Abstract
The enzyme poly-ADP-ribose-polymerase (PARP) mediates DNA-repair and rearrangements of the nuclear chromatin. Generally, PARP activity is thought to promote cell survival and in recent years a number of PARP inhibitors have been clinically developed for cancer treatment. Paradoxically, PARP activity is also connected to many diseases including the untreatable blinding disease Retinitis Pigmentosa (RP), where PARP activity appears to drive the pathogenesis of photoreceptor loss. We tested the efficacy of three different PARP inhibitors to prevent photoreceptor loss in the rd1 mouse model for RP. In retinal explant cultures in vitro, olaparib had strong and long-lasting photoreceptor neuroprotective capacities. We demonstrated target engagement by showing that olaparib reduced photoreceptor accumulation of poly-ADP-ribosylated proteins. Remarkably, olaparib also reduced accumulation of cyclic-guanosine-monophosphate (cGMP), a characteristic marker for photoreceptor degeneration. Moreover, intravitreal injection of olaparib in rd1 animals diminished PARP activity and increased photoreceptor survival, confirming in vivo neuroprotection. This study affirms the role of PARP in inherited retinal degeneration and for the first time shows that a clinically approved PARP inhibitor can prevent photoreceptor degeneration in an RP model. The wealth of human clinical data available for olaparib highlights its strong potential for a rapid clinical translation into a novel RP treatment.
Similar content being viewed by others
Introduction
The enzyme poly(ADP-ribose) polymerase (PARP) is one of the key mediators of DNA damage repair1 and generally seen as a beneficial factor in cell physiology. However, PARP activity is also connected to a variety of human diseases, essentially in two different ways: 1) in cancer, the repair of DNA damage allows cells to survive and possibly contributes to cancerogenesis; 2) in neurodegenerative diseases, excessive activation of PARP may deplete cellular substrates and lead to a specific form of programmed cell death, termed PARthanatos2. Thus, PARP seems to be localized at a cross-road of cell physiology and pathology. The tight control of its activity is a major focus in recent therapy developments.
Retinitis pigmentosa (RP) is a group of hereditary retinal degenerative diseases in which rod photoreceptors die due to a genetic mutation, whereas cone photoreceptors disappear secondarily, once rods are gone. While the initial disease symptoms (i.e. night blindness) are comparatively mild, the secondary loss of cones ultimately leads to complete blindness. The disease affects approximately 1 in 3,000 to 7,000 people3 and is characterized by strong genetic heterogeneity with causative mutations in more than 65 genes. In 4–8% of human RP cases, the disease is caused by mutations in the genes encoding for cGMP specific phosphodiesterase 6 (PDE6)4,5. The non-functional enzyme fails to hydrolyze cGMP, causing its accumulation4,6. Animal models like the retinal degeneration 1 (rd1) mouse, which harbors a mutated Pde6b gene7, have advanced the understanding of the cellular processes underlying retinal degeneration. Notably, elevated cGMP levels in dying photoreceptors were found to correlate with increased activity of PARP8,9.
PARP is an important mediator of base excision repair. It has three zinc finger domains that differentially recognize DNA double strand breaks and single strand breaks10. DNA damage activates PARP to catalyze extensive polymerization of ADP-ribose from NAD+ onto acceptor proteins, for instance histones and PARP itself11. The cofactor of PARP is nicotinamide adenine dinucleotide (NAD) and sustained PARP activity following excessive DNA damage decreases NAD+ levels in a dose-dependent manner12. Consequently, ATP levels will fall because NAD+ is needed for glycolysis and the Krebs cycle13. Berger proposed a mechanism, known as the “PARP suicide hypothesis”, suggesting that excessive activation of PARP may account for rapid cell death before DNA repair can take place14. This kind of cell death, later named ‘parthanatos’ (derived from the Greek Θάνατος, “Death”) is associated with nuclear translocation of the mitochondrial protein apoptosis-inducing factor (AIF)15 and energy depletion16. Although NAD+ and ATP depletion appear to be relatively early events after PARP activation, cell death only takes place many hours later17, indicating that other downstream mediators may be present and epigenetic changes, e.g. cytosine methylation, are involved. This corresponds to similar observations in rd1 photoreceptors, both in terms of cell death timing18 and in dramatically altered gene expression found in rd1 retinas19. Moreover, the methylated and hydroxymethylated form of cytosine (5mC and 5hmC) accumulate in rd1 retinas20,21, implying dynamic changes in global epigenetic regulation during retinal degeneration.
The retina of mice in which PARP-1 was genetically deleted is morphologically and functionally normal, but resistant to PDE6 inhibition-induced retinal degeneration9, suggesting that PARP-1 in particular is responsible for photoreceptor degeneration. In a comparative study, excessive PARP activity was found to be a common denominator for photoreceptor cell death in ten different retinal degeneration models, including in the rd1 mouse22 highlighting the potential of PARP inhibitors for the treatment of genetically diverse groups of RP patients.
Here, we tested three recently developed PARP inhibitors for photoreceptor neuroprotective capacities. Among the inhibitors tested, the phthalazinone-based olaparib, an FDA approved drug for the treatment of ovarian cancer23 markedly reduced photoreceptor degeneration in vitro and in vivo. Our data confirms the importance of PARP activity for photoreceptor degeneration and suggests olaparib for a rapid clinical translation into a treatment for RP.
Results
Previously, we had found that the 1st generation PARP inhibitor PJ-34 afforded moderate but significant photoreceptor protection in rd1 retina8. Recently, several PARP inhibitors have been developed clinically and we decided to test three promising compounds for their photoreceptor protective capacities, initially in organotypic retinal explant cultures derived from rd1 animals. The PARP inhibitors tested were: R503, an experimental compound developed by the company Radikal Therapeutics; ABT-888 (Veliparib), a PARP inhibitor currently being used in several phase III clinical trials (NCT02264990, NCT02163694, NCT02152982); and olaparib (LynparzaTM), a drug approved in 2014 for the treatment of ovarian cancer positive for BRCA1/2 mutations. rd1 retinal explants cultured from post-natal (P) day 7 to 11 with either ABT-888 or R503 exhibited clear signs of toxicity at concentrations of 0.1 μM or 1 μM, respectively (Supplementary Fig. 1). We also observed a disruption of the normal retinal layering with both ABT-888 and R503, suggesting adverse effects on early post-natal retinal development. However, in this initial drug screening olaparib, a drug targeting in particular PARP-1 and PARP-224, appeared to show strong photoreceptor protective effects, calling for a more thorough evaluation of this compound.
PARP inhibition with olaparib rescues photoreceptor cell death in rd1 retinal explant cultures
The effect of olaparib was assessed by counting both surviving photoreceptor rows and dying TUNEL positive cells. Olaparib appeared to have a dose-dependent protective effect on retinal explant cultures with a maximum preservation of photoreceptor rows and a minimum number of TUNEL positive cells at a concentration of 100 nM olaparib. In wildtype (wt) cultures the number of photoreceptor rows and the percentage of TUNEL positive cells in 100 nM olaparib treated rd1 cultures approached the level of untreated wt, with no other adverse effects seen (Fig. 1). We used DMSO as a solvent for olaparib and since a recent study found toxic effects of DMSO in the retina25, we examined whether the DMSO concentrations used in control explants (0.6–30 μM) influenced photoreceptor survival. When the DMSO concentration of control groups for each experiment was calculated and plotted against the average number of photoreceptor rows and the percentages of TUNEL positive cells, no disturbances due to DMSO were found, indicating that the solvent had not influenced the course of degeneration (Supplementary Fig. 2).
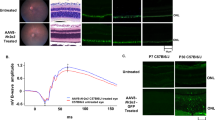
Olaparib rescues rd1 photoreceptors in short-term retinal explant cultures.
(a) Immunohistochemical staining revealed a dose-dependent effect of olaparib treatment with 100 nM as the most protective concentration and toxic effects of high concentrations. (b) Quantification of photoreceptor rows. (c) Quantification of the percentage of TUNEL positive cells. Bar graphs represent means ± SEM. n(wt, untreated) = 6; n(wt, 0.1 μM olaparib) = 6; n(rd1, untreated) = 9; n(rd1, 0.01 μM olaparib) = 5; n(rd1, 0.1 μM olaparib) = 9; n(rd1, 1 μM olaparib) = 7; n(rd1, 10 μM olaparib) = 7; n(rd1, 20 μM olaparib) = 4; n(rd1, 50 μM olaparib) = 6. **p < 0.01 by Kruskal-Wallis test for multiple analysis; scale bar is 20 μm.
Olaparib decreases PARylation and cGMP levels in rd1 retinal explant cultures
The efficacy of PARP inhibition was assessed using an immunostaining for PAR residues in individual photoreceptor cells. There was a significant decrease in the numbers of photoreceptors showing PAR accumulation in 100 nM olaparib treated rd1 retinal cultures, while 100 nM olaparib did not affect the numbers of PAR positive cells in wt cultures (Fig. 2a,b). Remarkably, higher concentrations of olaparib did not further reduce the PAR signal, indicating that PARP isoforms other than PARP-1 or PARP-2 may also have contributed to the total PAR accumulation found in rd1 photoreceptors. Western blot analysis in principle confirmed the immunohistochemistry results. For this, rd1 and wt in vivo retina were used as positive and negative controls, respectively, showing a strong increase in PARylated proteins in rd1 retina in vivo, in line with earlier publications8,9. Cultured, in vitro retina showed an overall lower level of protein PARylation than in vivo samples, together with a numerical reduction of PARylation in rd1 retinal explants treated with olaparib in vitro (Fig. 2c; Fig. S5). However, since the number of cells showing strong PARylation at any given time-point is relatively low (approx. 1% of ONL cells; >0.5% of all cells in the retina), the western blot analysis at the whole tissue level failed to show a statistically significant effect. Therefore, for all later analysis, we focused on methods allowing for cellular resolution (i.e. TUNEL assay, PAR immunostaining).
Effect of PARP inhibition with olaparib on PARylation and cGMP levels in rd1 retinal cultures.
(a,b) The higher the concentration of olaparib, the smaller was the percentage of PAR positive cells. (c) Western blot analysis confirmed the previously reported strong PARylation difference between rd1 and wildtype (wt) retinas8,9. Furthermore, there was a strong reduction of PARylation in rd1 cultures treated with 100 nM olaparib compared to control cultures. (d) Immunohistochemical stainings showed an increase of cGMP level in rd1 compared to wt. (e) Quantification of the cGMP signal revealed a significant reduction due to treatment with olaparib. Bar graphs represent means ± SEM. n(wt, untreated) = 6; n(wt, 0.1 μM olaparib) = 6; n(rd1, untreated) = 9; n(rd1, 0.01 μM olaparib) = 5; n(rd1, 0.1 μM olaparib) = 9; n(rd1, 1 μM olaparib) = 7; n(rd1, 10 μM olaparib) = 7; n(rd1, 20 μM olaparib) = 4; n(rd1, 50 μM olaparib) = 6; n(WB, in vivo, rd1) = 1; n(WB, in vivo, wt) = 1; n(WB, in vitro, rd1, untreated) = 3; n(WB, in vitro, rd1, olaparib) = 3. **p < 0.01 by Kruskal-Wallis test for multiple analysis; scale bars in (a and d) are 20 μm.
Previous studies indicated that increased cGMP levels due to PDE6 dysfunction are reduced in PARP-1 knockout (KO) retina9. Thus, the effect of pharmacological PARP inhibition on cGMP levels was assessed and, remarkably, the strong increase of cGMP levels in rd1 was significantly reduced upon olaparib treatment (Fig. 2d,e).
Olaparib shows sustained protective effects
To evaluate the long-term effects of olaparib on rd1 retinal explant cultures, the treatment paradigm was extended to P17. Since cone photoreceptors are fully differentiated at this age, for these experiments, we used rd1 mice carrying the TN-XL biosensor26 (i.e. rd1TN-XL) to directly visualize cone survival. Photoreceptor rows were increased after treatment (Fig. 3a,b), whereas the percentage of TUNEL positive cells in olaparib treated cultures was decreased (Fig. 3a,c). Cone density, on the other hand, was unaffected (Fig. 3a,d). Finally, the in vitro treatment was prolonged even further to P24. However, here, olaparib had no significant effect on photoreceptor rows, TUNEL positive cells, and cone density (Supplementary Fig. 3), indicating that the treatment delayed photoreceptor degeneration, but could not entirely prevent it in the long-term.
Long-term protective effects of olaparib treatment in rd1TN-XL retinal cultures at P17.
(a) Immunohistochemical staining, at P17, showed improved photoreceptor viability in 0.1 μM olaparib treated cultures, when compared to untreated (untr.) control. (b) The number of photoreceptor rows was significantly increased after treatment. (c) There were less TUNEL positive cells in olaparib-treated cultures. (d) Cone density (EGFP signal) remained unchanged after treatment. Bar graphs represent means ± SEM. n(rd1, untreated) = 5; n(rd1, 0.1 μM olaparib) = 5. **p < 0.01 by two-sample Komolgorov-Smirnov test; scale bar is 20 μm.
The effect of PARP inhibition on DNA hypermethylation
Epigenetic changes are often reflected in alterations of the methylation level of cytosine in the DNA. Indeed, DNA methylation was recently found to be strongly increased in dying photoreceptor cells20,21 and this is correlated to a strong over-activation of PARP. To investigate whether DNA methylation and PARP activity were causally connected to each other, we examined global DNA methylation in the ONL of both wt and rd1 retinal explant cultures, by staining for 5-methyl-cytosine (5mC) and 5-hydroxy-methyl-cytosine (5hmC). At P13, at the peak of degeneration in rd1 retinas in vivo, high levels of 5mC co-localized completely with the TUNEL assay. 5hmC positive cells on the other hand showed a 90% overlap with the TUNEL signal (Fig. 4a–c). Similarly, in retinal cultures at P11, rd1 retinal explants showed a heavy increase in 5mC and 5hmC DNA methylation of photoreceptors compared to wt. However, inhibition of PARP with 100 nM olaparib did not significantly decrease the levels of 5mC or 5hmC (Fig. 4d,e), indicating that DNA methylation was either unrelated to or upstream of PARP activity.
Olaparib does not affect increased DNA methylation.
(a) Co-localization of TUNEL, 5mC, and 5hmC on rd1 retina at P13. (b) Of the TUNEL positive cells (n = 69), about half were also 5mC positive and nearly all cells were 5hmC positive. All 5mC positive cells (n = 29) were also TUNEL positive, whereas of all 5hmC positive cells (n = 57), there was a small fraction (n = 6) that did not show TUNEL signal. (c) Quantification of co-localization, in rows, e.g. of all TUNEL positive cells 50.6 ± 11.4 were 5mC and 72.2 ± 4.7 were 5hmC positive. (d) Immunohistochemical staining showed an increase of both 5mC and 5hmC levels in rd1 retinal cultures compared to wildtype (wt). (e) Quantification of histological methylation signal. In rd1 cultures there was no significant reduction after treatment with olaparib (5mC: p = 0.3517; 5hmC: p = 0.7858). Bar graphs represent means ± SEM. n(rd1 P13) = 4; n(wt, untreated) = 6; n(wt, olaparib) = 6; n(rd1, untreated) = 10; n(rd1, olaparib) = 8. Tested by two-sample Komolgorov-Smirnov test. Scale bars in (a and d) are 20 μm.
Olaparib protects rd1 photoreceptors in vivo
The in vitro data suggested olaparib as a promising compound for in vivo application in the rd1 mouse, with an effective dose to lie between 0.01–0.1 μM. Although olaparib is known to be well tolerated when given systemically23, we wanted to avoid the possibility of any systemic side-effects and therefore decided to use direct application to the eye via intravitreal injection. To guide and optimize the in vivo paradigm, we used the recently developed Quantitative Structure-Property Relationships (QSPR) mode27 to predict an intravitreal clearance for olaparib of 0.665 ml/h in rabbit eye, while in mouse eye, based on size scaling it is expected to be of 0.021 ml/h. Furthermore, the intravitreal half-life of olaparib (t1/2 = ln2 × vitreal volume/clearance) in mouse eye was estimated to be eight minutes. This estimate should be considered a theoretical minimum; the intravitreal half-life may be extended, for instance, if olaparib was bound to specific proteins in the vitreous.
We then chose a 1 μM olaparib solution for intravitreal injection, giving an effective concentration of 0.1 μM when assuming even intraocular distribution. This allowed being well below the toxic dose while remaining in an effective dose range for at least four half-lifes (i.e. at least 30 min). After a single intravitreal injection of 0.1 μM olaparib at P11, the injected eye’s retina showed a strong decrease in the numbers of dying ONL cells, at P13, as assessed with the TUNEL assay, when compared to the sham-injected contralateral eye (Fig. 5a,b). At P15 the olaparib treated eye still displayed a numerical decrease of dying cells in the ONL assessed over the whole retina, but this effect was no longer statistically significant. Similarly, PAR immunohistochemistry showed a decrease of PAR positive cells in the ONL of treated eyes at both P13 and P15; however, this effect did not attain statistical significance. Importantly, when P15 photoreceptor survival was analyzed along the dorso-ventral axis, the spider plot for the numbers of ONL photoreceptor rows showed a statistically significant increase in photoreceptor numbers in the dorsal retina (Fig. 5d).
Olaparib protects rd1 photoreceptors in vivo.
After a single intravitreal olaparib injection at P11 rd1 animals where analyzed for cell death (TUNEL assay) and photoreceptor survival at P13 and P15. (a) Retinas from sham-injected rd1 animals display a high number of dying, TUNEL positive cells in the ONL. At P13, in treated animals cell death is reduced, an effect that is still apparent at P15. Note the decrease in the overall size of the rd1 ONL and the number of photoreceptor rows at P15. (b) A subset of rd1 photoreceptors showed a strong immunoreactivity for PARylated proteins, their numbers appeared lower in P13 and P15 treated animals. (c) Top panel: Quantification of TUNEL positive cells in untreated vs. treated animals at P13 and P15. At P13 there were significantly less photoreceptors dying in treated rd1 retina. Bottom panel: Quantification of PAR positive cells in untreated vs. treated animals at P13 and P15. (d) Quantitative analysis of photoreceptor survival along the dorso-ventral axis. While untreated P15 rd1 retina displayed 3-6 photoreceptor rows, olaparib treated rd1 animals had up to two rows more photoreceptors in the mid-periphery, dorsal retina. n(rd1, sham-injected P13) = 6; n(rd1, treated P13) = 5; n(rd1, sham-injected P15) = 4; n(rd1, untreated P15) = 5. Tested by unpaired, two-tailed Student’s t-test. Scale bars in (a and b) are 50 μm.
Discussion
Excessive activation of PARP has been connected to hereditary photoreceptor degeneration in a large variety of relevant animal models22. Here, we show that olaparib, a 3rd generation PARP inhibitor that was recently approved for the treatment of ovarian cancer23, rescued mutant photoreceptors both in vitro and in vivo at nanomolar concentrations. These results highlight olaparib as a candidate drug for the rapid clinical translation into a treatment for currently still untreatable hereditary retinal degeneration.
In many retinal degeneration animal models the causative genetic mutations lead to dysregulated cGMP levels22. In rd1 rod photoreceptors cGMP levels rise because of their non-functional phosphodiesterase 6 (PDE6)6,28 and this is closely correlated to over-activation of PARP and photoreceptor PAR accumulation8,9. Interestingly, olaparib inhibition of PARP – which is thought to be downstream of cGMP-signaling – could significantly lower abnormally high cGMP levels. This finding corresponds to a similar observation on reduced photoreceptor cGMP levels in retina obtained from PARP-1 KO mice9. Yet, PARP inhibition in isolated coronary arterioles had the exact opposite effect on cGMP: There the PARP inhibitors ABT-888 and INO1001 increased the activity of nitric oxide synthase (NOS) and soluble guanylyl cyclase (sGC) to result in a net increase of cGMP production (Choi et al. 2012). However, in photoreceptors neither NOS nor sGC29 are expressed so that this pathway to raise cGMP is unavailable. Instead in photoreceptors, PARP activity and cGMP-signaling may be connected in at least two possible ways: (1) via PARP-dependent regulation of gene expression30, which could have a bearing on GC or on GC regulating enzymes31. (2) via clearance of cGMP, which can be shuttled by ATP-binding cassette family (ABC) pumps to the extracellular space32. ATP depletion due to excessive PARP activity would impair the ATP-driven removal of cGMP, whereas PARP inhibition would rescue this effect. The latter possibility might also explain why the effect of PARP inhibition on PAR accumulation (down to approx. one third) is more pronounced than the effect on cGMP accumulation (down to approx. 70%). In the future, it may be interesting to study these negative feedback effects to identify further mediators of the degenerative processes.
Epigenetics likely play an important role in programmed cell death in the retina33,34. Via alterations of PARP-DNA complexes and corresponding changes in DNA replication and transcription, PARP inhibitors could bring about indirect changes in epigenetic signatures that may be independent of their direct effect on PARP catalytic activity35. Furthermore, the activation of PARP may be related to the upstream activity of histone deacetylases (HDAC)33,36. While both HDAC and PARP influence histone and chromatin structure, epigenetic processes may also target the DNA structure. One repressor mark is 5mC, which is known to recruit proteins that can mediate the activation of co-repressor complexes to target promoters37. In rd1 retinal degeneration, both 5mC and 5hmC were found to be increased and colocalized with TUNEL staining20,21. 5hmC is generated from 5mC by the activity of ten eleven translocation (Tet), and can be further processed to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) by Tet family members. 5hmC appears to function as an active mark at enhancers, and thus 5mC and 5hmC might have reciprocal roles in the dynamic regulation of DNA methylation37. Therefore, a balance of 5mC and 5hmC is likely to be important for the homeostasis of postmitotic neurons, where 5hmC is particularly abundant38. The loss of photoreceptors in rd1 retinas has been found to be accompanied by high levels of 5mC and 5hmC and both modifications seem to colocalize with TUNEL and moreover with PAR20. This suggests that DNA hypermethylation plays an important role in retinal cell death. While all 5mC positive cells were also TUNEL positive, this was not the case for all of the 5hmC positive cells. This would imply that DNA hypermethylation could follow a specific sequence with the dying cell first turning 5hmC and then 5mC positive. Since olaparib treatment did not reduce 5 mC and 5hmC levels, cytosine methylation is likely to be upstream of excessive PARP activity, or may be the result of a parallel process following dysregulated cGMP levels.
Several previous studies have suggested PARP inhibition as a therapeutic strategy to treat RP8,9,22; however, now it is important to identify PARP inhibitors suitable for long-term use in a chronic human disease. In recent years, numerous PARP inhibitors have undergone clinical trials – mostly for cancer therapy – and a large amount of human tolerability and efficacy data is available today39. This in turn should facilitate and accelerate clinical trials and repurposing of PARP inhibitors for RP. While for cancer treatment PARP inhibition aims to disrupt DNA repair so as to cause cell death, in retinal neurodegeneration, pathological over-activation of PARP needs to be prevented. Olaparib is a novel PARP inhibitor with increased specificity for PARP-1 and -224 that was approved for the treatment of ovarian cancer positive for BRCA1/2 mutations (FDA reference ID: 3675412) in 2014. In our retinal explant cultures olaparib showed a significant reduction of PARylation and cell death and, conversely, an increase in photoreceptor survival already at 0.1 μM. Moreover, the therapeutic range was large as only concentrations about 200 fold higher showed toxic effects. While another PARP inhibitor, PJ-34, was found to reduce cell death in the ONL by 20% at the most protective concentration of 6 μM8, 0.1 μM olaparib was able to decrease cell death by 50%. Moreover, the protective effects were still visible at P17 in vitro, with no obvious detrimental effects until P24. The in vivo application of olaparib, however, still faces the problem of sustained delivery to the photoreceptors. While PARP inhibition at P11 caused a transient decrease of TUNEL positive, dying cells at P13, this effect on cell death was no longer significant at P15, even though the numbers of surviving photoreceptors were still higher at this time-point. Thus, in the future it will be important to identify drug delivery vehicles40 that will allow for a long-term intravitreal release of olaparib with as few as possible applications.
Here, we show that the clinically approved PARP inhibitor olaparib significantly increases photoreceptor survival in rd1 retinal explant cultures, in both the short- and long-term. Additionally, PARP inhibition significantly reduced cGMP levels, while it did apparently not affect DNA methylation. Remarkably, a single intravitreal dosing of olaparib significantly preserved rd1 photoreceptors up to four days post-injection. The importance of these findings lies in the fact that olaparib is already clinically used and regarded as safe when given systemically to patients. This may allow for a rapid clinical translation and the development of olaparib treatment for RP and related neurodegenerative diseases of the retina, either in a local treatment to the eye (e.g. via intravitreal injection) or even in systemic treatments.
Methods
Animals
C3H rd1 and wildtype (wt) mice41 were used. For cone examination in long-term experiments, HR2.1:TN-XL x rd1 (rd1TN-XL) animals were used that stably express the TN-XL biosensor in cones29. Animals were housed under standard white cyclic lighting, had free access to food and water, and were used irrespective of gender. Animal protocols compliant with §4 of the German law of animal protection were reviewed and approved by the Tübingen University committee on animal protection (Einrichtung für Tierschutz, Tierärztlichen Dienst und Labortierkunde; Registration No.: 08/12/2015, 22/05/2014) and by the Service de la consommation et des affaires vétérinaires du Canton de Vaud (VD1367.5). All the experiments were performed in accordance with the ARVO statement for the use of animals in ophthalmic and visual research.
Retinal Explant Cultures
Retinal explant cultures were prepared as previously published42. Briefly, the eyes were enucleated and incubated for 15 min at 37 °C in pre-warmed 0.12% proteinase K (Sigma-Aldrich, Hamburg, Germany; P-6556) in basal R16 medium (Thermo Scientific, Rockfort, Illinois, USA; 07490743 A). To stop enzymatic activity, eyes were rinsed in basal R16 medium with 10% Sera Plus Fetal Calf Serum (FCS; PAN Biotech GmbH, Aidenbach, Germany; P30-3701) and then washed in serum-free basal medium. Cornea, lens, sclera, and choroid were removed carefully, with only the RPE remaining attached to the retina. Finally, the retina was cut at four sides so it could spread flat like a clover-leaf, with RPE facing the membrane of the cell culture insert (0.45 μm; Merck Millipore, Tullagreen, Ireland; PIHA03050). The culture medium was changed every other day during 6, 12, or 19 culturing days. Retinal explants were left without treatment for two days (until P7), followed by olaparib treatment (10 nM to 50 μM; Biomol, Hamburg, Germany; BPS-27003). Olaparib was prepared in dimethyl sulfoxide (DMSO; Sigma-Aldrich, Hamburg, Germany; D-8779) and diluted in R16 serum free culture medium with supplements. For controls, the same amount of DMSO was diluted in culture medium.
Assessing the intravitreal clearance of olaparib
The intravitreal clearance (CLivt) of olaparib was calculated in silico using the QSPR model27. The chemical structure of olaparib was retrieved from ACD/Dictionary from ACDlabs software (version 12, Advanced Chemistry Development, Inc., Toronto, Canada) and 30 molecular descriptors were generated: pKa for the most acidic molecular form, pKa for the most basic form, LogD at pH 5.5 and 7.4, LogP, MW, PSA (polar surface area), FRB (freely rotatable bonds), HD (hydrogen bond donors), HA (hydrogen bond acceptors), Htot (HD + HA), rule of 5, molar refractivity, molar volume, parachor, index of refraction, surface tension, density, polarizability, C ratio, N ratio, NO ratio, hetero ratio, halogen ratio, number of rings and number of aromatic, 3-, 4-, 5- and 6-membered rings. The PCA score plot of the training set of the model including olaparib was inspected (Supplementary Fig. 4). Olaparib was found to be within the applicability domain of the model and thus predictable by the model. The intravitreal clearance value of olaparib was then calculated using the QSPR model: LogCLivt = −0.25269–0.53747 (LogHD) + 0.05189 (LogD7.4), with the corresponding values of HD and LogD7.4 of olaparib. Half-life was calculated using equation t1/2 = ln2 Vd/CL, where Vd is the volume of distribution and CL is the intravitreal clearance.
The half-life obtained in rabbit eyes was scaled down to mice eyes using the following rationale: Small lipophilic compounds are cleared from the vitreous mainly through the RPE43. The CLivt of small lipophilic compounds in mice is expected to be 30 times smaller than in rabbits. This is based on the equation CL = P × S, where P is the drug permeability in the RPE and S is the surface area of the RPE. The RPE surface areas in mice and rabbits are 16.5 mm2 44 and 520 mm2 ; ref. 45, respectively. Assuming similar permeability of mouse and rabbit RPE, the intravitreal clearance in mouse would be 0.021 ml/h (rabbit value: 0.665 ml/h). The expected half-life in the vitreous can thus be calculated using the equation: t1/2 = ln2 Vd/CL (Vd = anatomical volume of the vitreous; in mouse 5 μl) giving an expected half-life of olaparib in the mouse vitreous of about eight minutes.
In vivo treatment on rd1 animals
Mice were anesthetized by an intraperitoneal injection of ketamine (100 mg/kg) and xylazine (5 mg/kg), their pupils dilated with tropicamide eye drops (Mydriaticum Stulln; Pharma Stulln GmbH, Germany), and eye lids anesthetized locally with Novesin (Omnivision, Puchheim, Germany). Olaparib was diluted in DMSO and single intravitreal injections were performed at P11 on one eye. As contralateral control, the other eye was sham injected with the same concentration of DMSO (0.0003%) in sterile PBS. The in vitro data suggested 0.1 μM olaparib as the most effective concentration. To obtain this concentration in vivo, intravitreal injections were performed with 0.5 μl of a 1 μM olaparib solution. Assuming an average mouse eye volume of 5 μl and even compound distribution, this gave an effective intraocular concentration of 0.1 μM olaparib. Twelve rd1 mice were used for intravitreal injections, six of these animals were sacrificed two days after treatment (P13), while the other six were sacrificed at P15. The eyes were enucleated immediately, fixed and prepared for cryosectioning.
Fixation and sectioning
Eyes obtained from the in vivo study were fixed in 4% paraformaldehyde (PFA) for 75 min, while explant cultures were fixed for 45 min. After fixation, tissues were washed for 10 min in PBS. For cryoprotection, they were incubated in 10% sucrose solution for 10 min, 20% sucrose solution for 20 min, and 30% sucrose solution for at least 30 min. The retinas were frozen in Tissue-Tek O.C.T. Compound (Sakura Finetek Europe, Alphen aan den Rijn, Netherlands; 4583)-filled boxes. 12 μm tissue sections were prepared on a Leica CM3050S Microtome (Leica Biosystems, Wetzlar, Germany), thaw-mounted onto Superfrost Plus Object slides (R. Langenbrinck, Emmendingen, Germany; 03–0060).
TUNEL assay
The terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL) assay was performed on cryosections from treated/untreated wt and rd1 retinas, using an in situ cell death detection kit conjugated with fluorescein isothiocyanate (Roche Diagnostics, Mannheim, Germany; 11 684 795 910). The sections were mounted in Vectashield with 4’,6-diamidino-2-phenylindole (DAPI) as a nuclear counterstain (Vector Laboratories, Burlingame, California, USA; H-1200).
PAR staining
3,3′-diaminobenzidine (DAB) staining was performed with quenching of endogenous peroxidase activity with 40% MeOH and 10% H2O2 in PBS for 20 min. The sections were incubated with 10% normal goat serum (NGS) in PBS containing 0.1% Triton X-100 for 1 h followed by anti-PAR antibody (1:200; Enzo Life Sciences, Lörrach, Germany; ALX-804-220-P100) incubation for 1 h. Incubation with the biotinylated secondary antibody (1:150, Vector Laboratories Inc., Burlingame, CA, USA; BA-9200; in 5% NGS in PBST) for 1 h was followed by application of Vector ABC-Kit (Vector Laboratories, Burlingame, California, USA; PK-4000) for 1 h. DAB staining solution (0.05 mg/ml NH4Cl, 200 mg/ml glucose, 0.8 mg/ml nickel ammonium sulphate, 1 mg/ml DAB, 0.1 vol. % glucose oxidase in PB) was applied evenly, incubated for exactly 60 s and immediately rinsed with PB to stop the reaction. The sections were mounted in Aquatex (Merck, Darmstadt, Germany; 1.08562.0050).
Immunofluorescence staining
Tissue sections were blocked and permeabilised in 10% BSA and 10% normal serum in PBS containing 0.1% Triton X-100 and incubated overnight in primary antibody in blocking solution. Primary antibody sources and dilutions are listed in Table 1. To increase the visibility of cones, TN-XL biosensor was enhanced by staining against its EGFP domain. Secondary antibodies were anti-sheep and -rabbit IgG’s, respectively, coupled to Alexa488 (1:350; Life Technologies, Carlsbad, California, USA). The sections were mounted in Vectashield (Vector Laboratories) with DAPI to visualize cell nuclei. Serial sections processed similarly, but without primary antibody, were used to control for non-specific background.
Microscopy and cell counting
The mounted cultures were analyzed using Zeiss Axio Imager Z1 ApoTome microscope, AxioCam MRm camera and Zeiss AxioVision 4.7 software in Z-stack (3 slices per picture; slice distance: 14 μm) and mosaic mode at 20 × magnification. For quantitative analysis, positive cells in the entire ONL of four cross-sections per culture were counted manually. The percentage of positive cells was calculated by dividing the absolute number of positive cells by the total number of ONL cells. which was assessed by dividing ONL area by the size of a photoreceptor nucleus (17.3 μm2), as measured via DAPI staining. Photoreceptor rows were assessed by counting the individual nuclei lining up in one ONL column, every 200 μm and averaging the counts. Cone density was calculated by counting GFP positive somata per 100 μm of ONL.
Graphs were prepared in GraphPad Prism 6 (GraphPad Software, La Jolla, CA, USA); Adobe Photoshop CS5, and Corel DRAW X3 were used for image processing.
Western Blot
Retinal tissue from wt and rd1 mice were homogenized in modified RIPA lysis buffer (50 mM trizma base, 150 mM NaCl, 19 mM Na4O7P2, 1 mM EDTA, 1 vol% Triton-X-100, 1 mM DTT and 0.1 vol% of protease inhibitor cocktail III EDTA-free (EMD Millipore Corp., Billerica, Massachusetts, USA); pH = 7.4) with a Precellys homogenisator (Bertin Technologies, Montigny le Bretonneux, France). For separation of proteins, 25 μg protein per well were loaded onto a 12% SDS-PAGE gradient gel, and run at 120 V. Subsequently, the proteins were transferred to PVDF membranes (Merck Millipore, Tullagreen, Ireland). Roti block buffer (Roth, Karlsruhe, Germany) was applied for 3 h at room temperature. Membranes were incubated in primary antibodies against PAR (1:1000; see above) or actin (1:400; Abcam, Milton, UK; Ab1801) in buffer containing PBST and 5% dried milk (Carl Roth GmbH, Karlsruhe, Germany) overnight at 4 °C. Membranes were washed with PBST and incubated with secondary antibodies labelled with IRDye680 RD (LI-COR Biotechnology GmbH, Bad Homburg, Germany; 926–68070) or IRDye800 CW (LI-COR Biotechnology GmbH, Bad Homburg, Germany; 926–32211) for 1 h at room temperature. LI-COR Odyssey Sa Infrared Imaging System (LI-COR Biotechnology GmbH, Bad Homburg, Germany) was used for detection of fluorescent protein bands, which were quantified using ImageJ (National Institute of Health, Washington, USA).
Statistics
Statistical analysis was performed using GraphPad Prism 6 software and Kruskal-Wallis test for multiple analyses or two-sample Komolgorov-Smirnov test. For one-to-one group comparisons Student’s t-test as implemented in Microsoft Excel software (Microsoft Corporation, Seattle, WA, USA) was used.
Additional Information
How to cite this article: Sahaboglu, A. et al. Olaparib significantly delays photoreceptor loss in a model for hereditary retinal degeneration. Sci. Rep. 6, 39537; doi: 10.1038/srep39537 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
De Vos, M., Schreiber,V. & Dantzer, F. The diverse roles and clinical relevance of PARPs in DNA damage repair: current state of the art. Biochem. Pharmacol. 84, 137–146 (2012).
Wang, Y. et al. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos). Sci. Signal. 4, ra20 (2011).
Chizzolini, M. et al. Good epidemiologic practice in retinitis pigmentosa: from phenotyping to biobanking. Curr. Genomics 12, 260–266 (2011).
Dryja, T. P., Rucinski, D. E., Chen, S. H. & Berson, E. L. Frequency of mutations in the gene encoding the alpha subunit of rod cGMP-phosphodiesterase in autosomal recessive retinitis pigmentosa. Invest Ophthalmol. Vis. Sci. 40, 1859–1865 (1999).
McLaughlin, M. E., Sandberg, M. A., Berson, E. L. & Dryja, T. P. Recessive mutations in the gene encoding the beta-subunit of rod phosphodiesterase in patients with retinitis pigmentosa. Nat. Genet. 4, 130–134 (1993).
Farber, D. B. & Lolley, R. N. Cyclic guanosine monophosphate: elevation in degenerating photoreceptor cells of the C3H mouse retina. Science 186, 449–451 (1974).
Pittler, S. J., Keeler, C. E., Sidman, R. L. & Baehr, W. PCR analysis of DNA from 70-year-old sections of rodless retina demonstrates identity with the mouse rd defect. Proc. Natl. Acad. Sci. USA 90, 9616–9619 (1993).
Paquet-Durand, F. et al. Excessive activation of poly(ADP-ribose) polymerase contributes to inherited photoreceptor degeneration in the retinal degeneration 1 mouse. J. Neurosci. 27, 10311–10319 (2007).
Sahaboglu, A. et al. PARP1 gene knock-out increases resistance to retinal degeneration without affecting retinal function. PLoS. ONE. 5, e15495 (2010).
Ikejima, M. et al. The zinc fingers of human poly(ADP-ribose) polymerase are differentially required for the recognition of DNA breaks and nicks and the consequent enzyme activation. Other structures recognize intact DNA. J Biol. Chem. 265, 21907–21913 (1990).
Langelier, M. F., Planck, J. L., Roy, S. & Pascal, J. M. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science 336, 728–732 (2012).
Goodwin, P. M., Lewis, P. J., Davies, M. I., Skidmore, C. J. & Shall, S. The effect of gamma radiation and neocarzinostatin on NAD and ATP levels in mouse leukaemia cells. Biochim. Biophys. Acta 543, 576–582 (1978).
Bernofsky, C. Physiology aspects of pyridine nucleotide regulation in mammals. Mol. Cell Biochem. 33, 135–143 (1980).
Berger, N. A. Poly(ADP-ribose) in the cellular response to DNA damage. Radiat. Res. 101, 4–15 (1985).
Fatokun, A. A., Dawson, V. L. & Dawson, T. M. Parthanatos: mitochondrial-linked mechanisms and therapeutic opportunities. Br. J Pharmacol. 171, 2000–2016 (2014).
Conrad, M., Angeli, J. P., Vandenabeele, P. & Stockwell, B. R. Regulated necrosis: disease relevance and therapeutic opportunities. Nat. Rev. Drug Discov. 15, 348–366 (2016).
Hong, S. J., Dawson, T. M. & Dawson, V. L. Nuclear and mitochondrial conversations in cell death: PARP-1 and AIF signaling. Trends Pharmacol. Sci. 25, 259–264 (2004).
Sahaboglu, A. et al. Retinitis Pigmentosa: Rapid neurodegeneration is governed by slow cell death mechanisms. Cell Death & Disease 4, (2013).
Azadi, S., Paquet-Durand, F., Medstrand, P., van Veen, T. & Ekstrom, P. A. Up-regulation and increased phosphorylation of protein kinase C (PKC) delta, mu and theta in the degenerating rd1 mouse retina. Mol. Cell Neurosci. 31, 759–773 (2006).
Wahlin, K. J. et al. Epigenetics and cell death: DNA hypermethylation in programmed retinal cell death. PLoS One. 8, e79140 (2013).
Farinelli,. P. et al. DNA methylation and differential gene regulation in photoreceptor cell death. Cell Death Dis. 5, e1558 (2014).
Arango-Gonzalez, B. et al. Identification of a common non-apoptotic cell death mechanism in hereditary retinal degeneration. PLoS One. 9, e112142 (2014).
Ledermann, J. A. PARP inhibitors in ovarian cancer. Ann. Oncol. 27, Suppl 1, i40–i44 (2016).
Steffen, J. D., Brody, J. R., Armen, R. S. & Pascal, J. M. Structural Implications for Selective Targeting of PARPs. Front Oncol. 3, 301 (2013).
Galvao, J. et al. Unexpected low-dose toxicity of the universal solvent DMSO. FASEB J 28, 1317–1330 (2014).
Wei, T. et al. Generation and functional characterization of a transgenic mouse expressing a Ca2+ biosensor in cone photoreceptors. J. Neurosci. 32, 6994–6981 (2012).
Del Amo, E. M., Vellonen, K. S., Kidron, H. & Urtti, A. Intravitreal clearance and volume of distribution of compounds in rabbits: In silico prediction and pharmacokinetic simulations for drug development. Eur. J Pharm. Biopharm. 95, 215–226 (2015).
Bowes, C. et al. Retinal degeneration in the rd mouse is caused by a defect in the beta subunit of rod cGMP-phosphodiesterase. Nature 347, 677–680 (1990).
Wei, T. et al. Light-driven calcium signals in mouse cone photoreceptors. J Neurosci. 32, 6981–6994 (2012).
Cohen-Armon, M. et al. DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: a link to histone acetylation. Mol. Cell 25, 297–308 (2007).
Azadi, S., Molday, L. L. & Molday, R. S. RD3, the protein associated with Leber congenital amaurosis type 12, is required for guanylate cyclase trafficking in photoreceptor cells. Proc. Natl. Acad. Sci. USA 107, 21158–21163 (2010).
Jedlitschky, G., Burchell, B. & Keppler, D. The multidrug resistance protein 5 functions as an ATP-dependent export pump for cyclic nucleotides. J Biol. Chem. 275, 30069–30074 (2000).
Sancho-Pelluz, J. et al. Excessive HDAC activation is critical for neurodegeneration in the rd1 mouse. Cell Death & Disease 1, 1–9 (2010).
Trifunovic, D. et al. HDAC inhibition in the cpfl1 mouse protects degenerating cone photoreceptors in vivo. Hum Mol. Genet. (2016).
Murai, J. et al. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 72, 5588–5599 (2012).
Trifunovic, D. et al. Neuroprotective Strategies for the Treatment of Inherited Photoreceptor Degeneration. Curr. Mol. Med. 12, 598–612 (2012).
Breiling, A. & Lyko, F. Epigenetic regulatory functions of DNA modifications: 5-methylcytosine and beyond. Epigenetics. Chromatin. 8, 24 (2015).
Kriaucionis, S. & Heintz, N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930 (2009).
Sonnenblick, A., de, A. E., Azim, H. A., Jr. & Piccart, M. An update on PARP inhibitors–moving to the adjuvant setting. Nat. Rev. Clin. Oncol. 12, 27–41 (2015).
Meyer, C. H., Krohne, T. U., Charbel, I. P., Liu, Z. & Holz, F. G. Routes for Drug Delivery to the Eye and Retina: Intravitreal Injections. Dev. Ophthalmol. 55, 63–70 (2016).
Sanyal,. S. & Bal, A. K. Comparative light and electron microscopic study of retinal histogenesis in normal and rd mutant mice. Z. Anat. Entwicklungsgesch. 142, 219–238 (1973).
Caffe,. A. R. et al. Mouse retina explants after long-term culture in serum free medium. J Chem. Neuroanat. 22, 263–273 (2001).
Del Amo, E. M. & Urtti, A. Rabbit as an animal model for intravitreal pharmacokinetics: Clinical predictability and quality of the published data. Exp. Eye Res. 137, 111–124 (2015).
Jeon, C. J., Strettoi, E. & Masland, R. H. The major cell populations of the mouse retina. J. Neurosci. 18, 8936–8946 (1998).
Reichenbach, A. et al. Development of the rabbit retina. V. The question of ‘columnar units’. Brain Res. Dev. Brain Res. 79, 72–84 (1994).
Acknowledgements
We thank Marius Ueffing for scientific advice and discussions, and Norman Rieger and Klaudija Masarini for excellent technical assistance. We thank Andrew Salzman for helpful discussions and the experimental PARP inhibitor R503. This work was supported by the European Union (DRUGSFORD; HEALTH-F2-2012-304963), Deutsche Forschungsgemeinschaft (DFG; PA1751/7-1 and SA3040/1-1), the Charlotte and Tistou Kerstan Foundation (SAH001/2015).
Author information
Authors and Affiliations
Contributions
M.B. carried out the in vitro retinal explant culture studies and carried out the analysis for in vitro part and drafted the manuscript. A.S. performed the in vivo studies, and carried out the analysis. Y.A. and E.S. participated to the analysis of in vivo studies. A.U. and E.A. performed in silico calculation of clearance of olaparib. A.S., F.P.D., and E.Z. participated in the design, analysis and interpretation of the study. A.S. and F.P.D. conceived the study, participated in its design and coordination, and helped to write the manuscript. All authors read and approved the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Sahaboglu, A., Barth, M., Secer, E. et al. Olaparib significantly delays photoreceptor loss in a model for hereditary retinal degeneration. Sci Rep 6, 39537 (2016). https://doi.org/10.1038/srep39537
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep39537
This article is cited by
-
Poly(ADP-ribose) polymerase inhibition: past, present and future
Nature Reviews Drug Discovery (2020)
-
Drug repurposing studies of PARP inhibitors as a new therapy for inherited retinal degeneration
Cellular and Molecular Life Sciences (2020)
-
Poly ADP ribosylation and extracellular vesicle activity in rod photoreceptor degeneration
Scientific Reports (2019)
-
A retinal model of cerebral malaria
Scientific Reports (2019)
-
Ceramide Induces the Death of Retina Photoreceptors Through Activation of Parthanatos
Molecular Neurobiology (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.