Abstract
The role of osteoclastic miRNAs in regulating osteolytic bone metastasis (OBM) of breast cancer is still underexplored. Here, we examined the expression profiles of osteoclastogenic miRNAs in human bone specimens and identified that miR-214-3p was significantly upregulated in breast cancer patients with OBM. Consistently, we found increased miR-214-3p within osteoclasts, which was associated with the elevated bone resorption, during the development of OBM in human breast cancer xenografted nude mice (BCX). Furthermore, genetic ablation of osteoclastic miR-214-3p in nude mice prevent the development of OBM. Conditioned medium from MDA-MB-231 cells dramatically stimulated miR-214-3p expression to promote osteoclast differentiation. Mechanistically, a series of in vitro study showed that miR-214-3p directly targeted Traf3 to promote osteoclast activity and bone-resorbing activity. In addition, osteoclast-specific miR-214-3p knock-in mice showed remarkably increased bone resorption when compared to the littermate controls, which was attenuated after osteoclast-targeted treatment with Traf3 3′UTR-containing plasmid. In BCX nude mice, osteoclast-targeted antagomir-214-3p delivery could recover the TRAF3 protein expression and attenuate the development of OBM, respectively. Collectively, inhibition of osteoclastic miR-214-3p may be a potential therapeutic strategy for breast cancer patients with OBM. Meanwhile, the intraosseous TRAF3 could be a promising biomarker for evaluation of the treatment response of antagomir-214-3p.
Similar content being viewed by others
Introduction
Osteolytic metastasis is the most common form of bone metastasis in patients with breast cancer1. The metastatic cancer cell could cause exaggerated osteoclast formation and excessive bone resorption, leading to osteolytic bone metastasis (OBM)2,3. It is becoming evident that the interaction between cancer cells and the bone microenvironment are important for the development of OBM. However, our knowledge on the underlying molecular mechanism responsible for the aberrantly elevated osteoclastic bone resorption during the development of OBM is still limited.
MicroRNAs (miRNAs) are ~22-nucleotide noncoding RNAs involved in the post transcriptional regulation of gene expression to coordinate a broad spectrum of biological processes4,5,6,7,8. Although accumulating evidence demonstrates that miRNAs also participate in regulating osteoclast differentiation and activity9,10,11,12,13,14,15, the role of osteoclastic miRNAs in regulating the osteoclastic bone resorption under cancer conditions and its contribution to the development of OBM are largely underexplored. More importantly, there is still a lack of miRNAs directly identified from human bone specimens from breast cancer individuals to contribute to the pathophysiological regulation of OBM.
In the present study, we showed that miR-214-3p was significantly upregulated in bone specimens from breast cancer patients with OBM as well as in osteoclasts from nude mice with human breast cancer xenografts (BCX) during OBM development, which was accompanied by the elevated bone resorption. Loss of miR-214-3p gene within osteoclasts prevented the development of OBM in nude mice with BCX. A series of in vitro and in vivo mechanistic studies demonstrated that miR-214-3p could directly target Traf3 to promote osteoclastic bone resorption. Finally, we found that osteoclast-targeted antagomir-214-3p treatment could attenuate OBM in nude mice with BCX. Collectively, our data suggest that inhibition of osteoclastic miR-214-3p may be a potential therapeutic strategy for breast cancer with OBM.
Results
High miR-214-3p in osteoclasts correlates with elevated bone resorption during the development of OBM
We examined the expression profiles of the miRNAs that have been previously reported to promote osteoclastogenesis16 and the mRNA expression of bone resorption marker genes (TRAP and CTSK) in bone specimens from breast cancer patients and cancer-free individuals with fracture (Fig. 1, Supplementary Fig. 1 and Supplementary Table 1). Among the osteoclastogenic miRNAs, the intraosseous miR-214-3p level in OBM patients was significantly higher than that in breast cancer patients without OBM and cancer-free individuals, respectively (Fig. 1a). Meanwhile, the intraosseous mRNA levels of TRAP and CTSK in OBM patients was also higher than those in breast cancer patients without OBM and cancer-free individuals, respectively (Fig. 1b). Consistently, the serum TRAP-5b levels in OBM patients were significantly higher than those in the relative control groups (Supplementary Table 1). The correlation analysis further revealed that the miR-214-3p level was positively correlated with the mRNA levels of bone resorption marker genes in bone specimens pooled from breast cancer patients with/without OBM (Fig. 1c). These data suggest that the highly expressed intraosseous miR-214-3p were closely associated with the elevated bone resorption in OBM patients. Given that a recent study reported that miR-214-3p targets phosphatase and tensin homolog (PTEN) to promote osteoclastogenesis17, we also examined the protein expression of PTEN in the bone specimens. Surprisingly, no obvious differences in the PTEN protein levels were found between the breast cancer patients with or without OBM (Supplementary Fig. 2).
(a) The miR-214-3p level and (b) mRNA levels of TRAP and CTSK in bone specimens from breast cancer patients and cancer-free individuals with fracture (Control). (c) The correlation analysis between miR-214-3p level and TRAP (or CTSK) mRNA level in bone tissue. Note: miR-214-3p levels were normalized to U6 and osteoclast marker genes mRNA levels were normalized to Gapdh. *P < 0.05, ***P < 0.001.
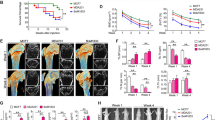
Given that miR-214-3p is evolutionarily conserved in human and rodents18, we sought to investigate the relationship between osteoclastic miR-214-3p and bone resorption during the development of OBM in mouse models. To mimic the pathological condition of OBM, the nude mice were grafted with human breast cancer MDA-MB-231 cells (hereafter BCX mice) through intraventricular injection and sacrificed at 1, 5 and 8 weeks after xenografts19, respectively. In the mouse model, the bone marrow cells positive for osteoclast-associated receptor (OSCAR), a marker specifically expressed in the cell surfaces of pre-osteoclasts and mature osteoclasts20, were isolated by magnetic-activated cell sorting (MACS). The mRNA expression of the osteoclast-related marker genes (Rank, Ctsk and Trap5b) in OSCAR+ cells were remarkably higher than those in OSCAR− cells (Supplementary Fig. 3). The osteoclastic miR-214-3p level within OSCAR+ cells, the value of Oc.S/BS and the serum bone resorption marker CTX-1 level were all gradually elevated in BCX mice during the development of OBM from week 1 to week 8 after xenografts (Fig. 2a). Moreover, the correlation analysis indicated that the osteoclastic miR-214-3p level was positively correlated with Oc.S/BS and serum CTX-1 levels in this mouse model (Fig. 2b). Collectively, these data imply a close association between the elevated osteoclastic miR-214-3p level and the excessive bone resorption during the development of OBM.
(a) The osteoclastic miR-214-3p levels (upper left), Oc.S/BS in proximal tibia (upper middle), serum CTX-I levels (upper right) and the representative images of TRAP staining in proximal tibia (bottom) were examined (n = 12 for each group). Scale bar: 100 μm. Arrow indicates TRAP-positive cells. The levels of miR-214-3p were normalized to the mean value of control group. (b) The correlation analysis between osteoclastic miR-214-3p level and Oc.S/BS and between osteoclastic miR-214-3p level and serum CTX-I levels was performed, respectively. Note: miR-214-3p levels were normalized to U6 and osteoclast marker genes mRNA levels were normalized to Gapdh. *P < 0.05 when compared to Week 1 in BCX mice.
Loss of miR-214-3p within osteoclasts prevents osteolytic bone metastasis
To investigate the role of osteoclastic miR-214-3p in the development of OBM, we constructed the heterozygous mice carrying the mutant allele with LoxP sites harboring the genomic region of the 106-bp pre-miR as previously reported21. Then, the mice were intercrossed with nude mice to obtain the miR-214-3pflox/− nude mice, which were then crossed with the Ctsk-Cre mice to generate the osteoclast-specific miR-214-3p knockout nude mice (Ctsk; miR-214-3pflox/flox, hereafter CKO mice) and the relative controls (miR-214-3pflox/flox, hereafter WT mice). The miR-214-3p level and the mRNA levels of Ctsk and Trap were all significantly lower in osteoclasts (OSCAR+ cells isolated from bone marrow by MACS) from CKO mice as compared to WT controls (Fig. 3a,b). To examine the effect of osteoclast-specific miR-214-3p depletion on osteolytic bone metastasis, the CKO mice and WT controls were grafted with human breast cancer cells through intraventricular injection and sacrificed at 8 weeks after xenografts. The data showed that CKO mice with BCX had remarkably lower level of serum CTX-1 and lower value of Oc.S/BS and N.Oc/B.Pm as compared to WT controls (Fig. 3c–e). Bone metastasis was markedly diminished in the CKO mice with BCX while no significant difference in the metastasis to the non-bone tissues were found between CKO mice and WT controls with BCX (Fig. 3f–h). In addition, numerous osteolytic bone lesions were detected in WT nude mice with BCX, whereas the osteolytic lesions were hardly detected in the CKO mice with BCX (Fig. 3i,j). Thus, the above findings suggest that loss of miR-214-3p within osteoclasts could prevent osteolytic bone metastasis.
Q-PCR analysis of the miR-214-3p levels (a) and the TRAP and CTSK mRNA levels (b) in osteoclasts and non-osteoclasts isolated from bone marrow cells by MACS. (c) ELISA assay of serum CTX-I levels. (d) The representative images of TRAP staining in distal femur. Scale bar: 100 μm (e) Bone histomorphometric analysis for Oc.S/BS and N.Oc/B.Pm in proximal tibia. (f) The representative bioluminescence imaging showing the degree of metastasis. (g) The number of nude mice with/without bone metastasis in the forelimb, hind limb and spine area detected by bioluminescence imaging. (h) The bioluminescence signal intensity at the sites of bone metastasis (left) and non-bone metastasis (right). (i) The representative micro-CT images showing the osteolytic bone lesion at distal femur. Scale bar: 1 mm (j) The number of osteolytic lesion at distal femur. Note: n = 8 for each group. (a,b) CKO: osteoclast-specific miR-214-3p knockout nude mice, WT: Wild-type nude mice. (c–j) WT: WT nude mice with BCX, CKO: CKO nude mice with BCX. Data were means ± s.d. **P < 0.01.
MiR-214-3p directly targets Traf3 to regulate osteoclast activity in vitro
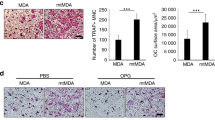
We next sought to understand how osteoclastic miR-214-3p regulates the osteoclast activity and bone resorption during the development of OBM. The mouse bone marrow-derived macrophages (BMMs) were treated with or without the condition medium (CM) from the cultured MDA-MB-231 cells in the absence of 5 ng/ml RANKL. As expected, the miR-214-3p level and the mRNA levels of Ctsk and Trap gradually increased during the osteoclast differentiation. Notably, the time-dependent increase in the expression of miR-214-3p, Ctsk and Trap were more pronounced in the presence of CM from breast cancer (Supplementary Fig. 4a). Further study showed that the mRNA levels of Trap and Ctsk were upregulated by agomir-214-3p and downregulated by antagomir-214-3p in the RAW264.7 cells treated with RANKL and CM (Supplementary Fig. 4b). Consistently, the number of TRAP-staining positive cells and bone-resorbing activity of osteoclasts was suppressed by antagomiR-214 and promoted by agomir-214 in RAW264.7 cells treated with RANKL and CM (Supplementary Fig. 4c,d). Unexpectedly, no changes in the protein expression of PTEN during the course of osteoclastogenesis (Supplementary Fig. 4e). Given that our data from breast cancer patients also showed that the intraosseous PTEN protein expression seemed to be not affected under pathological conditions of OBM, we next sought to elucidate the mechanisms by which miR-214-3p regulates osteoclastic bone resorption. We used miRBase (version 21) and TargetScan (release version 6.2) to predict the candidate target genes of miR-214-3p, wherein we found that Traf3 mRNA contains a miR-214-3p binding site in its 3′ untranslated region (3′UTR) (Fig. 4a). A previous study has demonstrated that TRAF3 could limit the RANKL-induced osteoclastogenesis by suppressing canonical and noncanonical NF-κB signaling22. Therefore, we sought to investigate the connection between miR-214-3p and TRAF3 during osteoclastogenesis. We observed a gradual increase in the miR-214-3p levels together with a decrease in the amount of TRAF3 protein in RAW 264.7 cells during RANKL-induced osteoclastogenesis (Fig. 4b). Moreover, the protein level but not the mRNA level of TRAF3 was downregulated in osteoclasts treated with agomir-214 treatment, whereas agomir-214-3p mutant containing the mutated binding sites treatment had minimum effect on either the protein or mRNA of TRAF3 (Fig. 4c). To determine whether miR-214-3p could directly target Traf3, the RAW264.7 cells were transfected with luciferase reporters containing either a wild-type (WT) Traf3 3′UTR or a mutant Traf3 3′UTR (Fig. 4d). Agomir-214-3p, but not the agomir-NC and agomir-214-3p mutant, substantially inhibited the luciferase reporter activity of the WT Traf3 3′ UTR, whereas the luciferase reporter activity of the mutant Traf3 3′ UTR was not repressed by agomir-214-3p (Fig. 4e). Moreover, the luciferase activity of the WT Traf3 3′ UTR was notably upregulated after the endogenous levels of miR-214-3p were reduced by antagomir-214-3p treatment (Fig. 4f).
(a) Sequence alignments between miR-214-3p and candidate binding sites in the 3′UTR of Traf3. (b) Schematic diagram illustrating the design of luciferase reporters with the WT Traf3 3′ UTR (WT 3′ UTR) or the site-directed mutant Traf3 3′ UTR (3′ UTR-Mut). The sequences of agomir-214-3p and a agomir-214-3p mutant (agomir-214-Mut) are also shown. Luc, luciferase. (c) The relationship between miR-214-3p level (top) and the mount of TRAF3 protein (bottom) during osteoclast differentiation in RAW264.7 cells. (d) The effects of agomir-214-3p and agomir-214-Mut on Traf3 mRNA levels (top) and the amount of TRAF3 protein (bottom) in RAW 264.7 cells. (e) The effect of agomir-214-3p and mutated agomir-214-3p on luciferase activity in RAW 264.7 cells transfected with either the WT the mutant Traf3 3′ UTR reporter (left) or Traf3 3′ UTR reporter (right). (f) The effect of antagomir-214-3p on luciferase activity in RAW 264.7 cells transfected with either the WT Traf3 3′ UTR reporter or the mutant Traf3 3′ UTR reporter. (g) Real-time PCR analysis of Trap and Ctsk mRNA levels (top) and western blot analysis of TRAF3 protein (bottom) in RAW264.7 cells after blockade of miR-214-3p binding to Traf3 by overexpression of WT Traf3 3′UTR (UTR). (h) Trap and Ctsk mRNA levels in RAW264.7 cells after the indicated treatment. The results from a scrambled control siRNA (siRNA-NC) and a mock transfection are also shown. (i) Western blot analysis of whole cell lysates of osteoclasts transfected with antagomir-214-3p and cultured with RANKL for 5 days. Note: All data are the mean ± s.d. from three independent experiments. *P < 0.05, NS, not significant.
We then transfected the RAW264.7 cells with WT Traf3 3′ UTR to block the binding of endogenous miR-214-3p to Traf3. The Trap and Ctsk mRNA levels were both remarkably lower in RAW264.7 cells transfected with the WT Traf3 3′ UTR when compared to the cells transfected with vector alone. Accordingly, the level of TRAF3 protein in RAW 264.7 was upregulated after transfection with the Traf3 3′ UTR (Fig. 4g). Furthermore, when Traf3 expression was knocked down by siRNA, the Trap and Ctsk mRNA consistently remained at a low level under the treatment of agomir-214-3p, antagomir-214-3p and the corresponding negative controls, respectively (Fig. 4h). Moreover, we examined the effect of miR-214-3p on the protein expression level of NFATc1, NIK and p65 (RelA) in RAW 264.7 undergone RANKL-induced osteoclast differentiation. The protein levels of NFATc1, NIK and p65 were all downregulated by antagomir-214-3p when compared to the antagomir-negative control (Fig. 4i). However, no obvious changes in the protein levels of NFATC1, NIK, RelA and RelB after treatment with agomir-214 during osteoclastogenesis when compared to the agomir-negative control (Data not shown).
Elevated miR-214-3p in osteoclast targets Traf3 to promote osteoclastic bone resorption in vivo
Next, we investigated the role of osteoclastic miR-214-3p in regulating bone resorption in vivo using our previously established osteoclast-specific miR-214-3p knock-in (OC-214) mice that overexpressing miR-214-3p under the control of Cre-mediated recombination driven by the Ctsk promoter23,24. Thereafter, we compared the levels of osteoclast activity and bone resorption between the OC-214 mice and WT littermate controls at 8 weeks of age. The mRNA expression of Trap and Ctsk in bone specimens were remarkably higher in OC-214 mice when compared to those in WT mice (Fig. 5a). The serum CTX-1 level was significantly higher in OC-214 mice than that in WT mice (Fig. 5b). The intraosseous TRAF3 protein levels were notably lower in OC-214 mice when compared to those in WT mice (Fig. 5c). In addition, significantly more pit area formed in the bone slices cultured with the bone marrow-derived osteoclasts from OC-214 mice when compared to those from WT mice (Fig. 5d). All these data suggest that the elevated expression of miR-214-3p within osteoclast could promote osteoclastic bone resorption in OC-214 mice. To investigate whether the elevated miR-214-3p within osteoclasts could promote osteoclastic bone resorption via targeting Traf3 in vivo, we evaluated the degree of bone resorption in the OC-214 mice weekly administrated with or without TRAF3 3′UTR-containing plasmid (20 μg per mice) encapsulated in the our previously developed osteoclast-targeted delivery system25. The treatment with Traf3 3′UTR-containing plasmid but not mutant Traf3 3′UTR-containing plasmid could significantly attenuate the increase in bone resorption-related parameters, the decrease in bone mass and the deterioration in trabecular architecture in OC-214 mice (Fig. 5e–h). Collectively, it indicates that the elevated miR-214-3p in osteoclast could target Traf3 to promote osteoclastic bone resorption in vivo.
(a) Q-PCR analysis of the mRNA levels of osteoclast marker genes in bone tissue. (b) ELISA analysis of serum CTX-1 level from OC-miR-214-3p and WT mice. (c) Western blot analysis of the protein expression of TRAF3 in bone tissues. (d) In vitro bone resorption activity assay showing the bone resorption pit surface of the osteoclasts differentiated from bone marrow macrophages (BMMs) derived from either OC-miR-214-3p mice or WT mice. (e) The micro-CT analysis for the BMD and BV/TV of the proximal tibia from the indicated group after treatment. (f) The representative micro-CT images for the trabecular micro-architecture in proximal tibia. Scale bar: 1 mm. Note: n = 8~10 for each group. (g) Bone histomorphometric analysis for Oc.S/BS and N.Oc/B.Pm in proximal tibia. (h) The representative TRAP staining images for the trabecular bone in proximal tibia. Scale bar: 100 μm. Arrow indicates TRAP+ cells. Note: n = 6 for each group. Data were means ± s.d. *P < 0.05, **P < 0.01. OC-214: OC-miR-214-3p mice; WT: age-matched littermates; BS: mice that were sacrificed before treatment as baseline; PBS: mice treated with PBS as blank control; Veh: mice treated with (D-Asp)8-liposome alone; 3′UTR-Mut: mice treated with (D-Asp)8-liposome-mutant Traf3 3′UTR plasmid; 3′UTR: mice treated with (D-Asp)8-liposome-Traf3 3′UTR plasmid.
Osteoclast-targeted inhibition of miR-214-3p attenuates osteolytic bone metastasis
We next evaluated the therapeutic effect of inhibiting osteoclastic miR-214-3p on osteolytic bone metastasis. We performed pulsed administration of the antagomir-214-3p encapsulated within osteoclast-targeting delivery system (AMO) in BCX nude mice as described in Fig. 6a. The optimal dosage of antagomir-214-3p was determined in our pilot study, in which almost 80% knockdown efficiency of osteoclastic miR-214-3p was achieved at a dose of 10 mg/kg. In addition, over 50% knockdown was maintained for 8 days after a single dose at 10 mg/kg (Supplementary Fig. 5). Six weeks after the first treatment, the intraosseous miR-214-3p level in BCX nude mice treated with AMO was significantly lower than that in BCX nude mice without AMO treatment (Supplementary Fig. 6a,b), suggesting the efficient inhibition of miR-214-3p in BCX nude mice after AMO treatment. No significant difference in the indexes of liver function (ALT and AST) and kidney function (BUN) were found between the BCX nude mice with or without AMO treatment (Supplementary Table 2). In addition, the intraosseous TRAF3 protein levels in BCX nude mice received AMO treatment were remarkably higher than those in BCX nude mice treated without AMO, whereas the intraosseous PTEN protein levels were not affected by different treatments (Supplementary Fig. 6c). On the other hand, the serum CTX-1 level in BCX nude mice with AMO treatment was also remarkably lower than that in BCX nude mice without AMO treatment (Fig. 6b). Consistently, the values of Oc.S/BS and N.Oc/B.Pm were both significantly lower in those BCX nude mice administrated with AMO when compared to those in BCX nude mice without AMO treatment (Fig. 6c,d). On the other hand, AMO treatment markedly diminished bone metastasis in BCX nude mice (Fig. 6e,f and Supplementary Fig. 6d). In addition, the osteolytic metastatic lesions in tibiae were remarkably alleviated in BCX nude mice after AMO treatment (Fig. 6g,h). These data suggest that therapeutic inhibition of miR-214-3p within osteoclast could attenuates osteolytic bone metastasis.
(a) A schematic diagram illustrating the experimental design. (b) ELISA analysis of serum CTX-1 level in mice from the indicated group after treatment. (c) The representative images for TRAP staining in the proximal tibiae bone sections from the indicated group after treatment. Arrow heads indicate osteoclasts. Scale bar: 50 μm. (d) The bone histomorphometric analysis for Oc.S/BS and N.Oc/B.Pm in the proximal tibiae bone sections from the indicated group after treatment. (e) The representative bioluminescence imaging showing the degree of metastasis in mice from the indicated group after treatment. (f) The bioluminescence signal intensity at the sites of bone metastasis (left) and non-bone metastasis (right) in the mice from the indicated group after treatment. (g) Representative micro-CT images showing the osteolytic bone lesion at proximal tibiae from the indicated group after treatment. Arrowheads indicate the osteolytic bone lesions. Scale bar = 1 mm. (h) The number of osteolytic lesion at proximal tibiae from the indicated group after treatment. Note: n = 12 for each group. The data were presented as the mean ± s.d., **P < 0.01. Control: nude mice without breast cancer xenografts, Model: breast cancer xenografted nude mice (BCX mice) administrated with PBS, Veh: BCX mice administrated with (D-Asp)8-liposome, NC: BCX mice administrated with (D-Asp)8-liposome-antagomir negative control, AMO: BCX mice administrated with (D-Asp8)-liposome-antagomir-214-3p.
Discussion
In this study, we found that elevated miR-214-3p within osteoclast could target TRAF3 to promote osteoclastic bone resorption during the development OBM. Our results suggest that therapeutic inhibition of miR-214-3p in osteoclasts may be a potential therapeutic strategy for OBM with dominant osteoclastic bone resorption.
Previous studies have documented that miRNAs could participate in regulating osteoclastic bone resorption under pathological settings such as osteolytic bone metastasis19,26. However, these miRNAs were detected from animal studies or in vitro experiments. The current study provides the first clinical evidence of a miRNA directly identified from bone specimens of patients with OBM, which contributes to the pathogenesis of OBM. The data from OBM patients as well as murine models concordantly showed a close association between the aberrantly elevated osteoclastic miR-214-3p expression and the increased bone resorption during the progression of OBM. In addition, we have previously shown that miR-214-3p could promote osteoclast differentiation in vitro. In this study, we further found that the CM from breast cancer cells could dramatically stimulate the expression of miR-214-3p and induce the osteoclast differentiation in vitro, which could be blocked/promoted by antagomir-214-3p/agomir-214-3p treatment. These data suggest that the metastatic breast cancer cells could stimulate miR-214-3p expression in osteoclast precursors to promote osteoclast formation during the development of OBM. On the other hand, it has been proved that miR-214-3p could target p53 to enhance the invasion ability of breast cancer cells27, while depletion of miR-214-3p could block the dissemination of breast cancer cells28, suggesting an important role of miR-214-3p in mediating breast cancer metastasis. Impressively, we found that osteoclast-specific ablation of miR-214-3p could substantially prevent the development of OBM, as evidenced by the low level of CTX-I and bone resorption-related parameters as well as the diminished metastasis to bone in the CKO nude mice after BCX. According to the ‘viscous cycle’ theory1,29, the interplay between cancer cells and the osteoclasts at the metastasis microenvironment were crucial for the development of OBM. Since we have previously demonstrated that the osteoclast could secrete exosomal miR-214-3p to the bone microenvironment24, it was reasonable to expect that the aberrant elevated miR-214-3p levels in osteoclasts may, in turn, contribute to affect the colonized cancer cells at the bone metastatic site.
We further found that miR-214-3p could target TRAF3 to stimulate the osteoclastic bone resorption during the development of OBM. TRAF3 is an important regulator of cytokine production and type I interferons30, which is also involved in regulating RANKL-induced osteoclast formation22. Consistently, we observed a time-dependent decrease in the TRAF3 protein level, which was accompanied by a gradual increase in the miR-214-3p level, during osteoclastogenesis in vitro. In addition, the TRAF3 protein levels in osteoclasts were downregulated after transfection with agomir-214 but not the nonsense agomir control or mutant agomir-214, whereas no differences in the Traf3 mRNA levels were found among different treatments, suggesting a role of miR-214-3p in the posttranscriptional regulation on TRAF3 protein expression. More importantly, the evidences from the luciferase reporter assay and the gain- and loss-of-function experiments indicate that TRAF3 could be a functional target of miR-214-3p. The above in vitro findings were further supported by the data from the OC-214 mice showing that osteoclast-targeted treatment with Traf3 3′UTR plasmid could remarkably attenuate the excessive bone resorption in OC-214 mice. However, the limitation of this study was that we didn’t examine the effect of miR-214-3p agomir or antagomir on osteoclast differentiation from human peripheral blood mononuclear cells. On the other hand, a recent study shows that miR-214-3p could promote osteoclastogenesis and increase osteoclast activity by targeting PTEN during osteoclast differentiation from bone marrow monocytes (BMMs)17. Considering PTEN could regulate bone resorption during the development of OBM, the protein expression of PTEN in the bone specimens was detected. However, we unexpectedly observed little change in the protein expression of PTEN between the breast cancer patients with or without OBM. This could be explained by the previous study showing that the phosphorylated PTEN but not the total PTEN levels decreased during RANKL-induced osteoclast differentiation31. Importantly, the BCX nude mice received AMO treatment had a higher TRAF3 protein expression in bone specimens when compared to those without AMO treatment, whereas no obvious changes in the PTEN protein levels were found between the mice with and without AMO treatment. Collectively, it strongly suggests that miR-214-3p could regulate the amount of TRAF3 proteins to contribute to the excessive osteoclastic bone resorption during the development of OBM.
Although controversial, emerging evidence showed that, pharmacological inhibition of osteoclast resorption may increase the survival of patients with OBM32. In breast cancer patients, treatment with bisphosphonates decreases skeletal events, such as pathological fracture or bone pain, by 30%~40%33. Bisphosphonates also block the formation of bone tumors, and osteoclast inhibition prevents spontaneous osteolytic tumors in HTLV-1 Tax transgenic mice34. Thus far, there is no established non-coding RNA target that has been translated into clinical antiresorptive therapy. Considering the distinct role of miR-214-3p described here, we also evaluated the therapeutic effect of inhibiting miR-214-3p within osteoclasts on the development of OBM in BCX nude mice using antagomir-214-3p encapsulated in our established osteoclast-targeting delivery system25. The results showed that osteoclast-targeting antagomir-214-3p treatment could dramatically diminish bone metastatic lesions in BCX nude mice, respectively, which could be explained by the decreased bone resorption after knocking down the miR-214-3p level within osteoclasts by antagomir-214. In addition, it has been demonstrated that OBM are always associated with the uncoupled bone formation to bone resorption, i.e. increased osteoclast activity with reduced osteoblast activity. Given that the osteoclastic miR-214-3p could transfer to osteoblast to inhibit bone formation24, the current strategy of inhibiting osteoclastic miR-214-3p may also exert anabolic effect on bone formation. In addition, the intraosseous TRAF3 protein levels was markedly restored after antagomir-214-3p treatment, suggesting that the protein expression of TRAF3 in bone could be a promising biomarker for evaluation of the treatment response of antagomir-214-3p in future translation.
Materials and Methods
Bone specimens from patients
We collected the small pieces of dissociated bones from fractured patients during surgery. The postmenopausal women with breast cancer suffering fractures were enrolled. OBM were identified by X-ray or CT imaging. The age-matched postmenopausal women without malignant status were also included. All written informed consent was obtained from each patient or family member before inclusion in this study. All experiments were performed in accordance with relevant guidelines and regulations and all clinical procedures were approved by the Ethics Committees of Shenzhen 8th People Hospital, Shenzhen People’s Hospital and Guanghua Integrtive Medicine Hospital, respectively.
Agomir-214, antagomir-214 and Traf3 siRNA
Agomir-214-3p, agomir-negative control (agomir-NC), agomir-214-3p-mutant (agomir-214-Mut), antagomir-214-3p (antago-214) and antagomir-214-3p negative control (antago-NC) were purchased from Guangzhou RiboBio Co., Ltd. The Traf3 siRNA and negative control (siRNA-NC) were purchased from Invitrogen.
Mouse Genetic mouse model
The miR-214-3p flox/− mice were generated as previously reported21. Briefly, a mutant allele with loxP sites flanking the genomic region encompassing the 106-bp pre-miR and a neomycin resistance cassette were constructed and electroporated into embryonic stem (ES) cells. Positive targeting clones were identified by polymerase chain reaction (PCR) and southern blotting. The targeted ES clones were microinjected into C57BL/6 blastocysts, and male chimeras were mated to C57BL/6 females to obtain miR-214-3p flox-neo/+ mice, which were then intercrossed with flp-deleter mice (JAX, 009086) to remove neo cassette in order to generate miR-214-3p flox/+ mice. The female miR-214-3p flox/+ mice in C57BL/6 background were intercrossed with the male BALB/c nu/nu mice to obtain the miR-214-3pflox/+ nude mice. The female Ctsk-cre mice in C57BL/6 background were also intercrossed with the male BALB/c nu/nu mice to generate the Ctsk-cre nude mice. Thereafter, the miR-214-3p flox/+ nude mice were crossed with the Ctsk-cre nude mice to obtain the Ctsk;miR-214-3p flox/flox (CKO) nude mice and the miR-214-3p flox/flox (wild-type, WT) controls. On the other hand, the OC-miR-214-3p mice were generated according to our published protocols24. Firstly, we generate the Rosa26-PCAG-STOPfl-mmu-miR-214-3p-knock-in mice. In brief, a cassette containing the following components was constructed to target the Rosa26 locus: FRT-LoxP-stop codons-three SV40 poly(A) sequences-LoxP-mmu-miR-214-3p-WPRE-bGH poly(A)-AttB-PGK promoter-FRT-Neo-PGK poly(A)-AttP. The targeting vector is constructed, fully sequenced and electroporated into C57BL/6 embryonic stem (ES) cells. Positive targeting clones were identified by polymerase chain reaction (PCR) and southern blotting. The targeted ES clones were microinjected into BALB/c blastocysts to obtain chimeric mice following standard procedures. Thereafter, the chimeric mice were intercrossed with C57BL/6 mice to obtain F1 heterozygote mice and then backcrossed with C57BL/6 mice to expand the enough number of heterozygote Rosa26-PCAG-STOPfl-mmu-miR-214-knock-in mice. Thereafter, we crossed the Rosa26-PCAG-STOPfl-mmu-miR-214-knock-in mice with heterozygous Ctsk-cre mice to obtain OC-miR-214 mice. The littermates were used as wild-type control. The Ctsk-cre mouse was a generous gift from Prof. Xu Jiake in the West Australia University. It is the offspring of the Ctsk-cre mouse strain established by the Shigeaki Kato’s lab23. All experiments were performed in accordance with relevant guidelines and regulations and all experimental procedures were approved by the licensing Committees of Animal Ethics and Experimental Safety of Hong Kong Baptist University.
Bone resorption activity assay in vitro
For osteoclast resorptive function analysis, the osteoclast differentiation of bone marrow macrophages/RAW264.7 cells was conducted in OsteoAssay bone plates (Lonza) at a density of 1 × 104cells/well35. Either agomir-214-3p (200 μM) or antagomir-214-3p (200 μM) were transfected on day 0 and day 5. Bone slices were ultra-sonicated in 1 mol/L NH4OH to remove adherent cells and stained with 0.1% toluidine blue solution. Bone slice images were captured using electron microscopy. Three fields were randomly selected for each bone slice for further analysis. Pit areas were quantified using Image Pro Plus 6.2 software (Media Cybernetics Inc.).
Luciferase Assay
Traf3 mRNA 3′UTR containing the miR-214-3p-binding sequences for the mouse Traf3 gene was amplified by PCR from mouse genomic DNA. The PCR product was then sub-cloned into the XbaI site downstream of the stop codon in the pGL3 control vector (Promega). The QuickChange site-directed mutagenesis kit (Stratagene) was used to introduce mutations into the seed region of pGL3-Traf3, giving pGL3- Traf3-mutant, following the manufacturer’s instructions. HEK293 and RAW264.7 cells were seeded in 96-well plates and co-transfected with either 50 nM agomir-NC agomir-214-3p/agomir-214-3p-Mut or antagomir-NC/antagomir-214-3p, 100 ng of luciferase vector (pGL3 constructs) and 25 ng Renilla vector (pRL-TK) using Lipofectamine 2000 (Invitrogen). 24 h after transfection, luciferase activity was measured using the dual-luciferase reporter assay system (Promega). Luminescent signals were quantified by luminometer (Glomax, Promega), and each value from the firefly luciferase construct was normalized by Renilla luciferase assay.
Bone metastasis analyses for xenograft nude mice
The luciferase-labelled human breast cancer cell line (MDA-MB-231-luc-D3H2LN) was purchased from Caliper Life Sciences Inc., and cultured with DMEM medium (high glucose) with 10% fetal bovine serum (Gibco), 0.1 mM MEM non-essential amino acids and 2 mM L-glutamine. After trypsin digestion, the cell suspension was injected into the left cardiac ventricle of 6-week-old female nude mice at 1 × 105 cells per mouse in 100 μl PBS19, which was confirmed by in vivo biophotonic imaging immediately after injection (Supplementary Fig. 6a). Bone metastases were monitored and quantified weekly after injection by biophotonic imaging using a Caliper Xenogen Spectrum instrument. At 6 to 8 weeks after xenograft, the mice were sacrificed after biophotonic imaging and the limb bone were collected for microCT scanning and analysis for the the osteolytic metastatic lesions. All experiments were performed in accordance with relevant guidelines and regulations and all experimental procedures were approved by the licensing Committees of Animal Ethics and Experimental Safety of Hong Kong Baptist University.
Total RNA extraction, reverse transcription and quantitative real-time PCR analysis
Total RNA extraction was extracted using the RNeasy Mini Kit (Qiagen, 74106) according to the manufacturer’s protocol. For mRNA expression, RNA was reverse transcribed into cDNA using a QuantiTect Reverse Transcription Kit according to an established protocol. The primer for mRNAs (Traf3, Trap, Ctsk and Gapdh) were purchased from Shanghai Integrated Biotech Solutions Co., Ltd. For miRNA expression, the TaqMan primer-probe combinations for miRNA-214-3p and RUN6B were products of Ambion. Real-Time PCR reactions were performed using the StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA, USA), and the signal was converted into numerical values by StepOne software v2.1 (Applied Biosystems, Foster City, CA, USA). All reactions were performed in triplicate and the cycle threshold (CT) value in each reaction well was recorded. The relative quantification of miR-214 expression was calculated using the 2−∆∆CT method.
Western blot analysis
Proteins were extracted from the ultracentrifugation pellets and separated on a denatured SDS-polyacrylamide gel before transfer to a polyvinylidene difluoride membrane (0.45 um; Millipore, Bedford, MA, USA). Afterwards, the membranes were blocked with 5% non-fat dry milk in Tris-buffered saline with Tween 20 (TBST) for 1 hour. The membranes were probed with primary anti-TRAF3 (1:200, Santa Cruz, sc-949), anti-PTEN (1:1000, Cell Signaling, #9559), anti-p65 (1:200, Santa Cruz, sc-372), anti-NIK (1:200, Santa Cruz, sc-7211) or anti-NFATc1 (1:1000, Cell Signaling, #8032), respectively. To check the amount of proteins transferred to nitrocellulose membrane, GAPDH was used as control and detected by an anti-GAPDH (1:500; Santa Cruz, sc-25778) antibody. The membranes were then washed three times with TBST and incubated for 1 hour with HRP-conjugated secondary antibodies (1:2000; Santa Cruz, sc-2004). The antigen-antibody complexes were visualized using SuperSignal® West Dura Extended Duration Substrate (Thermo Scientific, Prod # 34075). Supplementary Fig. 7 depicts uncropped western blots.
Magnetic-activated cell sorting
Bone marrow cells were flushed from the diaphysis of the femur and dissociated to a single-cell suspension by pipetting up and down and passed through 30 μm nylon mesh to remove cell clumps which may clog the column. Cells were centrifuged at 300 g for 10 min and 107 total cells were re-suspended in 200 μl of ice-cold buffer (DPBS without Ca2+ and Mg2+, with 0.5% bovine serum albumin, and 2 mM EDTA). 10 ug/ml Rat anti-Mouse OSCAR (R&D Systems) was added to the cell suspension and incubated at 4 °C for 1 h. After washing and centrifuge, the cells were re-suspended in 80 μl of ice-cold buffer. 20 μl of Anti-Rat IgG MicroBeads (Miltenyi Biotec) was added per 107 total cells, incubated for 15 min at 4 °C. Cells were washed with 1–2 ml of buffer per 107 cells, centrifuged at 300 g for 5 min, and re-suspended in 500 μl of buffer. A MACS column (Miltenyi Biotec) was placed in the magnetic field of a suitable MACS Separator (Miltenyi Biotec) and prepared by rinsing with 3 ml of buffer. The column was washed by 3 ml of buffer for three times. Flow-through of unlabeled cells (OSCAR− cells) were collected. The column was placed over a collection tube, 5 ml buffer was pipetted onto the column, and the magnetically labeled cells (OSCAR+ cells) were flushed out by firmly pushing the plunger into the column. OSCAR+ and OSCAR− cells were collected for downstream analysis. To perform phenotypical analysis, the mRNA levels of osteoclast-related marker genes (Rank, Ctsk and Trap5b) in OSCAR+/OSCAR- cells were examined by RT-PCR.
Micro-CT analyses
Tibiae were collected, and the attached soft tissue was removed thoroughly and fixed in 4% paraformaldehyde. The fixed tibiae were analyzed using micro-CT (vivaCT 40) and software IPL V5.15 at Scanco Medical AG, Swtizerland. Totally 200 slices with a voxel size of 15 μm was chosen at the region of distal femur beginning at the growth plate extending along the tibiae diaphysis to evaluate bone mass, geometry, and trabecular microarchitecture. The whole trabecular bone were isolated for three-dimension reconstruction (Sigma = 1.2, Supports = 2 and Threshold = 180) to calculate the following parameters: bone mineral density (BMD), bone volume/tissue volume (BV/TV), trabecular thickness (Tb.Th), and trabecular number (Tb.N).
Bone histomorphometric analyses
Tibiae specimens were fixed in 4% PFA for 24 hours and decalcified in fresh 10% EDTA for 4 weeks following paraffin embedding. Thereafter, the trabecular sections at the proximal tibiae and tartrate resistant acid phosphatase (TRAP) staining were performed. Bone static histomorphometric analyses for osteoclast surface (the percent of trabecular bone surface covered by osteoclasts, Oc.S/BS) and osteoclast number per bone perimeter (N.Oc/B.Pm) were made using professional image analysis software (Image J, NIH, USA) under microscope (Leica image analysis system, Q500MC). The bone histomorphometric parameters were calculated and expressed according to standardized nomenclature for bone histomorphometry.
Synthesis of osteclast-targeting delivery system encapsulating antagomiR-214-3p or Traf3 3′UTR plasmid
The antagomir-214-3p, antagomir-NC and plasmid containing Traf3 3′UTR or mutant Traf3 3′UTR were encapsulated into the osteoclast-targeting delivery system (D-Asp8-liposome), respectively, using our previously established protocol25. Briefly, the lipids of DOTAP, DOPE, Chol, DSPE-mPEG2000, and DSPE-PEG2000-MAL at a molar ratio of 42:15:38:3:2 dissolved in chloroform were dried into a thin film followed by hydration in PBS. The liposomal solution was then extruded through two membranes (0.2 and 0.1 um) in five times, respectively. Then, the preformed liposome was reacted with D-Asp8 peptide with an N-terminal acetylcysteine residue for 2 h at 4 °C. Subsequently, the reacted mixture was purified by Sepharose CL-4B column to ensure removal of the un-conjugated D-Asp8 peptide. The liposomal dissolution in 0.5 ml aliquot was mixed with 0.5 ml distilled water containing mannitol and then lyophilized. Finally, the above lyophilized liposome with 15 umol lipids were rehydrated by adding 0.5 ml DNase-Free water containing antagomir-214-3p (750 ug) or Traf3 3′UTR plasmid (120 ug) and incubated for 20 min at room temperature. The encapsulation procedure was performed immediately before use and then sterilized by passing through the sterile filter.
Statistical Analyses
In general, statistical differences among groups were analyzed by one-way analysis of Variance (ANOVA) with a post-hoc test (after normalization to baseline/control group) to determine group differences in the study parameters. Especially, the rescue treatment data were compared by means of two-way ANOVA (genetic background and pharmacological treatment) with full factorial design. When ANOVA revealed differences and when treatment presented homogeneity of variance, we performed Tukey’s HSD test for multiple comparison. For heterogeneous variance, we employed Games-Howell multiple comparison tests. All statistical analyses were performed with SPSS software at version 19.0 (SPSS Inc., Chicago, IL, USA).
Additional Information
How to cite this article: Liu, J. et al. Osteoclastic miR-214 targets TRAF3 to contribute to osteolytic bone metastasis of breast cancer. Sci. Rep. 7, 40487; doi: 10.1038/srep40487 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Yin, J. J., Pollock, C. B. & Kelly, K. Mechanisms of cancer metastasis to the bone. Cell Res 15, 57–62, doi: 10.1038/sj.cr.7290266 (2005).
Rachner, T. D., Khosla, S. & Hofbauer, L. C. Osteoporosis: now and the future. Lancet 377, 1276–1287, doi: 10.1016/S0140-6736(10)62349-5 (2011).
Zaidi, M. Skeletal remodeling in health and disease. Nature medicine 13, 791–801, doi: 10.1038/nm1593 (2007).
Rigoutsos, I. & Furnari, F. Gene-expression forum: Decoy for microRNAs. Nature 465, 1016–1017, doi: 10.1038/4651016a (2010).
Sun, K. & Lai, E. C. Adult-specific functions of animal microRNAs. Nature reviews. Genetics 14, 535–548, doi: 10.1038/nrg3471 (2013).
Kloosterman, W. P. & Plasterk, R. H. The diverse functions of microRNAs in animal development and disease. Developmental cell 11, 441–450, doi: 10.1016/j.devcel.2006.09.009 (2006).
Ambros, V. The functions of animal microRNAs. Nature 431, 350–355, doi: 10.1038/nature02871 (2004).
Kosik, K. S. MicroRNAs and cellular phenotypy. Cell 143, 21–26, doi: 10.1016/j.cell.2010.09.008 (2010).
Cheng, P. et al. miR-148a regulates osteoclastogenesis by targeting V-maf musculoaponeurotic fibrosarcoma oncogene homolog B. J Bone Miner Res 28, 1180–1190, doi: 10.1002/jbmr.1845 (2013).
Kagiya, T. & Nakamura, S. Expression profiling of microRNAs in RAW264.7 cells treated with a combination of tumor necrosis factor alpha and RANKL during osteoclast differentiation. Journal of periodontal research 48, 373–385, doi: 10.1111/jre.12017 (2013).
Zhang, J. et al. Interferon-beta-induced miR-155 inhibits osteoclast differentiation by targeting SOCS1 and MITF. FEBS letters 586, 3255–3262, doi: 10.1016/j.febslet.2012.06.047 (2012).
Gong, M. et al. miR-335 inhibits small cell lung cancer bone metastases via IGF-IR and RANKL pathways. Molecular cancer research: MCR 12, 101–110, doi: 10.1158/1541-7786.MCR-13-0136 (2014).
Rossi, M. et al. miR-29b negatively regulates human osteoclastic cell differentiation and function: implications for the treatment of multiple myeloma-related bone disease. Journal of cellular physiology 228, 1506–1515, doi: 10.1002/jcp.24306 (2013).
Li, C. J. et al. MicroRNA-188 regulates age-related switch between osteoblast and adipocyte differentiation. The Journal of clinical investigation 125, 1509–1522, doi: 10.1172/JCI77716 (2015).
Li, H. et al. A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. The Journal of clinical investigation 119, 3666–3677, doi: 10.1172/JCI39832 (2009).
Tang, P., Xiong, Q., Ge, W. & Zhang, L. The role of microRNAs in osteoclasts and osteoporosis. RNA Biol 11, 1355–1363, doi: 10.1080/15476286.2014.996462 (2014).
Zhao, C. et al. miR-214 promotes osteoclastogenesis by targeting Pten/PI3k/Akt pathway. RNA biology 12, 343–353, doi: 10.1080/15476286.2015.1017205 (2015).
Wang, X. et al. miR-214 targets ATF4 to inhibit bone formation. Nature medicine 19, 93–100, doi: 10.1038/nm.3026 (2013).
Krzeszinski, J. Y. et al. miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature 512, 431–435, doi: 10.1038/nature13375 (2014).
Kim, N., Takami, M., Rho, J., Josien, R. & Choi, Y. A novel member of the leukocyte receptor complex regulates osteoclast differentiation. J Exp Med 195, 201–209 (2002).
Aurora, A. B. et al. MicroRNA-214 protects the mouse heart from ischemic injury by controlling Ca(2)(+) overload and cell death. The Journal of clinical investigation 122, 1222–1232, doi: 10.1172/JCI59327 (2012).
Xiu, Y. et al. Chloroquine reduces osteoclastogenesis in murine osteoporosis by preventing TRAF3 degradation. The Journal of clinical investigation 124, 297–310, doi: 10.1172/JCI66947 (2014).
Nakamura, T. et al. Estrogen prevents bone loss via estrogen receptor alpha and induction of Fas ligand in osteoclasts. Cell 130, 811–823, doi: 10.1016/j.cell.2007.07.025 (2007).
Li, D. et al. Osteoclast-derived exosomal miR-214-3p inhibits osteoblastic bone formation. Nat Commun 7, 10872, doi: 10.1038/ncomms10872 (2016).
Liang, C. et al. Aptamer-functionalized lipid nanoparticles targeting osteoblasts as a novel RNA interference-based bone anabolic strategy. Nature medicine 21, 288–294, doi: 10.1038/nm.3791 (2015).
Ell, B. et al. Tumor-induced osteoclast miRNA changes as regulators and biomarkers of osteolytic bone metastasis. Cancer Cell 24, 542–556, doi: 10.1016/j.ccr.2013.09.008 (2013).
Wang, F., Lv, P., Liu, X., Zhu, M. & Qiu, X. microRNA-214 enhances the invasion ability of breast cancer cells by targeting p53. Int J Mol Med 35, 1395–1402, doi: 10.3892/ijmm.2015.2123 (2015).
Orso, F. et al. miR-214 and miR-148b targeting inhibits dissemination of melanoma and breast cancer. Cancer Res, doi: 10.1158/0008-5472.CAN-15-1322 (2016).
Chirgwin, J. M. & Guise, T. A. Molecular mechanisms of tumor-bone interactions in osteolytic metastases. Crit Rev Eukaryot Gene Expr 10, 159–178 (2000).
Tseng, P. H. et al. Different modes of ubiquitination of the adaptor TRAF3 selectively activate the expression of type I interferons and proinflammatory cytokines. Nat Immunol 11, 70–75, doi: 10.1038/ni.1819 (2010).
Jang, H. D., Noh, J. Y., Shin, J. H., Lin, J. J. & Lee, S. Y. PTEN regulation by the Akt/GSK-3beta axis during RANKL signaling. Bone 55, 126–131, doi: 10.1016/j.bone.2013.02.005 (2013).
Lipton, A. et al. Advances in treating metastatic bone cancer: summary statement for the First Cambridge Conference. Clinical cancer research: an official journal of the American Association for Cancer Research 12, 6209s–6212s, doi: 10.1158/1078-0432.CCR-06-1213 (2006).
Body, J. J. Bisphosphonates in the treatment of metastatic breast cancer. J Mammary Gland Biol Neoplasia 6, 477–485 (2001).
Gao, L. et al. HTLV-1 Tax transgenic mice develop spontaneous osteolytic bone metastases prevented by osteoclast inhibition. Blood 106, 4294–4302, doi: 10.1182/blood-2005-04-1730 (2005).
La, V. D., Tanabe, S. & Grenier, D. Naringenin inhibits human osteoclastogenesis and osteoclastic bone resorption. Journal of periodontal research 44, 193–198, doi: 10.1111/j.1600-0765.2008.01107.x (2009).
Acknowledgements
We thank the technical staff from Institute for Advancing Translational Medicine in Bone & Joint Diseases, Hong Kong Baptist University for providing critical comments and technical support. This study was supported by the Ministry of Science and Technology of China (2013ZX09301307 to A.L.), the Hong Kong General Research Fund (HKBU479111 to G.Z., HKBU478312 to G.Z., HKBU262913 to G.Z., HKBU261113 to A.L. CUHK14112915 to B.Z. and CUHK489213 to B.Z.), the Natural Science Foundation Council of China (81602556 to D.L.; 81272045 to G.Z. 81272045 to B.G., 81401833 to B.G. and 81470072 to X.H.), the Research Grants Council & Natural Science Foundation Council of China (N_HKBU435/12 to G.Z.), the Croucher Foundation (Gnt#CAS14BU/CAS14201 to A.L.), the Interdisciplinary Research Matching Scheme (IRMS) of Hong Kong Baptist University (RC-IRMS/12-13/02 to A.L. and RC-IRMS/13-14/02 to G.Z.), the Hong Kong Baptist University Strategic Development Fund (SDF) (SDF13-1209-P01 to A.L.), the Hong Kong Research Grants Council (RGC) Early Career Scheme (ECS) (489213 to G.Z.), the Inter-institutional Collaborative Research Scheme of Hong Kong Baptist University (RC-ICRS/14-15/01 to G.Z.), the Faculty Research Grant of Hong Kong Baptist University (FRG1/13-14/024 to G.Z. FRG2/12-13/027 to G.Z. and FRG2/14-15/010 to G.Z.), the China Academy of Chinese Medical Sciences (Z0252 and Z0293 to A.L.). The statistical analysis was performed by a contract service from Bioinformedicine (http://www.bioinformedicine.com/index.php).
Author information
Authors and Affiliations
Contributions
G.Z. and A.L. supervised the whole project. J.L., D.L. and L.D. performed the major experiments and wrote the manuscript in equal contribution. C.L., B.G., C. Lu, X.H., H.C., B.H., B.L., F.L., J.Lu, L.W., A.S. and F.J. provided the technical support. C.W. Lu, S.P., Z.Z., B.Z., X.P. and L.X. provided their professional expertise.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Liu, J., Li, D., Dang, L. et al. Osteoclastic miR-214 targets TRAF3 to contribute to osteolytic bone metastasis of breast cancer. Sci Rep 7, 40487 (2017). https://doi.org/10.1038/srep40487
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep40487
This article is cited by
-
The novel circRNA circ_0045881 inhibits cell proliferation and invasion by targeting mir-214-3p in triple-negative breast cancer
BMC Cancer (2024)
-
In vivo validation of the functional role of MicroRNA-4638-3p in breast cancer bone metastasis
Journal of Cancer Research and Clinical Oncology (2024)
-
TRAF6 as a potential target in advanced breast cancer: a systematic review, meta-analysis, and bioinformatics validation
Scientific Reports (2023)
-
Secreted microRNAs in bone metastasis
Journal of Bone and Mineral Metabolism (2023)
-
Breast Cancer Exosomal microRNAs Facilitate Pre-Metastatic Niche Formation in the Bone: A Mathematical Model
Bulletin of Mathematical Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.