates resuspended in 15 μl of peptone water. Mixes were spotted on BAB plates. After 24 hours at 37 °C, dilutions of the resuspended spots were plated on BAB with and without the appropriate antibiotic (50 μg ml−1 Str) and incubated for 3 to 5 days. Transformation frequencies were calculated as the number of resistant colonies per recipient cfu.
Western blot analysis of ComB8 and ComB10
Proteins from total cell extracts were separated by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were incubated in blocking buffer (1X phosphate buffered saline [PBS] and 0.1% Tween 20 with 5% nonfat dry milk) overnight at 4 °C. Incubation with antibodies against either ComB8 or ComB10 (both provided by R. Haas, ref. 16) were carried out for 1 h at room temperature, followed by incubation with horseradish peroxidase (HRP)- conjugated secondary antibody under the same conditions. Proteins were revealed by adding ECL Prime Western blotting detection reagents (GE Healthcare) and read using the G-Box system (Syngene). Analysis of the correlation between ComB10 and ComB8 expression in all strains studied in this paper is significant (r2 = 0.6872 with a p value of 0,000459 (Fig. S1A).
PCR DNA labelling
We used a 405 bp PCR fragment of the rpsl gene (hp1197) from 26696 mutated to give a Streptomycin resistance phenotype (LR133).
Interaction of fluorescent DNA with H. pylori
To optimize images and to prevent cellular aggregation bacteria were recovered from exponential liquid cultures. 200 ng of ATTO labelled tDNA was mixed with 20 μl of cells (105 cells) in BHI medium. The suspension was incubated for 10 min at 37 °C. The cells were then pelleted, washed once with 100 μl of BHI and resuspended in 20 μl of BHI.
Microscopy assay on fixed bacteria
Samples from bacteria interacting with fluorescent ATTO-DNA were spotted on coated gelatin glass coverslips and fixed for 1 h with 4% formaldehyde. Slides were washed twice with PBS containing 0.1% glycine and once with PBS. Cell membranes were stained with FM64-X (Invitrogen) at a 1:500 dilution in PBS for 10 min and DNA was stained with 1 μg/ml 4,6-diamidino-2-phenylindole (DAPI). Coverslips were mounted in Dako fluorescent mounting medium. Image acquisition was performed with a Leica SPE confocal microscope (Wetzlar, Germany) using an ACS APO 63X (NA = 1.3) objective. Image treatment and analysis were performed using Leica and ImageJ software programs. All images presented correspond to a Z maximum intensity projection.
Statistical analysis
Pairwise comparisons were performed using Mann-Whitney test, and correlations by Pearson test were measured on GraphPad Prism software.
Additional Information
How to cite this article: Corbinais, C. et al. ComB proteins expression levels determine Helicobacter pylori competence capacity. Sci. Rep. 7, 41495; doi: 10.1038/srep41495 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Axon, A. Helicobacter pylori and public health. Helicobacter 19 Suppl 1, 68–73 (2014).
Sitas, F. Twenty five years since the first prospective study by Forman et al. (1991) on Helicobacter pylori and stomach cancer risk. Cancer Epidemiol 41, 159–164 (2016).
De Francesco, V. et al. Worldwide H. pylori antibiotic resistance: a systematic review. J Gastrointestin Liver Dis 19, 409–414 (2010).
de Reuse, H. & Bereswill, S. Ten years after the first Helicobacter pylori genome: comparative and functional genomics provide new insights in the variability and adaptability of a persistent pathogen. FEMS Immunol Med Microbiol 50, 165–176 (2007).
Suerbaum, S. & Josenhans, C. Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat Rev Microbiol 5, 441–452 (2007).
Wang, G., Humayun, M. Z. & Taylor, D. E. Mutation as an origin of genetic variability in Helicobacter pylori. Trends Microbiol 7, 488–493 (1999).
Kraft, C. & Suerbaum, S. Mutation and recombination in Helicobacter pylori: mechanisms and role in generating strain diversity. Int J Med Microbiol 295, 299–305 (2005).
Nedenskov-Sorensen, P., Bukholm, G. & Bovre, K. Natural competence for genetic transformation in Campylobacter pylori. J Infect Dis 161(2), 365–366 (1990).
Suerbaum, S. et al. Free recombination within Helicobacter pylori. Proc Natl Acad Sci USA 95, 12619–12624 (1998).
Falush, D. et al. Recombination and mutation during long-term gastric colonization by Helicobacter pylori: estimates of clock rates, recombination size, and minimal age. Proc Natl Acad Sci USA 98, 15056–15061 (2001).
Corbinais, C., Mathieu, A., Kortulewski, T., Radicella, J. P. & Marsin, S. Following transforming DNA in Helicobacter pylori from uptake to expression. Mol Microbiol 101, 1039–1053 (2016).
Stingl, K., Muller, S., Scheidgen-Kleyboldt, G., Clausen, M. & Maier, B. Composite system mediates two-step DNA uptake into Helicobacter pylori. Proc Natl Acad Sci USA 107, 1184–1189 (2010).
Hu, Y., Zhang, M., Lu, B. & Dai, J. Helicobacter pylori and Antibiotic Resistance, A Continuing and Intractable Problem. Helicobacter (2016).
Johnston, C., Martin, B., Fichant, G., Polard, P. & Claverys, J. P. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol 12, 181–196 (2014).
Mortier-Barriere, I. et al. A key presynaptic role in transformation for a widespread bacterial protein: DprA conveys incoming ssDNA to RecA. Cell 130, 824–836 (2007).
Hofreuter, D., Odenbreit, S. & Haas, R. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol Microbiol 41, 379–391 (2001).
Karnholz, A. et al. Functional and topological characterization of novel components of the comB DNA transformation competence system in Helicobacter pylori. J Bacteriol 188, 882–893 (2006).
Chandran, D. V. & Waksman, G. Structural Biology of Bacterial Type IV Secretion Systems. Annu Rev Biochem 84, 603–629 (2015).
Yeh, Y. C., Lin, T. L., Chang, K. C. & Wang, J. T. Characterization of a ComE3 homologue essential for DNA transformation in Helicobacter pylori. Infect Immun 71, 5427–5431 (2003).
Claverys, J. P., Prudhomme, M. & Martin, B. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu Rev Microbiol 60, 451–475 (2006).
Seitz, P. & Blokesch, M. Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microbiol Rev 37, 336–363 (2013).
Baltrus, D. A. & Guillemin, K. Multiple phases of competence occur during the Helicobacter pylori growth cycle. FEMS Microbiol Lett 255, 148–155 (2006).
Moore, M. E., Lam, A., Bhatnagar, S. & Solnick, J. V. Environmental determinants of transformation efficiency in Helicobacter pylori. J Bacteriol 196, 337–344 (2014).
Kruger, N. J., Knuver, M. T., Zawilak-Pawlik, A., Appel, B. & Stingl, K. Genetic Diversity as Consequence of a Microaerobic and Neutrophilic Lifestyle. PLoS Pathog 12, e1005626 (2016).
Marsin, S., Mathieu, A., Kortulewski, T., Guerois, R. & Radicella, J. P. Unveiling novel RecO distant orthologues involved in homologous recombination. PLoS Genet 4, e1000146 (2008).
Marsin, S. et al. Genetic dissection of Helicobacter pylori AddAB role in homologous recombination. FEMS Microbiol Lett 311, 44–50 (2010).
Dorer, M. S., Fero, J. & Salama, N. R. DNA damage triggers genetic exchange in Helicobacter pylori. PLoS Pathog 6, e1001026 (2010).
Prudhomme, M., Attaiech, L., Sanchez, G., Martin, B. & Claverys, J. P. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313, 89–92 (2006).
Slager, J., Kjos, M., Attaiech, L. & Veening, J. W. Antibiotic-induced replication stress triggers bacterial competence by increasing gene dosage near the origin. Cell 157, 395–406 (2014).
Charpentier, X., Kay, E., Schneider, D. & Shuman, H. A. Antibiotics and UV radiation induce competence for natural transformation in Legionella pneumophila. J Bacteriol 193, 1114–1121 (2011).
Gagne, A. L. et al. Competence in Streptococcus pneumoniae is a response to an increasing mutational burden. PLoS One 8, e72613 (2013).
Stevens, K. E., chang, D. J., Zwack, E. E. & Sebert, M. E. Competence in Streptococcus pneumoniae is regulated by the rate of ribosomal decoding errors. MBio 2, (2011).
Wang, G. & Maier, R. J. Critical role of RecN in recombinational DNA repair and survival of Helicobacter pylori. Infect Immun 76, 153–160 (2008).
Garcia-Ortiz, M. V. et al. Unexpected role for Helicobacter pylori DNA polymerase I as a source of genetic variability. PLoS Genet 7, e1002152 (2011).
Mathieu, A., O’Rourke, E. J. & Radicella, J. P. Helicobacter pylori genes involved in avoidance of mutations induced by 8-oxoguanine. J Bacteriol 188, 7464–7469 (2006).
Huang, S., Kang, J. & Blaser, M. J. Antimutator role of the DNA glycosylase mutY gene in Helicobacter pylori. J Bacteriol 188, 6224–6234 (2006).
Kulick, S., Moccia, C., Kraft, C. & Suerbaum, S. The Helicobacter pylori mutY homologue HP0142 is an antimutator gene that prevents specific C to A transversions. Arch Microbiol 189, 263–270 (2008).
Akada, J. K., Shirai, M., Takeuchi, H., Tsuda, M. & Nakazawa, T. Identification of the urease operon in Helicobacter pylori and its control by mRNA decay in response to pH. Mol Microbiol 36, 1071–1084 (2000).
Levine, S. M. et al. Plastic cells and populations: DNA substrate characteristics in Helicobacter pylori transformation define a flexible but conservative system for genomic variation. FASEB J 21, 3458–3467 (2007).
Bubendorfer, S. et al. Genome-wide analysis of chromosomal import patterns after natural transformation of Helicobacter pylori. Nat Commun 7, 11995 (2016).
Pinto, A. V. et al. Suppression of homologous and homeologous recombination by the bacterial MutS2 protein. Mol Cell 17, 113–120 (2005).
Keyamura, K., Sakaguchi, C., Kubota, Y., Niki, H. & Hishida, T. RecA protein recruits structural maintenance of chromosomes (SMC)-like RecN protein to DNA double-strand breaks. J Biol Chem 288, 29229–29237 (2013).
Amundsen, S. K. et al. Helicobacter pylori AddAB helicase-nuclease and RecA promote recombination-related DNA repair and survival during stomach colonization. Mol Microbiol 69, 994–1007 (2008).
Loughlin, M. F., Barnard, F. M., Jenkins, D., Sharples, G. J. & Jenks, P. J. Helicobacter pylori mutants defective in RuvC Holliday junction resolvase display reduced macrophage survival and spontaneous clearance from the murine gastric mucosa. Infect Immun 71, 2022–2031 (2003).
Dorer, M. S., Sessler, T. H. & Salama, N. R. Recombination and DNA repair in Helicobacter pylori. Annu Rev Microbiol 65, 329–348 (2011).
Moccia, C. et al. The nucleotide excision repair (NER) system of Helicobacter pylori: role in mutation prevention and chromosomal import patterns after natural transformation. BMC Microbiol 12, 67 (2012).
Tomb, J. F. et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388, 539–547 (1997).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6, 343–345 (2009).
Acknowledgements
We thank Rainer Haas for the generous gift of the antibodies against ComB proteins. We thank the IRCM microscopy facility and Lamya Irbah for technical support and advice. This work was supported by grants to J.P.R. from the Agence Nationale pour la Recherche (ANR-09-BLAN-0271) and CEFIPRA (5203-5) and a doctoral fellowship to C.C. from the Region Ile de France (Maladies Infectieuses DIM120089). The authors declare no competing financial interests. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Author information
Authors and Affiliations
Contributions
C.C., A.M., T.K., J.P.R. and S.M. designed the experiments. C.C., A.M., P.D., D.B., M.P.A. and S.M. performed the experiments. C.C., A.M., J.P.R. and S.M. analysed the results. J.P.R. and S.M. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Corbinais, C., Mathieu, A., Damke, P. et al. ComB proteins expression levels determine Helicobacter pylori competence capacity. Sci Rep 7, 41495 (2017). https://doi.org/10.1038/srep41495
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep41495
This article is cited by
-
Methylome evolution suggests lineage-dependent selection in the gastric pathogen Helicobacter pylori
Communications Biology (2023)
-
Profiling of the Helicobacter pylori redox switch HP1021 regulon using a multi-omics approach
Nature Communications (2023)
-
Random transposon mutagenesis identifies genes essential for transformation in Methanococcus maripaludis
Molecular Genetics and Genomics (2023)
-
Identification of low oxygen-tolerating bacteria in prostate secretions of cancer patients and discussion of possible aetiological significance
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.