Abstract
A delicate balance between proliferating and differentiating signals is necessary to ensure proper growth and neuronal specification. By studying the developing zebrafish brain, we observed a specific and dynamic expression of a microtubule destabilizer gene, stathmin-like 4 (stmn4), in the dorsal midbrain region. The expression of stmn4 was mutually exclusive to a pan-neuronal marker, elavl3 that indicates its role in regulating neurogenesis. We showed the knockdown or overexpression of stmn4 resulted in premature neuronal differentiation in dorsal midbrain. We also generated stmn4 maternal-zygotic knockout zebrafish by the CRISPR/Cas9 system. Unexpectedly, only less than 10% of stmn4 mutants showed similar phenotypes observed in that of stmn4 morphants. It might be due to the complementation of the increased stmn1b expression observed in stmn4 mutants. In addition, time-lapse recordings revealed the changes in cellular proliferation and differentiation in stmn4 morphants. Stmn4 morphants displayed a longer G2 phase that could be rescued by Cdc25a. Furthermore, the inhibition of Wnt could reduce stmn4 transcripts. These results suggest that the Wnt-mediated Stmn4 homeostasis is crucial for preventing dorsal midbrain from premature differentiation via the G2 phase control during the neural keel stage.
Similar content being viewed by others
Introduction
During embryonic development, cells proliferate and differentiate to assume different lineages needed for a mature organism. Precise controls on transitions between proliferation and differentiation are essential for proper patterning within compartments, in particular brain development. However, the regulatory mechanisms still remain largely unexplored. Zones of delayed neurogenesis are those areas confined and conspicuous during neurogenesis in vertebrates1,2. Among this, neuron-free midbrain-hindbrain boundary (MHB) or intervening zones is crucial for patterning and growth of midbrain-hindbrain3,4. Mutual interactions between Wnts and fibroblast growth factors (Fgfs) originated from the MHB are proposed to control patterning, cell fate decision and proliferation5,6,7. The Fgf gradient thus induces the expression of a hairy gene her5 which prevents neuron differentiation invading from anterior brain via cell cycle regulation8 and inhibition of proneural gene expressions9. Although we know some of the core molecular switches mediating cell cycle exit during neuronal differentiation10 yet how the altered mitosis could direct cell fate determination is still far from clear.
Microtubules are the basal components of mitotic spindles that drive cell division during cell cycle progression. They are highly dynamic and are constantly switching between growth and shrinkage phases11. The microtubule cytoskeleton is also vital during neurogenesis by mediating neuronal migration or neurite growth. It thus implies that microtubule dynamics may be involved in maintaining the neural progenitor cell pools presumably through their associated proteins. Diverse microtubule-associated proteins, including stathmin family phosphoproteins, assist in particular neurogenesis phase by mediating microtubule dynamicity. Stathmin family genes, comprised of stathmin, SCG10, SCLIP, and RB3 with its splice variants RB3’ and RB3”, share a highly conserved stathmin-like domain (SLD), which binds with two tubulin heterodimers to form a “T2S” complex12. Mostly by the sequestering of free tubulins, stathmins are regarded as microtubule destabilizers and intracellular signaling relay centers. In addition to stathmin, other stathmin member proteins possess an additional N-terminal extension. With palmitoylations onto two conserved cysteine residues and conserved sequence AYKEMKEL in domain A of the N-terminal extension, stathmin phosphoproteins could be tethered to Golgi membranes or other vesicle-like structures13.
Even with differential expressions, all four stathmins are highly enriched in the nervous system not only during development but also adult stages. Through modulating neurite outgrowth or axon branching, stathmins are recognized as neuronal differentiation hallmarks. In addition, cellular migration involving tissue patterning is associated to stathmins. Stathmins have also been reported to influence neuronal proliferation14.
We have previously demonstrated distinct expression of 4 of 7 stathmin genes in the central nervous system during development in zebrafish embryos15. Among them, stathmin-like 4 (stmn4) called our attention due to its restricted expression within the neuron-free midbrain during a critical period of brain development. We demonstrate here that the homeostasis of stmn4 expression is indispensable for neuron progenitor cell pool maintenance in the dorsal midbrain. Either overdose or reduction of stmn4 expression led to premature appearance of post-mitotic neurons. Although the premature differentiation was only observed in a small portion of stmn4 mutants generated, it might be due to the complementation of increased stmn1b expression. With stochastically labeling, single-cell resolution analysis further confirmed the link between Stmn4 dynamics and proliferation within intervening zones. A longer G2 phase observed in stmn4-deficient embryos appears to be critical to drive neuron progenitors exit from cell cycle and subsequent neurogenesis. Thus, we conclude that Stmn4 is vital for preventing premature neurogenesis to keep progenitor cell pools within dorsal midbrain during early brain development.
Results
stmn4 is highly expressed before neuronal differentiation in dorsal midbrain
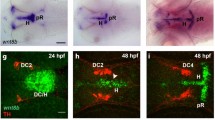
To investigate the role of Stmn4 in dorsal midbrain development, we performed whole-mount in situ hybridization (WISH) to examine its expression patterns in the developing brain of zebrafish embryos from 11–24 h post fertilization (hpf). stmn4 was not expressed until 11 hpf (Fig. 1a, data not shown for those embryos before 11 hpf). It first appeared at the dorsal midbrain region and then ventral tegmentum (Fig. 1b–e) until 20 hpf. The expression of stmn4 in the dorsal midbrain decreased abruptly, but persisted in the ventral tegmentum at 24 hpf (Fig. 1f). The stmn4 transcripts were also found in spinal neurons (Fig. 1b–f). Since neurogenesis of dorsal midbrain occurs late in embryogenesis, we hypothesized that Stmn4 may be inhibitory to neurogenesis. To test the hypothesis, we performed double WISH against stmn4 and ELAV like neuron-specific RNA binding protein 3 (elavl3). The elavl3 is the current annotated name for huc, a pan-neuronal marker, and will be used hereafter. Notable mature neurons first arouse in telencephalon and ventral tegmentum at 12 hpf (Fig. 1g), expanded into trigeminal sensory neurons (TgSNs) at 14 hpf (Fig. 1h) and then epiphysis (e) at 16 hpf (Fig. 1i). In the dorsal midbrain, the stmn4 expression domain was distinct from the surrounding elavl3 expression (Fig. 1g–j, arrowheads). It suggested that stmn4 may prohibit neuronal differentiation to maintain the neuronal progenitor pools in the dorsal midbrain.
Stmn4 prohibited elavl3 expression in the dorsal midbrain during early embryogenesis.
(a–f) Representative photographs of whole-mount in situ hybridization against stmn4 in zebrafish embryos at designated stages are displayed. stmn4 were highly expressed in the midbrain regions (boxed). A magnified photograph of the boxed regions is shown beneath each panel. (g–j) Representative photographs of double whole-mount in situ hybridization against stmn4 (purple) and elavl3 (red) at designated stages are presented. Arrowheads point to the dorsal midbrain. Scale bars are shown in the panels (a,g) on the right for each row unless specified in (f). Embryonic stages are shown in hours post fertilization (hpf) at the lower left corner in each panel. e: epiphysis; t: telencephalon; TgSN: trigeminal ganglion sensory neurons; vt: ventral tegmentum.
Characterization of Stmn4
All stathmin family genes possess a SLD and an additional N-terminal domain except for stathmin1b (stmn1b) (Fig. 2a). The Stmn4 contains two conserved cysteine residues, C20 and C22, which can be palmitoylated to enhance the recruitment of Stmn4 to Golgi apparatus. We mutated the C20 and C22 residues of Stmn4 to alanine and found that the recruitment of Stmn4 to Golgi apparatus is significantly reduced in Hela cells (Fig. S1). In addition, the Stmn4 SLD has the first three conserved serine residues, S59, S71 and S81, but does not have the fourth serine reside found in their mammalian homologues (Fig. S2a). Phosphorylations of these serine residues hamper the microtubule depolymerizing ability of stathmins16. Thus, we changed all these serine residues of Stmn4 to alanine to make it a non-phosphorylatable constitutively active form denoted as CA-Stmn4, which presumably has a higher microtubule depolymerizing activity. The CA-Stmn4 plasmids were injected into 1-cell stage zebrafish embryos, subjected to immunohistochemistry against histidine and alpha-tubulin to reveal Stmn4 and microtubule, respectively, and examined under confocal microscopy. As shown in Fig. S3(b,c), the percentage of polymerized microtubules in each blastomere appears to be less and negatively correlated to the expression of ca-stmn4. On the other hand, the percentage of depolymerized microtubules in each blastomere is higher and positively correlated to the expression of ca-stmn4 (Fig. S3b,d). The overexpression of ca-stmn4 driven by a CMV promoter interferes gastrulation, a process mediated by microtubule dynamics17. It further validates the higher microtubule destabilizing activity of CA-Stmn4 (Fig. S2b). Collectively, these results clearly demonstrated that the zebrafish Stmn4 contains its basic characteristics and is functionally similar to their mammalian homologues.
stmn4 knockdown resulted in premature expression of elavl3 in zebrafish embryos.
(a) Target sites are indicated for two antisense translational blocking morpholino oligo nucleotides (tMO1 and tMO2) and one small hairpin RNA (shRNA) at the start codon, 5′ and 3′ untranslated region (UTR), respectively. (b) Embryos were injected with 200 pg of PCS2+ vector with an insertion of a fragment of stmn4 containing both tMO1 and tMO2 binding sites as indicated in (a) without (control) or with tMO1 or tMO2 (stmn4 MO) as indicated, examined and photographed at the bud stage. The percentages of embryos with green fluorescence were calculated and compared to the control groups as shown in (g). Tg(Huc:kaede) embryos were injected without (c, Ctrl) or with different amounts of stmn4 tMO1 or tMO2 (d, stmn4 MO), P53 MO and stmn4 expression vectors, cultured to 16 hpf and examined under confocal microscopy. A stack of 10 layers (2.5 μm for one layers) from the top of midbrain were combined and shown. (e–i) Wild type embryos were treated as described previously, cultured to 10-somite stage, fixed and subjected to WISH against elavl3. Representative photographs are presented (e,f). Scale bars are only shown on the left panel in a row. A premature elevation of elavl3 expression was observed in embryos injected with stmn4 MO (f) compared that of control. The percentages of embryos with elevated elavl3 expression in the dorsal midbrain were calculated and shown in (g–i). Photographs are at the same scale in each row and scale bars are only shown in the left panels. Experiments were repeated three times. Data was analyzed by one-way ANOVA and presented as mean +/−s.e.m. *P < 0.05, **P < 0.005, ***P < 0.0005. The numbers of embryos used are shown at the top or bottom of each bar.
Knockdown of stmn4 causes precocious neuronal differentiation in dorsal midbrain
To understand whether Stmn4 is involved in maintenance of neural progenitor state in the dorsal midbrain, two antisense translation-blocking morpholino oligonucleotides (MOs) against stmn4 were designed to target start codon (tMO1) and 5′ untranslated region (UTR, tMO2) as indicated in Fig. 2a. We examined the efficiency of MO by co-injecting the pCS2+ XLT-GFP vector with an insertion of stmn4 fragment containing both MO target sites without or with designated dosages of MOs. The control group showed mosaic GFP fluorescence in most embryos that was in clear contrast to the notably reduced fluorescence in MO-injected embryos, which will be referred as morphants hereafter (Fig. 2b). The percentage of embryos expressing GFP was dose-dependently reduced in morphants of both MOs (Fig. 2g). Both tMO1 and tMO2 morphants were grossly normal throughout gastrulation (data not shown).
To address the regulation of neurogenesis by Stmn4, we used a transgenic Tg(HuC:Kaede) line which expressing green Kaede fluorescence in differentiated neurons18. At 16 hpf, Kaede-labeled differentiated neurons were mainly found in epiphysis and trigeminal ganglion neurons (TgSNs) but not in the dorsal midbrain (area between epiphysis and TgSN) of control embryos (Fig. 2c). In contrast, differentiated neurons appeared in dorsal midbrain in up to 80% of stmn4 morphants (Fig. 2d,h). Similar results were obtained by WISH analysis against elavl3 (Fig. 2e,f). The percentage of embryos with precocious elavl3 expression was MO dose-dependent in both stmn4 morphants (Fig. 2h). tMO2 appeared to be more effective than tMO1, so it was used for the rest of experiments.
To figure out whether the precocious elavl3 expression observed in stmn4 morphants was specifically due to the depletion of endogenous Stmn4, a plasmid containing the coding region sequence of stmn4 was co-injected with 5′ UTR targeting tMO2 into embryos and found significant (P < 0.05) but partial rescue of phenotypes. In addition, the injection of stmn4 plasmids had no significant effect on precocious elavl3 expression at the dosages tested (Fig. 2i). We also tested whether the effect of stmn4 MOs might be caused by non-specific activation of P5319. The stmn4 MO-induced expression of elavl3 was partially alleviated by co-injecting p53 MO but not significant (Fig. 2h). Taken together, these results indicated the depletion of Stmn4 could specifically induce the precocious elavl3 expression in dorsal midbrain during early development.
Targeted mutation of stmn4 causes mild precocious neuronal differentiation in dorsal midbrain and the increase in stmn1b expression
To further confirm the function of Stmn4 genetically, we generated stmn4−/− mutants in the transgenic Tg(HuC:Kaede) background using the Clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9-mediated approach20. We designed several guide RNAs (gRNAs) targeting different sites on the stmn4 gene. In brief, different gRNAs were separately co-injected with Cas9 mRNAs into 1-cell stage embryos. Survived embryos were raised to adulthood and then crossed to wildtype fish to obtain F1 embryos. The F1 embryos were raised to two months old and subjected to the Sanger sequencing analysis. Zebrafish fin genomic DNAs were used as templates for polymerase chain reaction (PCR) using a pair of primers (F1 and R1 as denoted in Fig. 3a). The PCR products were then sequenced to identify mutant founders. One of the gRNAs targeting the Exon 3 (sequence shown in Fig. 3a) effectively created deletions in 5 different mutant alleles (stmn4Δ3-12) of the stmn4 gene (Fig. 3b). The identified F1 founders were then in-crossed within each allele to acquire F2 generation. To identify homozygous mutants in F2 fish, we found a BfaI site (CTAG) within all the deletions and a second BfaI site at the beginning of Exon 5. The BfaI sites were thus useful for the rapid screening of mutant alleles by restriction digestion (Fig. 3a). We then raised the identified mutant founders to two month old, collected their fin genomic DNAs and subjected to restriction digestion assay to separate wildtype, heterozygous and homozygous stmn4 mutant fish (Fig. 3c). To perform the restriction digestion assay, zebrafish fin genomic DNAs were used as templates for PCR using a pair of primers (F2 and R2 as denoted in Fig. 3a) to give rise a 1235 bp PCR product. Taking the stmn4Δ5 allele as an example, the PCR product was cleaved by BfaI into three fragments (459, 405 and 371 bp) in samples from wildtype embryos. In clear contrast, two fragments (830 and 405 bp) were present in samples from homozygous mutants (−/−) due to the loss of first BfaI site. However, due to the presence of both wildtype and mutant PCR products, lower amounts of 459 and 371 bp fragments also existed in samples from heterozygous mutants (Fig. 3c). We also confirmed the validity of the restriction digestion by sequencing randomly selected 1235-bp PCR products (data not shown). Two of those mutant alleles containing 4 (stmn4Δ4) or 5 bp deletion (stmn4Δ5) near the protospacer adjacent motif (PAM) site caused premature stop codons and presumably should result in a truncated Stmn4 with 14 N-terminal and 28 or 36 missense (in red) amino acids, respectively, as shown in Fig. 3d. Both alleles lacked all Stmn4 functional domains and their F2 homozygous mutants were raised to adulthood. The F2 homozygous mutants of stmn4Δ4 and stmn4Δ5 alleles were in-crossed to produce maternal zygotic mutant embryos. Although mutants were generated in the transgenic Tg(HuC:Kaede) background, however the fluorescence intensity of Kaede was too low after repeated crossing (data not shown). Therefore, we used WISH against elavl3 to examine the premature neurogenesis in the dorsal midbrain. Unexpectedly, most of the mutant embryos from both alleles developed normally. Only 5 of 89 (6.7%) in stmn4Δ4 mutant embryos and 7 of 110 (7.8%) stmn4Δ5 mutant embryos showed premature elavl3 expression in dorsal midbrain (Fig. 3f). The lack of phenotype in most embryos from the stmn4 mutant alleles implied possible artifacts observed in the previous MO-based studies. To address this issue, we tested the response of embryos from both mutant alleles to the stmn4 tMO2 and found that the stmn4 tMO2-induced premature elavl3 expression in dorsal midbrain was notably and significantly lower in stmn4Δ4 and stmn4Δ5 mutant embryos, respectively, compared to that in wildtype embryos (Fig. 3f). This indicated that the observed premature elavl3 expression in dorsal midbrain was specifically induced by stmn4 tMO2. Furthermore, the F0 embryos-injected with the Exon 3 gRNAs revealed brain malformation and massive death similar to that observed in stmn4 morphants (data not shown). We also observed lower but notable precocious elavl3 expression in dorsal midbrain in 10 of 58 Exon 3 gRNAs/Cas9-treated embryos (Fig. 3e). These results suggested possible functional complementation by other stahmin isoforms or gene compensation21 that might mask the phenotypes resulted from the loss of stmn4. Thus, we examined the gene expression of other stathmin genes in stmn4Δ5−/− mutants at 12 and 18 hpf and found that the expression of stmn1b but not other stathmin genes was significantly (P < 0.05) and notably elevated at 12 and 18 hpf, respectively (Fig. 3g).
Targeted mutation of stmn4 increased stmn1b expression and caused mild precocious neuronal differentiation in dorsal midbrain.
(a) A partial genomic structure of stmn4 shows the Exon 3 targeting gRNA site and sequence (PAM site in red). The two BfaI sites (arrowheads) and the sites complementary to the forward (F) and the reverse (R) primers (arrows) used for cloning for sequencing and restriction digestion assay are shown. (b) The sequence flanking the deletion site stmn4 mutants (stmn4Δ3-12) are shown. The deleted nucleotides are denoted by dashes. The cut sites of Bfa1 are indicated by arrowheads. (c) Restriction digestion by BfaI. The genomic DNAs from tail fins of wild type (+/+), heterozygous (+/−) or homozygous (−/−) stmn4Δ5 were isolated to be used as templates for PCR using the forward and reverse primers indicated in (a). The amplicons were digested and run in agarose gels and stained. A representative gel image is shown. The sizes in bp for selected DNA marker bands and digested fragments are shown. (d) Two stmn4 mutant alleles (stmn4Δ4 and stmn4Δ5) have a premature stop codons that resulted in truncated Stmn4 proteins as shown lacking all known functional domains. (e) Partial activation of precocious elavl3 expression in dorsal midbrain of F0 Tg(Huc:Kaede) embryos by stmn4 CRISPR/Cas9 compared to that of control embryos (Ctrl). (f) The % of premature elavl3 expression was determined in wildtype (C), stmn4Δ4 (Δ4) and stmn4Δ5 (Δ5) embryos-injected with indicated concentration of stmn4 tMO2. (g) Relative expression of stmn1b, stmn2a and stmn3 were measured in wildtype (C), morphant (M), stmn4Δ4 (Δ4) and stmn4Δ5 (Δ5) embryos at 12 and 18 hpf by qPCR. Data was analyzed by one-way ANOVA and presented as mean +/− s.e.m. *P < 0.05, **P < 0.005, ***P < 0.0005.
Stmn4 homeostasis is essential for neural progenitor cell maintenance in dorsal midbrain
To further confirm the stmn4 MO-induced precocious elavl3 expression in dorsal midbrain, we utilized shRNA targeting the 3′ UTR of stmn4 (Fig. 2a) by employing the UAS-miR-30 backbone in combination with the Gal4-UAS system. We used two vectors for the Gal4-UAS induction system. One vector was constructed with a heat shock protein 70 (hsp70) promotor and a Gal4 gene22. Another vector contains 5X UAS elements, a tdTomato and a stmn4 shRNA which is situated at the 3′ UTR of tdTomato (modified from a previously reported vector23). We injected both vectors into 1-cell embryos, cultured to the bud stage and then heat shocked for 2 h to avoid the possible interference it may cause during early embryogenesis (Fig. 4b). The hsp70 promoter can be activated by heat shock to drive gal4 expression. Gal4 protein can then bind to the 5XUAS elements to trigger the expression of tdTomato and stmn4 shRNA. The stmn4 shRNA of 21 nucleotides was designed to target nucleotide 614 at the 3′ UTR of stmn4 (Fig. 2a). The expression of shRNAstmn4_614 effectively reduced the expression of stmn4 to 0.25% in 1-day-old embryos compared to that in control embryos by quantitative PCR (qPCR) analysis. All embryos were fixed at 16 hpf and subjected to immunohistochemistry probed with anti-Kaede and mCherry antibodies. We used the anti-mCherry antibody because it can cross recognize tdTomato. The dorsal mid brain area was depicted within the region between the epiphysis (arrow) and the TgSNs (green dashed circles) as shown in the first row of Fig. 4c. We examined the dorsal mid brain area under confocal microscopy to screen 10 slices (25 μm thick in total) and to see whether the expression of shRNAstmn4_614 (as indicated by the expression of tdTomato in red) can induce the precocious elavl3 expression (as indicated by the expression of Kaede in green). In three trials, we observed the stacked images from all slices and found 63.2% of 37 embryos exhibited precocious elavl3 expression in dorsal midbrain that confirmed the phenotypes of stmn4 morphants. We further counted fluorescent cells within dorsal midbrain. However, due to the respective background expression of tdTomato and Kaede, only those cells with respective fluorescence above a certain threshold was counted as positive cells (denoted with “*”). We observed an average of 10.8 tdTomato- and 4 Elavl3-positive cells in each vehicle (the same construct without stathmin) control embryos. In contrast, an average of 11.6 tdTomato- and 11.4 Elavl3-positive cells were found in each shRNAstmn4_614-treated embryos. Among those fluorescent cells, an average of 5.8 cells expressed both signals (Fig. 4c). These results clearly demonstrated at the cellular level that the loss of Stmn4 induces precocious neural differentiation.
Overexpression of CA-Stmn4 or depletion of stmn4 by shRNA induced elavl3 precocious expression in dorsal midbrain.
(a) Conditional gene expression system was introduced by combining Gal4-UAS system with heat shock 70 (hsp70) promoter. After heat shock, upstream activation sequence (UAS) elements will be recognized by Gal4 protein dimers (indicated by light blue) and then activate transcription of downstream genes including nuclear H2A-mCherry in red and Stmn4 intervening with a viral 2A peptide. (b) Flow chart showing the conditional expression system. We heat-shocked the Tg(HuC:Kaede) embryos injected with both plasmids described above until bud stage. After two hours of induction, embryos were analyzed until 16 hpf for elavl3 expression revealed by Kaede (green). (c) Dorsal view of midbrains between epiphysis (indicated by white arrows) and (TgSNs, circled by green dashed lines) are shown. Within one embryo, average number of precocious elavl3 (demonstrated by Kaede, green) expressing cells colocalized with (yellow) or without overexpressions of stmn4 either with mutations or its corresponding shRNAstmn4_614 (indicated by mCherry, red) in dorsal midbrain, are shown on the left-bottom corner of each panel. Scale bars are 50 μm.
On the other hand, we also performed gain of function assay of Stmn4 by overexpressing wildtype stmn4 or ca-stmn4 to test their effects on precocious elavl3 expression in dorsal midbrain. We employed a similar strategy by using the same hsp70-driven Gal4 vector and a vector with 5X UAS, H2A-mCherry, 2A peptide and wildtype Stmn4 or CA-Stmn4 for conditional expression (Fig. 4a). The 2A peptide was used to separate mCherry and Stmn424. Following similar protocols as used in the stmn4 shRNA experiments (Fig. 4b), we observed an average of (1) 8.67 mCherry- (white asterisks), 11 Elavl3-positive (white asterisks) and 5.33 double-positive cells (yellow asterisks) were found in each stmn4-treated embryos. (2) 14.5 mCherry-, 13 Elavl3-positive and 8 double-positive cells were found in each ca stmn4-treated embryos (Fig. 4c). In three trials, we observed from the stacked images that 54.8% of 36 stmn4- and 68.9% of 32 ca-stmn4-overexppressing embryos exhibited precocious elavl3 expression in dorsal midbrain. Collectively, these results demonstrated that either the loss or gain of Stmn4 can induce precocious neural differentiation and suggested an essential role of Stmn4 homeostasis in the maintenance of neuronal progenitor cells in the dorsal midbrain of zebrafish embryos.
Stmn4-mediated precocious neuronal differentiation occurs during the neural keel stage
To further investigate the progression of precocious neuronal differentiation in ca-stmn4-overexpressing cells, we conducted time-lapse recordings shortly after the heat shock at the 6-somite stage to 16 hpf in the dorsal midbrain of developing zebrafish embryos (Supplementary Videos 1 and 2). Snapshots of selected frames as designated are presented in Fig. 5c–l. In addition to two vectors used, the Medusa vector M2 containing doublecortin-GFP (DCX-GFP) was introduced to monitor the microtubule dynamics25. At the beginning, neural progenitor cells, stochastically highlighted by nucleus in red and microtubules in green (thread-like structure), were interdigitated with each other between the boundaries of neural tissues (Fig. 5c,h; dashed lines), forming a pseudostratified layer, which is called the “neural keel” stage (Fig. 5b; middle). Those cells oscillated between the boundaries and migrated medially (convergence) with final C-divisions (cross-midline division)26 giving the mirror-symmetric daughter cells and to form the midline as the key feature of the “neural rod” stage (Fig. 5b; bottom). At 16 hpf, few mature neurons (arrows, showing strong Elavl3 green fluorescence in cytosol) emerged in wildtype embryos (Supplementary Video 1; Fig. 5c–g; arrows). In contrast, differentiated neurons (arrows) appeared promptly at 14 hpf (neural keel stage) after heat shock in the embryos-injected with the ca-stmn4 plasmids (Supplementary Video 2; Fig. 5h–l). Typically, neurogenesis develops through asymmetric division which generates one daughter neuronal cell and the other remains as a progenitor cell. However, the vast majority of neuronal differentiation arose before cell division (71.4%), which suggested that the disturbance of Stmn4 homeostasis induces neuronal differentiation via an asymmetric division-independent mechanism. Besides, some progenitor cells underwent morphological changes with fragmented microtubules and eventually were apoptotic regardless of precocious elavl3 expression (Supplementary Video 2; Fig. 5h–l; arrowheads). It thus implies that overdose of CA-Stmn4 could disrupt cell viability through interruption of microtubule dynamics. Taken together, both precocious neurogenesis and apoptosis occurred during the neural keel stage in those ca-stmn4-overexpressing embryos.
Overexpression of CA-Stmn4 induced precocious expression of elavl3 in neuronal precursor cells during neural keel stage.
Time-lapse movies were conducted 6 minute per frame around the neural keel stage (13 hpf) after heat shock (6-somite stage). During neurulation (b), neural progenitor cells converge and migrate to the dorsal midline to establish neural rod (16 hpf) from a flattened neural plate (bud stage). Here, microtubules are revealed by doublecortin-GFP fusion protein (DCX-GFP, green, indicates thread-like microtubules structure depicting the cell shape). HuC:Kaede (green) expresses within cytoplasm. Compared to control embryo (c–g), more cells with elavl3 expression (arrows) and more apoptotic cells (arrowheads) appeared in embryo overexpressing CA-Stmn4 (mCherry, red) during the neural keel stage (h–l), characterized by a disorganized and thickening tissue (b). Corresponding time points are shown on the upper-right corner. Boundaries of neural tissues are disclosed by a pair of dash lines. All snapshots shown in dorsal side and anterior to the left. Scale bars equal 25 μm for all.
Knockdown of stmn4 prolongs G2 phase to trigger precocious neuronal differentiation in dorsal midbrain
To understand the mechanism underlying the early post-mitotic neuron cells appearance in stmn4 morphants, we investigated whether the deficiency of Stmn4 may alter cell cycle which is tightly associated with neuronal differentiation. We analyzed the length of G2 phase by using the percentage of labeled mitosis (PLM) method27,28. In brief, embryos were treated with short BrdU pulses at 16 hpf and then fixed sequentially at 1 to 3 h after treatments (Fig. 6a). Fixed embryos were subjected to immunohistochemistry probed with anti-BrdU and anti-phospho-Histone H3 (pH3) antibodies to reveal cells in S phase (green) and M phase (red). Then, the length of G2 phase was measured by the duration between the S and M phases (Fig. 6b,c). We noted that cell proliferation was obviously reduced in stmn4 morphants (n = 8.4 with 5 ng MO; n = 7.3 with 10 ng MO) compared to that in wildtype embryos (n = 12.6) by counting the number of pH3-positive cells within the dorsal midbrain. The percentages of pH3/BrdU positive cells were notably lower in stmn4 morphants compared to that in wildtype embryos. Extrapolating from the curves, we found that the average length of G2 phase (50% of BrdU positive cells with pH3 staining) was increased dose-dependently from 1 h and 29 min to almost 2 h. (Fig. 6b). The cell cycle is carefully administered by cyclins and cyclin-dependent kinases (CDKs). Among them, Cyclin B forms a complex with Cdk1 to guard the G2/M checkpoint through the regulatory phosphorylation cascade29. We found that precocious neuronal differentiation could be rescued by overexpressing cdc25a, an upstream regulatory element of CyclinB/Cdk1, in stmn4 morphants (Fig. 6c). It suggested that the control of G2/M transition by Stmn4 is key to its maintenance of neuronal progenitor cells.
Deficiency of stmn4 prolonged the G2 phase in a Cdc25a-dependent manner.
(a) 1-cell stage embryos were injected without (Ctrl) or with 5 and 10 ng stmn4 tMO2. A 15-min BrdU pulse (green arrowhead) was then given to embryos at 16 hpf. Part of embryos were fixed at an hour intervals from 17–19 hpf and subjected to double immunochemistry against α-BrdU (green) and α-pH3 (red). Fixed embryos were examined and photographed under confocal microscopy. Representative photos are presented for each treatment at designated times post the BrdU pulse. All images are shown in dorsal views with anterior to the top. Percentages of pH3+ and BrdU+ cells (yellow cells) within the pH3+ cells (yellow and red cells) were calculated as shown in (b) and analyzed by one-way ANOVA. (c) 1-cell stage Tg(Huc:Kaede) embryos were injected without or with 5 ng stmn4 tMO2 and indicated amounts of cdc25a plasmid. The percentages of embryos with premature elavl3 expression were examined and presented as described in Fig. 2.
Inhibition of Wnt signaling downregulates the expression of stmn4
Canonical Wnt signaling is one of the key signal to mediate midbrain development by regulating proliferation and morphogenesis with a bi-modal control upon FGF activity7,8. To understand whether Wnt morphogens may regulate stmn4, we suppressed Wnt activity by applying transgenic fish Tg(hsp70l:dkk1-GFP)w32 to overexpress dkk1, a Wnt signaling inhibitor, and to examine the expression of lef1 (a Wnt-activated downstream gene) and stmn4 (Fig. 7a). WISH analysis revealed that the expression of lef1 was reduced by dkk1 expression induced by heat shock compared to that of control embryos. It demonstrated the effective inhibition by Wnt signaling. More importantly, we observed the notable reduction of stmn4 expression in dorsal midbrain (Fig. 7a). This Wnt-dependent stmn4 expression was also shown by qPCR analysis in RNA extract of whole embryos (Fig. 7b). This indicated the Wnt signaling may regulate stmn4 expression at the transcriptional level.
Block of Wnt signaling inhibited the expression of stmn4.
Tg(hsp70l:dkk1-GFP) embryos were cultured to 10 hpf and then heat-shocked (HS) for 1 and 2 hr as indicated. Embryos were further cultured, fixed at 18 hpf and then subjected to WISH against stmn4 or lef1 (a) or qPCR analysis against stmn4 using actin as an internal control (b). Data represents mean +/− s.e.m. and analyzed by one-way ANOVA (N = 5), *P < 0.05.
Discussion
Balanced transitions between proliferation and differentiation are keys to proper patterning during brain development. Their regulations within compartments are spatial and temporal-dependent that is even more complicated and still remains largely unknown. The brain isthmus in between midbrain and hindbrain is an organizer directing the growth and differentiation and is an excellent platform to study this unresolved issue. In this study, we demonstrate a novel role of a microtubule destabilizer Stmn4 in maintaining neural progenitor cell pools in dorsal midbrain during early brain development. We are the first to discover the adequate expression of stmn4 is critical to prohibit the G2 phase of neural progenitors from lengthening that may cause early cell cycle exit and neurogenesis in the developing dorsal midbrain.
Stmn4 is less well studied comparing to other mammalian stathmins. Here, we first fully characterized the zebrafish Stmn4 owing features of tubulin sequestering and Golgi anchoring. However, since the cooperation of two subdomains within domain A of N-terminal extension controls the shuttling between Golgi apparatus and mitochondria13, whether palmitoylation-deficient Stmn4 could be recruited to mitochondria needs further investigation.
To study the in vivo function of Stmn4 in zebrafish, we demonstrated a possible role of Stmn4 in maintaining the neuronal progenitor pools by the perturbation of stmn4 expression or activity resulted in precocious neural differentiation. However, we also observed inconsistent phenotypes between morphants and mutants as reported by other studies21,30. It implies possible off-targeting effects of stmn4 MOs used. We revealed that the stmn4 tMO2 was less effective in triggering precocious neural differentiation in stmn4 maternal zygotic mutants that suggests the specific effects of stmn4 MOs21. In addition, CRISPR/Cas9 also caused similar phenotypes in F0 embryos. Although at a lower penetrance, it could be attributed to the mosaic expression frequently observed by the transient expression in F0 embryos. Collectively, these results suggest possible cellular compensations due to the loss of Stmn4. Other stathmin family isoforms might be functional complementary to Stmn4. The loss of Stmn4 might also be compensated by other molecules sharing similar properties or acting downstream of mutated factors21 that is a common features in lower vertebrates31. We addressed these issues by demonstrating the upregulation of stmn1b in stmn4Δ5−/− embryos. In addition, we also found that injection of stmn1b MO into stmn4Δ 5−/− embryos could partially induce precocious expression of elavl3 in dorsal midbrain (data not shown). It implicates that Stmn1b might complement the loss of Stmn4 to prevent premature neuronal differentiation in dorsal midbrain. Thus, double-knockout zebrafish of stmn4 and stmn1b are required for further studies. However, we still cannot rule out the possible involvements of other compensatory factors in regulating phenotypes of stmn4 mutants.
It is difficult to resolve the controversy for the inconsistent phenotypes between morphants and targeted mutants. So one needs to be cautious not to over interpreting data form each side. Although the knockout approach appears to be a more clear-cut gene inactivation, but it is often complicated by gene complementation and compensation as discussed previously. The knockdown approaches although still have residual gene activity and presumably should have weaker impact comparing to the knockout approach, but cellular compensation is less likely to be an issue and it still allows to see the phenotypes due to gene deficiency. However, the nonspecific secondary effects of MOs should be of concern, so we have applied different controls as shown in Fig. 2 to exclude the possible MO artifacts. In conjunction with the shRNA and constitutively activated Stmn4 studies that both showed the induction of premature neuronal differentiation at the cellular level, it suggests that Stmn4 is a critical regulator to keep the cell in the dorsal midbrain in a progenitor status for regional specification during early brain development in zebrafish.
Stathmin as a negative regulator for neurogenesis appears to be contradictory to previous findings that stathmins are positive regulators for neuronal differentiation. SCG10 is a neuronal differentiation marker for nerve growth factor (NGF)-induced cell differentiation in phenochromocytoma cells (PC12 cells)32. The expressions of SCLIP and RB3 are also notably increased33,34. STMN4 is expressed consistently at a high level during differentiation in multiple neuroblastoma cell lines35. In addition, proper exogenous expressions of stathmins promote not only neurite extension in rat hippocampal primary cultured neuron cells36,37 but also dendritic arborization in purkinje cells38. In spite of all described evidences disclosing the vital role of stathmins in neuron differentiation, we still assert that Stmn4 has a previously undescribed role as a neurogenesis blocker in the developing zebrafish dorsal midbrain. The discrepancy may be due to the facts that mainly adult neurons or somatic cell lines were used in previous studies. Those cells are derived from terminally differentiated cells. In contrast, we used a developing brain in an in vivo context. In addition, what we based on is the expression of earliest neuron differentiation marker Elavl3 which represents a much earlier signal comparing to the previous morphological observation in neurite formation in culture39. So what we found may represent a regulatory signal leading to the initiation of neurogenesis in dorsal midbrain.
To investigate how Stmn4 regulate the initiation of neurogenesis, we observed that proliferating cells labeled by BrdU or pH3 signals diminish significantly in stmn4-deficient embryos. Indeed, a strong correlation between stathmin expression and cellular proliferation had already been revealed in previous reports. First, high correlation between these two was confirmed in both normal and malignant cells40,41. Next, both overexpression and inhibition of stathmin expression give rise to mitotic arrest42. This is not surprising since mitotic spindles which are essential for chromosome aggregation and cell division should be carefully mediated by microtubule associated proteins such as stathmins. Since stathmin is crucial for mitosis progression, the lengthening of G2 phase in stmn4-deficient embryos observed in this study might be due to the difficulty of forming mitotic spindle for entering mitosis. Interestingly, we further observed that the stmn4 expression level in neuron progenitor cells should be properly controlled within a narrow window that less or higher expression of stmn4 both resulted in premature differentiation in dorsal midbrain. Similarly, neurite formation was also reported to be inhibited by knocking down SCG10 or SCLIP and enhanced by overexpressing a proper but not overdose of SCG10 or SCLIP in rat hippocampal primary cultured neuron cells36,37. For this dynamic instability, the abrupt switching behavior between microtubule growth and shrinkage, may provide some clues. Silva and Cassimeris proposed that stathmin may stand as a gatekeeper at the G2/M checkpoint via regulation of Plk1 and Aurora A activities which act upstream of Cdc25a phosphatase to facilitate cell cycle progression by withdrawing inhibitory phosphrylations from CDK1 partially through microtubule-dependent mechanisms in Hela cells43. This is in accord with our findings that Cdc25a could rescue the precocious differentiation phenotypes in stmn4 morphants. Interestingly, normal Plk1 activity requires stathmin expression within certain limits as well. It suggests that the homeostasis of Stmn4 within neuronal progenitor cells should be strictly controlled for their maintenance of cell cycle progression.
Boekhoorn et al.14 proposed that stathmin is involved in the maintenance of neural precursor cell types in the adult hippocampus14. In stathmin-knockout mice, the impaired cell proliferation leads to reduced number of nestin-positive or PSA-NCAM-positive proliferating cells accompanying with the increasing DCX-labeled differentiated neuron cells. Although further detailed characterization of neuronal lineages within dorsal midbrain still needs to be done, this strongly supports to our study that Stmn4 controls the neuronal cell fate through mediating Cdc25a particularly during development. The “Cell cycle length hypothesis” has been proposed and established to provide an explanation that cell cycle regulators could elicit effect on cell fate determination factors inversely10. Inhibitors of cdk/cyclin complex such as p21 or p27 are precisely regulated to lengthen G1 phase during development44 that allows proneural genes like neurogenin2 (Ngn2) to be transcribed and accumulated above thresholds to promote differentiation45. Furthermore, the lengthening of G1 phase not only avoids two compatible events regarding mitosis and morphogenesis46, but permit changes of epigenetic marks and nuclear architecture to determine cell fates47,48. While many researches were emphasized on the regulation of G1 phase, a few studies have begun to shed light on the G2 phase regulation lately49. In chordates, a longer G2 phase, which was introduced into epidermal cells flanking the neural plate, was found to be necessary for neural tube closure50. Restricted cdc25a expression was also proved to facilitate extension of body length and differentiation of muscle lineage cells in zebrafish51. Taken together, our results revealed that a short G2 phase, maintained by a proper level of Stmn4 expression in dorsal midbrain to guarantee active mitosis in neural progenitor cell pools. Despite that further investigation on cell cycle regulation (e.g., prolonged G2 phase may contribute to shorten G1 phase inversely27) still needs to be accomplished. We have offered strong support to the determination of cell fate via G2/M transition in addition to the well-studied G0/G1 regulation.
Several signaling pathways have been well investigated to play vital roles in midbrain development. Among this, the isthmus, which secretes morphogens such as Wnts and Fgfs, elicits effects on proliferation and neuron specification majorly through regulating Her5 activity9. A previous model stated that Wnt has a bi-modal role in activating FGF activity or impeding FGF activity via the induction of Sprouty genes which are known FGF inhibitors8. Combinations of positive- and negative-feedbacks loops ignite a retrograde FGF regulation on Her5 which preserves the isthmus as a neuron-free region. Our findings showing negative regulation of stmn4 expression upon inhibition of Wnt activity further supports that Stmn4 can be an alternative mitogenic factor downstream of the Wnt signaling pathway. Despite of the contradictory facts that merely STMN2 is a novel target of β-catenin/TCF signaling in hepatoma cells52, we found several potential TCF-bindings sites within the stmn4 promoter (data not shown) also implies that Wnts directly regulate stmn4 at the transcriptional level.
In sum, we propose a model as shown in Fig. 8 that during the neural keel stage, Wnts could regulate the expression of stmn4 in dorsal midbrain to control the length of G2 phase possibly via the coordination of G2/M checkpoint machinery. Deficiency or excess of Stmn4 would both lead to the retention of G2 phase that may allow accumulation of proneural gene transcripts to direct premature neuronal differentiation. Interestingly, zic2a and zic5, the proven targets of Wnt signaling pathway, display similar expression pattern in dorsal midbrain as stmn4 and regulate not only mitosis but also dorsolateral hinge-point (DLHP) formation53. Moreover, the late onset of proliferation defect (16–18 ss; neural rod stage) shown in the zic morphants indicates Stmn4 may compensate the loss of Zic proteins for assisting G2 phase during neural keel stage or they have complementary mitogenic role between neural and neural rod/neural tube stage. Last, while most cell cycle regulators participating in midbrain development situate in the G1 phase, e.g. p27Xic1 regulated by Her5 through FGF activity could inhibit cyclinD/Cdk complex or Wnt could directly regulate cyclin D1 and n-myc on transcription level, alternatively Stmn4 and Zic proteins (regulation of cyclin B1) influence the cell cycle progression via regulating the G2 phase. Regarding the rising evidences introducing G2 length control on cell fate determination, it will be fascinating to study how the regulation of G1 and/or G2 phases will cooperate spatially or temporally within the same platform, in particular midbrain development.
A proposed model describing interaction between proliferation and differentiation through Wnt-Stmn4 regulation during early neurogenesis.
At the neural keel stage, reciprocal interactions between Wnts (blue) and Fgfs (red) gradients underline the neuron-free intervening zone to trigger the expression of her5. Her5 then halts neurogenesis via inhibiting proneural genes and down-regulating p27 and p57, which hinder the progression of G1 phase through inactivating the Cdk/CyclinD complex. In addition, here we further propose that the Wnt signaling could keep the dorsal midbrain in a progenitor cell status via the induction of stmn4 expression to mediate the progression of G2 phase by activating Cdc25a. In contrast, the stmn4-deficient embryos have a lower Cdc25a activity that leads to a longer G2 phase and results in precocious neuronal differentiation (green) in the dorsal midbrain.
Methods
Ethics Statement
All experimental procedures on zebrafish were approved by the use of laboratory animal committee at National Taiwan University, Taipei, Taiwan (IACUC Approval ID: 103 Animal Use document No. 102) and carried out in accordance with the approved guidelines.
Zebrafish
Wild type AB zebrafish (Danio rerio) (from Taiwan Zebrafish Core Facility at National Health Research Center, Taiwan), Tg(HuC:Kaede) fish (from Dr. Hitoshi Okamoto) and Tg(hsp70l:dkk1-GFP)w32 fish (from Taiwan Zebrafish Core Facility at Academia Sinica, Taiwan) were maintained at 28.5 °C on a 14-h light/10-h dark cycle. Embryos collected from natural mating were cultured and staged according to Kimmel et al.54.
Plasmid constructions and Site-directed mutagenesis
Full-length coding sequences of stmn4 was cloned from zebrafish cDNAs into the pCS2+ vector with BamHI (New England Biolabs (NEB), Ipswich, Massachusetts) and XbaI (NEB) for ectopic expression. DNA fragment (~300 bp) containing both MO binding sites was inserted in-frame with XLT-GFP in pCS2+ vector for MO efficiency check. Mutagenic primer pairs designed with desired mutations were used to generate the mutated plasmids by extending the original vectors with incorporating mutations, described as QuikChange™ Site-Directed Mutagenesis Kit protocol. We established three mutations in the serine sites of the SLD, including S59A, S71A and S81A, and also one combined mutations residing in the N’-terminal region, C20, 22A. For visualization, mCherry without stop codon were subcloned in frame with the stmn4 gene in the pCS2+ vector with BamHI. For tagging Stmn4 directly, we cloned previous stmn4 with combinatorial mutations of serine residues into pET23a (Novagen, Merck Milipore) with BamHI and XhoI (NEB), possessing 6X His on C’ terminal end. The two component induction system was first generated by adding novel viral 2A peptide sequence, from rhinitis A virus (PTV1); referred to P2A, residing within primers designed for the insertion of stmn4 with 6X His tag into pT2MUASMCS22 with SalI (NEB) and EcoRI (NEB). The mCherry fused to the C-terminus of zebrafish histone variant H2A.F/Z from pME-H2AmCherry55 was further inserted into the N-terminal end of P2A with BglII (NEB) and SalI. The final constructs serves a convenient tool for conditional induction system when combining with Gal4 modules, with two translation-independent components which is easy to modify with several restriction sites to be selected. The shRNAstmn4_614 oligo (AAGACGTGAATGGTCACTTAC), designed against stmn4 3′ UTR by using the siRNA design tool available on GeneScript website, was generated through a two-step PCR with two overlapping primers forming BbsI restriction sites on both ends for cloning into miR30-based vector (SG1164) with CMV promoter (kind gift from Dr. Guo Su). This shRNA oligo could further shifted into UAS-based vectors (SG1180) with AccIII (NEB) and SnaBI23 (NEB).
Cell transfection and co-localization analysis
Plasmids at 0.5 μg were used for Hela cell transfection. To label Golgi apparatus, we co-transfected cells with pEYFP-Golgi (Clonetech, TAKARA BIO Inc., Shiga, Japan). Images were collected using an LSM 780 confocal laser-scanning microscope (Carl Zeiss, Oberkochen, Germany). Co-localization was analyzed for each individual cell and quantified by the Fiji (formerly ImageJ) software with Pearson’s coefficient56.
Embryo microinjections
Antisense morpholinos designed against stmn4 (tMO1: GTCTCGATATGCTGCCAAGGTCATG; tMO2: AGTTAAGAAGTCTTCACTCTGTCCT), p53 (tMO: 5′-GCGCCATTGCTTTGCAAGAATTG-3′) and random control MO (Random MO: NNNNNNNNNNNNNNNNNNNNNNNNN) were synthesized with a random based mixture at every position by Gene Tools. Plasmids for injection were prepared using the QIAGEN Plasmid Mini Kit (QIAGEN). Except for the corresponding dosages describing in the text, the doses we used in the conditional expression system were both 25 pg of pT2KhspGFF22 and pT2MUAS_H2AmCherry-2A-STMN4_3A or SG1180_STMN4 shRNA_614. All reagents were injected at the 1–2 cell stage.
Generation of stmn4 knockout lines by CRISPR/Cas9 system
Three guide RNAs (gRNAs, see sequences at Supplementary Table S1) were designed by the CHOPCHOP57 to target stmn4 gene. Preparation of gRNAs includes annealing of two oligonucleotides, fill-in of single strand DNA overhangs with T4 DNA polymerase (NEB) and in vitro transcription by Megascript kits (Thermo Fisher Scientific, Waltham, Massachusetts)58. Cas9 mRNA was transcribed from the linearized pCS2-Cas9 (a kind gift from Dr. Alex Schier) using SP6 mMessage mMchaine kits (Ambion). Approximately 300 pg Cas9 mRNA and 25 pg gRNA were co-injected into 1- to 2- cell stage wildtype embryos and raised up to adults (F0). To identify carrier fish with sense mutations, F0 fish were crossed with wildtype fish to obtain F1 offspring. Genomic DNAs of founder fish were acquired by incubation of tail-fin clip with lysis buffer (proteinase K (0.4 mg/ml) diluted in TE buffer) at 55 °C for 2 h, inactivated at 85 °C for 15 min and then purified to be PCR templates. PCR products amplified by F1 and R1 primer pairs (see sequences at Supplementary Table S2) were cloned by TA cloning and subjected to Sanger sequencing to identify potential alleles. At least five batches of offspring embryos from different founders were then raised up to adulthood and identified for allele characterization by sequencing. Heterozygous F1 adults with same allele were inbreed to obtain the F2 generation, which were further genotyped with digesting purified PCR fragments amplified by F2 and R2 primer pairs (see sequences at Supplementary Table S2) from fin-clip genomic DNAs by BfaI restriction enzyme. Maternal zygotic mutant embryos (F3) from in-cross of homozygous mutant fish (F2) were utilized for further experiments.
Whole-mount in situ hybridization and immunohistochemistry
DNA fragments of lef1 were cloned from zebrafish cDNAs by RT-PCR and subcloned into pGEMT-easy vectors for probe synthesis. elavl3 was obtained from Chin-Hwa Hu (National Taiwan Ocean University, Taiwan). stmn4 was used previously by Shih et al.15. Whole-mount in situ hybridization (WISH) was performed as described using digoxigenin (DIG)-labeled antisense RNA probe59. Double in situ hybridization with modifications60 was carried out utilizing Fluorescein (FITC) labeled elavl3 probe developed with the chromogenic substrate FastRed (Roche Applied Science) and DIG-labeled stmn4 probe developed with NBT/BCIP (Promega). Stained embryos were mounted in glycerol, observed under a Leica S8AP0 stereomicroscope (Leica Microsystems) and photographed using a Canon 7D DSLR camera (Canon). Whole-mount immunohistochemistry staining was conducted as previously described61 by using either Rabbit Kaede antibody (PM012M, MBL International corporation, 1:500), Rabbit 6X His antibody (GTX115045, Genetex, 1:500), mouse alpha tubulin antibody (T 9026, Sigma-Aldrich, Merck, 1:300) or rat mCherry antibody (M11217, Molecular probes, Thermo Fisher Scientific, 1:300). Goat Secondary antibodies were combined with primary antibodies respectively with goat anti-mouse or anti-rabbit IgG conjugated with Alexa Fluor 488 or Alexa Fluor 568 (Molecular probes, 1:500). Images were collected utilizing LSM 780 confocal laser-scanning microscope (Carl Zeiss).
Cell proliferation and G2 phase length analyses
Cell proliferation and G2 phase length were analyzed by specific experimental design described previously (Fig. 6a). Embryos were first treated with 10 mM 5-bromo-2′-deoxyuridine (BrdU, Sigma-Aldrich) pulses as described62 at 16 hpf and then fixed with fresh 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) every hour until 19 hpf. Double IHC was performed with mouse BrdU antibody (B2531, Sigma-Aldrich, 1:250) and rabbit P-H3 antibody (GTX128116, Genetex, 1:1000) to mark cells in S phase and in M phase with corresponding secondary antibodies (1:500). Images were collected utilizing LSM 780 confocal laser-scanning microscope (Carl Zeiss) with 20X lens scanning the dorsal midbrain. Every individual embryo was scanned reaching 25 μm deep from top (10 stacks, 2.5 μm per stack) and shown with represented z stack. Furthermore, G2 phase length was determined by the percentage of labeled mitosis (PLM) paradigm28 which reveal the progression of the BrdU and P-H3 co-labeled nuclei after BrdU incorporation. Due to its proportionality to G2 length, it is simple to predict the duration of G2 phase by 50% of co-labeled nuclei in average.
Confocal microscopy imaging, time-lapse recording and data analysis
Except for the MO efficiency check, all the other fluorescence photos were taken by LSM 780 confocal laser-scanning microscope (Carl Zeiss). For general scanning of the dorsal midbrain, stacks composed of 10 layers with 2.5 μm for each were performed. For detailed investigation of microtubule dynamics (Fig. S3b–d), thinner fraction of slices (1 μm) were scanned. All the collected data were further processed by the Fiji software56. Plugins “Skeletonized” and “AnalyzeSkeleton” were utilized to transform and analyze the 3D structure of microtubule patterns generated by confocal microscopy63. From the diversified raw data, polymerized microtubules and depolymerized microtubules were defined by the amount of slab voxels with more than 100 voxels giving rise to polymerized ones and less than 10 voxels to be depolymerized ones. For time-lapse recording, all the data were scanned using FV10i-DOC confocal laser-scanning microscopy (Olympus). Every frame photos were stacked images of 10 layers with 2.5 μm for each and taken every 6 minutes until 16 hpf.
Quantitative real-time PCR
RNA extracts from 18 hpf Tg(hsp70l:dkk1-GFP)w32 embryos or 1 dpf wildtype embryos co-injected with shRNAstmn4_614 and pT2KhspGFF were extracted by RNAzol (Molecular Research Center) and synthesized by M-MLV reverse transcriptase (Promega) with oligo-dT. Gene-specific primers were designed by Vector NTI (Invitrogen, Thermo Fisher Scientific). All the qPCR data were collected by Bio-Rad iQ5 (Bio-Rad) using QuatiFastSYBR Green PCR Master Mix (Qiagen).
Statistical analysis
All experimental values are presented as mean ± standard error and were analyzed by one-way ANOVA. The number in bottom or above the bar indicates the total sample number in one experimental condition. Groups denoted with different lettering refer to statistical significance (p < 0.05).
Additional Information
How to cite this article: Lin, M.-J. and Lee, S.-J. Stathmin-like 4 is critical for the maintenance of neural progenitor cells in dorsal midbrain of zebrafish larvae. Sci. Rep. 6, 36188; doi: 10.1038/srep36188 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Bally-Cuif, L., Goridis, C. & Santoni, M.-J. The mouse NCAM gene displays a biphasic expression pattern during neural tube development. Development 117, 543–552 (1993).
Palmgren, A. Embryological and morphological studies on the mid-brain and cerebellum of vertebrates. Acta Zoologica 2, 1–94 (1921).
Martinez, S. The isthmic organizer and brain regionalization. International Journal of Developmental Biology 45, 367–372 (2001).
Rhinn, M. & Brand, M. The midbrain–hindbrain boundary organizer. Current opinion in neurobiology 11, 34–42 (2001).
Chi, C. L., Martinez, S., Wurst, W. & Martin, G. R. The isthmic organizer signal FGF8 is required for cell survival in the prospective midbrain and cerebellum. Development 130, 2633–2644 (2003).
Lee, S., Danielian, P. S., Fritzsch, B. & McMahon, A. P. Evidence that FGF8 signalling from the midbrain-hindbrain junction regulates growth and polarity in the developing midbrain. Development 124, 959–969 (1997).
Panhuysen, M. et al. Effects of Wnt1 signaling on proliferation in the developing mid-/hindbrain region. Molecular and Cellular Neuroscience 26, 101–111 (2004).
Dyer, C. et al. A bi-modal function of Wnt signalling directs an FGF activity gradient to spatially regulate neuronal differentiation in the midbrain. Development 141, 63–72 (2014).
Geling, A., Plessy, C., Rastegar, S., Strähle, U. & Bally-Cuif, L. Her5 acts as a prepattern factor that blocks neurogenin1 and coe2 expression upstream of Notch to inhibit neurogenesis at the midbrain-hindbrain boundary. Development 131, 1993–2006 (2004).
Salomoni, P. & Calegari, F. Cell cycle control of mammalian neural stem cells: putting a speed limit on G1. Trends in cell biology 20, 233–243 (2010).
Brouhard, G. J. Dynamic instability 30 years later: complexities in microtubule growth and catastrophe. Molecular biology of the cell 26, 1207–1210 (2015).
Chauvin, S. & Sobel, A. Neuronal stathmins: A family of phosphoproteins cooperating for neuronal development, plasticity and regeneration. Progress in neurobiology 126, 1–18 (2015).
Chauvin, S., Poulain, F. E., Ozon, S. & Sobel, A. Palmitoylation of stathmin family proteins domain A controls Golgi versus mitochondrial subcellular targeting. Biology of the Cell 100, 577–591 (2008).
Boekhoorn, K., Goedknegt, E., Sobel, A., Lucassen, P. J. & Hoogenraad, C. C. The microtubule destabilizing protein stathmin controls the transition from dividing neuronal precursors to postmitotic neurons during adult hippocampal neurogenesis. Developmental neurobiology 74, 1226–1242 (2014).
Shih, D.-F., Chu, S.-L. & Lee, S.-J. Characterization and Expression Analysis of Stathmin Family Genes during Embryogenesis in Zebrafish, Danio rerio. Taiwania 59, 262–280 (2014).
Manna, T., Thrower, D. A., Honnappa, S., Steinmetz, M. O. & Wilson, L. Regulation of microtubule dynamic instability in vitro by differentially phosphorylated stathmin. Journal of Biological Chemistry 284, 15640–15649 (2009).
Solnica-Krezel, L. & Sepich, D. S. Gastrulation: making and shaping germ layers. Annual review of cell and developmental biology 28, 687–717 (2012).
Sato, T., Takahoko, M. & Okamoto, H. HuC: Kaede, a useful tool to label neural morphologies in networks in vivo. Genesis 44, 136–142 (2006).
Robu, M. E. et al. p53 activation by knockdown technologies. PLoS Genet 3, e78–e78 (2007).
Talbot, J. C. & Amacher, S. L. A streamlined CRISPR pipeline to reliably generate zebrafish frameshifting alleles. Zebrafish 11, 583–585 (2014).
Rossi, A. et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524, 230–233 (2015).
Asakawa, K. et al. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proceedings of the National Academy of Sciences 105, 1255–1260 (2008).
Dong, Z., Peng, J. & Guo, S. Stable gene silencing in zebrafish with spatiotemporally targetable RNA interference. Genetics 193, 1065–1071 (2013).
Provost, E., Rhee, J. & Leach, S. D. Viral 2A peptides allow expression of multiple proteins from a single ORF in transgenic zebrafish embryos. Genesis 45, 625–629 (2007).
Distel, M., Hocking, J. C. & Köster, R. W. In vivo cell biology using Gal4 mediated multicolour subcellular labelling in zebrafish. Communicative & integrative biology 4, 336–339 (2011).
Buckley, C. E. et al. Mirror-symmetric microtubule assembly and cell interactions drive lumen formation in the zebrafish neural rod. The EMBO journal 32, 30–44 (2013).
Peco, E. et al. The CDC25B phosphatase shortens the G2 phase of neural progenitors and promotes efficient neuron production. Development 139, 1095–1104 (2012).
Quastler, H. & Sherman, F. G. Cell population kinetics in the intestinal epithelium of the mouse. Experimental cell research 17, 420–438 (1959).
Morgan, D. O. The cell cycle: principles of control. (New Science Press, 2007).
Kok, F. O. et al. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Developmental cell 32, 97–108 (2015).
Spicer, O. S., Wong, T.-T., Zmora, N. & Zohar, Y. Targeted Mutagenesis of the Hypophysiotropic Gnrh3 in Zebrafish (Danio rerio) Reveals No Effects on Reproductive Performance. PloS One 11, e0158141 (2016).
Anderson, D. J. & Axel, R. Molecular probes for the development and plasticity of neural crest derivatives. Cell 42, 649–662 (1985).
Beilharz, E. J. et al. Neuronal activity induction of the stathmin-like gene RB3 in the rat hippocampus: possible role in neuronal plasticity. The Journal of neuroscience 18, 9780–9789 (1998).
Stein, R., Orit, S. & Anderson, D. J. The induction of a neural-specific gene, SCG10, by nerve growth factor in PC12 cells is transcriptional, protein synthesis dependent, and glucocorticoid inhibitable. Developmental biology 127, 316–325 (1988).
Sung, P.-J. et al. Identification and characterisation of STMN4 and ROBO2 gene involvement in neuroblastoma cell differentiation. Cancer letters 328, 168–175 (2013).
Morii, H., Shiraishi-Yamaguchi, Y. & Mori, N. SCG10, a microtubule destabilizing factor, stimulates the neurite outgrowth by modulating microtubule dynamics in rat hippocampal primary cultured neurons. Journal of neurobiology 66, 1101–1114 (2006).
Poulain, F. E. & Sobel, A. The “SCG10-LIke Protein” SCLIP is a novel regulator of axonal branching in hippocampal neurons, unlike SCG10. Molecular and Cellular Neuroscience 34, 137–146 (2007).
Ohkawa, N., Fujitani, K., Tokunaga, E., Furuya, S. & Inokuchi, K. The microtubule destabilizer stathmin mediates the development of dendritic arbors in neuronal cells. Journal of cell science 120, 1447–1456 (2007).
Park, H.-C. et al. Structural comparison of zebrafish Elav/Hu and their differential expressions during neurogenesis. Neuroscience letters 279, 81–84 (2000).
Hanash, S. M., Strahler, J. R., Kuick, R., Chu, E. & Nichols, D. Identification of a polypeptide associated with the malignant phenotype in acute leukemia. Journal of Biological Chemistry 263, 12813–12815 (1988).
Luo, X.-N. et al. Regulated expression of p18, a major phosphoprotein of leukemic cells. Journal of Biological Chemistry 266, 21004–21010 (1991).
Rubin, C. I. & Atweh, G. F. The role of stathmin in the regulation of the cell cycle. Journal of cellular biochemistry 93, 242–250 (2004).
Silva, V. C. & Cassimeris, L. Stathmin and microtubules regulate mitotic entry in HeLa cells by controlling activation of both Aurora kinase A and Plk1. Molecular biology of the cell 24, 3819–3831 (2013).
Blomen, V. & Boonstra, J. Cell fate determination during G1 phase progression. Cellular and Molecular Life Sciences 64, 3084–3104 (2007).
Britz, O. et al. A role for proneural genes in the maturation of cortical progenitor cells. Cerebral Cortex 16, i138–i151 (2006).
Mata, J., Curado, S., Ephrussi, A. & Rørth, P. Tribbles coordinates mitosis and morphogenesis in Drosophila by regulating string/CDC25 proteolysis. Cell 101, 511–522 (2000).
Stergachis, A. B. et al. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell 154, 888–903 (2013).
Zhu, J. et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell 152, 642–654 (2013).
Kimelman, D. Cdc25 and the importance of G2 control: insights from developmental biology. Cell Cycle 13, 2165–2171 (2014).
Ogura, Y. et al. Coordination of mitosis and morphogenesis: role of a prolonged G2 phase during chordate neurulation. Development 138, 577–587 (2011).
Bouldin, C. M., Snelson, C. D., Farr, G. H. & Kimelman, D. Restricted expression of cdc25a in the tailbud is essential for formation of the zebrafish posterior body. Genes & development 28, 384–395 (2014).
Lee, H.-S. et al. STMN2 is a novel target of β-catenin/TCF-mediated transcription in human hepatoma cells. Biochemical and biophysical research communications 345, 1059–1067 (2006).
Nyholm, M. K., Abdelilah-Seyfried, S. & Grinblat, Y. A novel genetic mechanism regulates dorsolateral hinge-point formation during zebrafish cranial neurulation. J Cell Sci 122, 2137–2148 (2009).
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B. & Schilling, T. F. Stages of embryonic development of the zebrafish. Developmental dynamics 203, 253–310 (1995).
Kwan, K. M. et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Developmental Dynamics 236, 3088–3099 (2007).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nature methods 9, 676–682 (2012).
Montague, T. G., Cruz, J. M., Gagnon, J. A., Church, G. M. & Valen, E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic acids research, gku410 (2014).
Gagnon, J. A. et al. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PloS One 9, e98186 (2014).
Thisse, C. & Thisse, B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nature protocols 3, 59–69 (2008).
Lauter, G., Söll, I. & Hauptmann, G. Multicolor fluorescent in situ hybridization to define abutting and overlapping gene expression in the embryonic zebrafish brain. Neural development 6, 1–13 (2011).
Inoue, D. & Wittbrodt, J. One for all–a highly efficient and versatile method for fluorescent immunostaining in fish embryos. PLoS One 6, e19713–e19713 (2011).
Amatruda, J. F. Analysis of the cell cycle in zebrafish embryos. The Zebrafish: Cellular and Developmental Biology: Cellular and Developmental Biology 76, 109 (2004).
Arganda-Carreras, I., Fernández-González, R. & Muñoz-Barrutia, A. & Ortiz-De-Solorzano, C. 3D reconstruction of histological sections: Application to mammary gland tissue. Microscopy research and technique 73, 1019–1029 (2010).
Acknowledgements
We thank Dr. Koichi Kawakami and Dr. Gembu Abe from National Institute of Genetics, Japan, for providing technical guidance and plasmids related to Tol2 transgenesis. We thank Dr. Reinhard Köster from TU Braunschweig for providing subcellular tracking plasmids. We thank Dr. Su Guo from University of California, San Francisco for providing shRNA plasmids. We thank Dr. David Kimelman from University of Washington for providing cdc25a plasmids. We thank Dr. Alex Schier from Harvard University for proving CRISPR/Cas9-related plasmids and protocols. We thank Dr. Chin-Hwa Hu and Taiwan Zebrafish core facility for providing transgenic fish, plasmids. We thank Ms. Yi-Chun Chuang from Technology Commons, National Taiwan University, for excellent technical assistance with confocal microscopy. The authors would also like to express great appreciation to the staffs of the Zebrafish Core at National Taiwan University for providing assistance in fish maintenance. The work was supported by grants from the Ministry of Science and Technology, Taiwan (NSC 95–2311-B-002-015-MY3, MOST 104-2313-B-002-033-MY3) and National Taiwan University (NTU CESRP-10R70602A5 and NTU ERP-10R80600) to SJL.
Author information
Authors and Affiliations
Contributions
M.-J.L. designed, performed all experiments and wrote the manuscript; S.-J.L. designed, supervised all experiments and wrote the manuscript. Both authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Lin, MJ., Lee, SJ. Stathmin-like 4 is critical for the maintenance of neural progenitor cells in dorsal midbrain of zebrafish larvae. Sci Rep 6, 36188 (2016). https://doi.org/10.1038/srep36188
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep36188
This article is cited by
-
Klf8 regulates left-right asymmetric patterning through modulation of Kupffer’s vesicle morphogenesis and spaw expression
Journal of Biomedical Science (2017)
-
A transcriptomic study of myogenic differentiation under the overexpression of PPARγ by RNA-Seq
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.