Abstract
Drought stress is a major abiotic stress affecting plant growth and development. In this study, we performed the first dynamic phosphoproteome analysis of Brachypodium distachyon L. seedling leaves under drought stress for different times. A total of 4924 phosphopeptides, contained 6362 phosphosites belonging to 2748 phosphoproteins. Rigorous standards were imposed to screen 484 phosphorylation sites, representing 442 unique phosphoproteins. Comparative analyses revealed significant changes in phosphorylation levels at 0, 6, and 24 h under drought stress. The most phosphorylated proteins and the highest phosphorylation level occurred at 6 h. Venn analysis showed that the up-regulated phosphopeptides at 6 h were almost two-fold those at 24 h. Motif-X analysis identified the six motifs: [sP], [Rxxs], [LxRxxs], [sxD], [sF], and [TP], among which [LxRxxs] was also previously identified in B. distachyon. Results from molecular function and protein-protein interaction analyses suggested that phosphoproteins mainly participate in signal transduction, gene expression, drought response and defense, photosynthesis and energy metabolism, and material transmembrane transport. These phosphoproteins, which showed significant changes in phosphorylation levels, play important roles in signal transduction and material transmembrane transport in response to drought conditions. Our results provide new insights into the molecular mechanism of this plant’s abiotic stress response through phosphorylation modification.
Similar content being viewed by others
Introduction
Drought is one of the most important sources of abiotic stress, as it limits growth, development, and yield of field crops. As the world population increases, and available water resources decrease, plant breeders have increasingly focused on the development of drought-tolerant crops with high water-use efficiency1. Therefore, it is vital to elucidate the molecular mechanisms of plant responses to drought stress.
Land plants have developed a variety of adaptation and defense strategies in response to internal and external stimuli, particularly for drought stress2. Molecular mechanisms in response to drought conditions are elusive, given the complexity of organisms and the limitations of experimental technology. It is commonly accepted that drought responses by plants include osmotic homeostasis or regulation, stress repair and defense, detoxification, and growth inhibition3. These processes are likely to follow signal transduction and cell cascades.
Results from recent studies have shown that three signal pathways are involved in drought response and defense: abscisic acid (ABA), reactive oxygen species (ROS), and Ca2+ signaling. ABA is an important signal molecule, as it causes the expression of regulation proteins and allows proteins to participate in the response to and defense from various stresses4,5. Ca2+ signaling is also a common signal pathway that responds to adverse signs in eukaryocytes6. ROS are involved in the response to oxidative stress, as it serves as a supplementary signal pathway to cope with drought stress7,8. In signal pathways, the signal processes generally involve protein phosphorylation and dephosphorylation by protein kinases and phosphatases, when subjected to drought stress.
Protein phosphorylation is closely related to a large number of processes, including metabolism, transcription and translation, protein degradation, homeostasis, cellular signaling and communication, and various stress responses9,10,11. Phosphorylation is a reversible and ubiquitous post-translational modification (PTM), which is a dynamic process regulated by kinases and phosphatases. In eukaryotes, phosphorylation occurs more frequently on serine (Ser) and threonine (Thr) than tyrosine (Tyr). Studies have shown that phosphorylation participates in many molecular events and biological processes, such as DNA transcription, stomatal closure12, stress response and defense, and energy metabolism13.
Recently, large-scale in vivo phosphoproteomic analyses have been performed to elucidate diverse response mechanisms in different plant species, such as rice14, Arabidopsis thaliana L.15, soybean16, maize17, and wheat18,19,20. Moreover, our group characterized the phosphoproteome of seedling leaves of B. distachyon21. However, the dynamic changes in the phosphoproteome in response to drought remain unknown.
B. distachyon, a temperate wild annual grass species typically grown in the Mediterranean and Middle East, has recently been used as a model plant for cereal crops and potential biofuel grasses22,23. It has a large number of desirable features: a small diploid genome (272 Mbp), short developmental cycle, efficient transgenosis, self-fertilizing and simple nutrient requirements24.
In recent years, the rapid development of genome sequencing in B. distachyon has facilitated further studies on transcriptomics, proteomics, and phosphoproteomics. In this study, we performed the first in vivo dynamic analysis of the phosphoproteome of B. distachyon seedling leaves at different time points (0, 6, and 24 h), using TiO2 enrichment and LC-MS/MS analysis coupled with label-free quantification. A large number of phosphorylated proteins involved in the drought stress response were identified, providing new information for understanding the molecular mechanisms of plant responses to drought stress.
Results
Phenotypic, physiological, and ultrastructural changes in seedling leaves under drought conditions
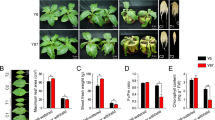
The dynamic changes in phenotypes and some important physiological indices of Brachypodium distachyon 21 (Bd21) seedling leaves under different drought conditions are shown in Figs 1 and S1. With the intensification of drought stress, leaves and roots displayed more obvious changes. At 24 h, leaves had a slight curl, while roots became thin compared to the control group. There were no significant differences at 6 h (Fig. 1A). Drought stress resulted in significant changes in the physiological index of seedling leaves (Figs S1 and1B). The results from five different periods (0, 6, 12, 24, and 48 h) showed that relative water content (RWC) and total chlorophyll content for the 20% PEG 6000 treatment displayed a continuous declining trend. In contrast, the levels of malondialdehyde (MDA) and proline (Pro) content continuously increased in response to drought stress. The 24 h treatment was a critical turning point, while 6 h was a transition period. Compared to the control group (0 h), MDA exhibited a significant (p < 0.05) and highly significant (p < 0.01) increase at 6 h (1.299-fold) and 24 h (1.939-fold), respectively. Proline content was significantly increased by 1.646-fold at 24 h (p < 0.05).
Phenotypic, physiological and ultrastructural changes in Bd21 leaves under 0, 6 and 24 h drought stress.
(A) Seedling phenotypic changes. (B) Leaf physiological changes: a. RWC; b. Chlorophyll content; c. MDA content; d. Proline content. Error bars indicate standard errors of three biological replicates. Statistically significant differences compared to the control were calculated based on an independent Student’s t-tests: *P < 0.05; **P < 0.001. (C) Leaf ultrastructural changes observed by TEM. a. Bd21 leaves ultrastructure; b. Nucleolus ultrastructure; c. Chloroplast ultrastructure; d. Mitochondria ultrastructure. Ch: Chloroplast; CW: Cell wall; Cy: Cytomembrane; Nu: Nucleolus; N, Nucleus; Mt: Mitochondria; P: Peroxysome.
To reduce experimental error, unified flag leaves of B. distachyon were selected for transmission electron microscope (TEM) observation. The ultrastructural changes between the non-stress and drought stress treatments of Bd21 seedling leaves are shown in Fig. 1C. The flag leaves in the control group had normal whole mesophyll cells and clear subcellular structures. Treatment with 20% PEG resulted in different degrees of damage to parts of the mesophyll cells and some organelles, including clear plasmolysis of the cell wall, misshapen chloroplasts and nucleolus, particularly at 24 h (Fig. 1C). With disrupted thylakoids, the number of starch granules increased, mitochondria became swollen, and the color faded. These results demonstrate that drought stress has significant effects on leaf ultrastructure.
Phosphoproteins identification and locations of phosphorylated sites
According to the dynamic changes in plant phenotype and leaf physiological traits under drought stress, leaves from three key time points (0, 6, and 24 h) were collected and used to study dynamic changes in the phosphoproteins based on phosphopeptides enrichment and LC-MS/MS and MaxQuant analyses (Figs S2 and S3; Table S1). All mass spectrometry proteomics data were deposited in the ProteomeXchange Consortium (http://proteomecentral. proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD003522. A total of 4924 phosphopeptides, containing 6362 phosphosites, were identified and belonged to 2748 phosphoproteins (Fig. S3a; Table S1). Moreover, 3843 (78.05%) phosphopeptides had only one phosphosite while 838 (17.02%) and 199 (4.4%) phosphopeptides contained two and three phosphosites, respectively. In particular, three phosphopeptides had six phosphosites (Fig. S3b). In Fig. S3c, a total of 5403 (84.92%), 932 (14.65%), and 27 (0.42%) phosphorylation sites were screened separately on Ser, Thr, and Tyr. According to the location probability of phosphorylated sites, 4356 (68.47%), 632 (9.93%), 781 (12.28%), and 593 (9.32%) phosphorylated sites belonged to class I (p ≥ 0.9), II (0.75 ≤ p < 0.9), III (0.5 ≤ p < 0.75), and IV (p < 0.5), respectively (Fig. S3d).
To increase reliability and accuracy, only phosphorylated sites belonging to the highest phosphorylation site localization probability (p ≥ 0.9, class I) were used for further analysis. Phosphopeptides with receivable Student’s t test (p < 0.05) consequence, significant intensity changes (≥2-fold), high phosphorylation site localization probability (≥0.9), and phosphorylation site score differences exceeding five were taken into consideration to identify significant changes in phosphorylation level. In the strict limitations screening progress, a total of 484 phosphorylation sites had significant changes in the phosphorylation level (SCPL), and divided into 0–6 h and 6–24 h. Comparisons of phosphorylation sites, phosphopeptides, and phosphorylated proteins are provided in Fig. 2a~c, Tables S2a and S2b, respectively. In two groups, there were 76 phosphorylation sites in common, representing 82 phosphorylated proteins.
A Venn diagram was used to analyze the significantly up-regulated and down-regulated phosphopeptides in both groups (Fig. 2b). The fold change in up-regulated phosphopeptides at 6 h was almost two-fold greater than at 24 h. Six and two phosphopeptides were up-regulated and down-regulated, respectively (Fig. 2d). Proteins involved in stress defense included a stress protein DDR48-like (gi|357163456) and a late embryogenesis abundant protein B19.3 (LEA, gi|721639026).
Expression pattern analysis of phosphoproteins with SCPL under drought stress
Principal component analysis (PCA) is a multivariate method of dimensionality that can be used to identify relationships among variables25. The PCA results from 442 SCPL phosphoproteins are shown in Fig. 3. Three groups of biological replication (NP1, NP2, and NP3) showed a high level of reproducibility (Fig. 3a). The treatment groups were significantly different from the control group. The treatment group at 6 h had the greatest differences from the other treatments. Some spots focused on the origin of coordinates and a few had scattered distribution in the periphery (Fig. 3b). Most of the scattered spots belonged to pattern I and pattern II in Fig. S5. Meanwhile, these scattered spots were common spots of 76 phosphorylation sites. These results indicate that Bd21 underwent significant changes in protein phosphorylation levels under drought stress.
PCA of SCPL phosphoproteins under drought stress.
(a) PCA of individual proteins samples in seedling leaves. Yellow circles (NP1) represent 0 h, green circles (NP2) represent 6 h and red circles (NP3) represent 24 h. (b) PCA of all SCPL phosphoproteins of seedling leaves. NP1, NP2 and NP3 represented the normalized intensities of SCPL phosphopeptides at 0 h, 6 h and 24 h, respectively. NP1-1, NP1-2 and NP1-3 mean the three biological replicates of NP1 (0 h). So does the NP2 and NP3.
To visualize the dynamic changes in the SCPL phosphopeptides, the Euclidean distance similarity metric was used to define the similarity, and hierarchical clusters were assembled using the complete linkage clustering method. Five hierarchical clusters (patterns I–V), corresponding to the different expression patterns, were defined in each period (Fig. S5; Table S3). Pattern I, containing 179 phosphorylation sites, was significantly up-regulated at 6 h, while pattern II, with 106 phosphorylation sites, showed up-regulated expression at 24 h. Furthermore, we classified the function of these two parts phosphorylated proteins in Figs S7a,b. These results further validate the previous PCA analysis, where the 6 h treatment had significantly more SCPL proteins compared to the other treatments.
Phosphopeptide screening and placement in functional categories
All of the identified SCLP proteins were used to predict the functional categories with GO annotation. They were involved in different biological processes, molecular functions, and cellular components (Fig. 4; Table S4). Regarding biological processes, primary metabolic processes accounted for 20% in Fig. 4a. The proportion of gene expression and single-organism cellular process was less than primary metabolic process, respectively. Nucleotide and nucleic acid binding occupied the main part of the molecular functions (Fig. 4b), indicating that nucleoproteins respond to drought stress positively and phosphorylation is one of the coping mechanisms. The cellular components of more than 30% of the phosphoproteins were localized to the membrane (Fig. 4c), indicating that they play a key role in metabolism and some phosphoproteins of water and ion transport were identified in Table 1. Furthermore, three aquaporins (AQPs) were analyzed by 3D structural analyses and sequence alignment in Fig. S6. These AQPs were aquaporin PIP1-5 (IPI1-5, gi|357137703), probable aquaporin IPIP2-7 (IPIP2-7, gi|357159722), and aquaporin NIP2-2 (NIP2-2, gi|357124731). In 3D structural analyses, only PIP1-5 phosphorylation site belonged to the α-helix. Others were on the random coil, which is in agreement with the secondary structure surrounding the phosphorylation site prediction results (Fig. 5b). Most of the SCPL phosphoproteins (nearly 90%) were random coils or turns, 7.6% were α-helixes, and the remaining proteins (1.6%) were β-pleated sheets (Fig. 5b; Table S5b).
Phosphorylation motif, protein secondary structure surrounding the phosphorylation site and conservation of the significantly changed phosphoproteins under drought stress.
(a) Significantly enriched phosphoproteins by Motif-X: left part represents amino acids surrounding the identified phosphorylated residues by Motif-X under drought stress; Right part represents phosphorylation of the related kinases according to the score height which are divided into different groups. (b) Protein secondary structure surrounding the phosphorylation site, including random coils or turns, α-Helix and β-pleated sheet. (c) Conservation analysis of the significant changed phosphoproteins.
Some SCPL proteins identified by Lv et al.26 under salt stress were also identified under drought stress in this study (Table 2). Their functional classification is provided in Fig. S7c. A total of 51 phosphorylated proteins were identified; 31% of them are related to transcription and translation, and approximately 18% participate in water and ion transport related to osmotic regulation. However, 29% of the proteins are associated with other processes and require further study (Fig. S7c).
Phosphorylation motif analysis of significantly changed phosphopeptides
Phosphorylation motifs of the phosphoproteins linked to kinase were ascertained by Motif-X and WebLogo online software tools (Fig. 5a; Table S5a). There were six motifs were identified, including five phosphorylation residues were enriched by Ser, (such as [sP], [Rxxs], [LxRxxs], [sxD], and [sF]) and only [TP] motif was enriched by Thr and Pro. Some luxuriant kinases were identified by hierarchical clusters. Both [sP] and [TP] were proline-directed motifs, recognized by cyclin-dependent kinase 5 (Cdk5)27. The motifs of [Rxxs] and [LxRxxs] were basic motifs recognized by protein kinase A (PKA)28. In addition, motif [sxD] was an acidic motif recognized by casein kinase-II (CKII)29. Only motif [sF] was easily recognized by protein kinase C30. [LxRxxs] is a newly identified motif in B. distachyon. Some key phosphoproteins, including heat shock protein 83 (Hsp 83, gi|357152022), zinc finger CCCH domain-containing protein 13 (gi|357117322), chloride channel protein CLC-c-like (gi|357149553), transcription factor bHLH128-like (gi|357126351), and calmodulin-binding transcription activator 3-like isoform X1 (gi|721658612), were involved in this motif.
Conservation analysis of phosphoproteins responsive to drought stress
Sequences of the SCPL phosphoproteins were used as queries to BLAST search the phosphoprotein databases constructed based on datasets in the Plant Protein Phosphorylation DataBase (P3DB)31, Medicago-Omics Repository (MORE)32, and PhosPhAt 4.033. Oryza. sativa, A. thaliana, Medicago truncatula, and others were compared to B. distachyon to determine the extent of conservation of phosphoproteins among different plant species. The thresholds were set at a score ≥80, E-value < 1E−10, and identity ≥30% (Fig. 5c; Table S5c). In total, among the 442 phosphoproteins identified in B. distachyon, 104 (23.48%), 86 (19.41%), and 49 (11.06%) had phosphorylated orthologs in three, two, and one species, respectively. Only 164 phosphoproteins had phosphorylated orthologs in all four species. A total of 40 (9.02%) phosphoproteins had no phosphorylated orthologs in any of these species.
Protein–protein interaction (PPI) analysis
To explore the relationships in the PPI network, 442 SCPL proteins identified in the current study were analyzed by STRING. A total of 307 EuKaryotic Orthologous Groups (KOGs) were used to construct the PPI network. To improve the reliability of PPI analysis, the confidence score was set at the highest level (≥0.900). Finally, a complex PPI network was completed through Cytoscape (Fig. S8; Table S6).
Numerous SCPL phosphoproteins were found to participate in transcription and translation of gene expression (Fig. S8). Some of the phosphoproteins belonged to nucleotide or nucleic acid binding phosphorylation proteins, in agreement with the functional categories (Fig. 4b). Moreover, some of the important protein kinases and phosphatases, including abscisic acid-inducible protein kinase (PKAB1, KOG0583), myotonin protein kinase 7–1 (MPK7-1, KOG0660), calcium-dependent protein kinase 26-like (CDPK, KOG0032), and the probable protein phosphatase 2C 47 (PP2C, KOG0698), played a key role in this PPI map, which participated in protein phosphorylation and dephosphorylation under drought stress. The proteins in this map were also involved in signal transduction, water and ion transport, cell structure and division, and photosynthesis and energy metabolism.
Verification of identified phosphoproteins by Pro-Q diamond staining and tandem mass spectrometry
To enhance the credibility of results, Pro-Q diamond staining and tandem mass spectrometry were used to validate the phosphoproteins identified in this study. The results from Pro-Q diamond and Coomassie brilliant blue (CBB) showed that 33 protein spots were phosphorylated at 6 and 24 h under drought stress (Fig. S4). MALDI-TOF/TOF-MS identification showed that these protein spots represented 24 phosphoproteins, further confirming the results from phosphoproteome analysis (Table S7). Four spots (1, 11, 14, and 24) belonged to three SCPL phosphoproteins: fructose-bisphosphatealdolase (chloroplastic, gi|357157399), ribulosebisphosphate carboxylase small chain PW9 (chloroplastic-like, gi|357146204), and ATPase beta subunit (chloroplast, gi|193075564). These phosphoproteins are related to photosynthesis and energy metabolism.
Discussion
Protein kinases/phosphatases participated in signal perception and transduction
The reversible phosphorylation modification event is regulated by protein kinase and phosphorylase, which could supervise multiple biological processes of organisms34. In this study, numerous protein kinases and phosphatases were SCPL, and played key roles in signal transduction (Figs 6 and S8). ABA is a crucial signal molecule in signal transduction responses to various abiotic stresses, including drought stress35. PKABA1 (gi|721630009) phosphorylated at Ser622 was identified in this study and found to be significantly up-regulated at 24 h (Table S2b). PKABA1, a member of the sucrose non-fermenting1-related protein kinase (SnRK) subfamily, acts as an intermediate in ABA suppression of GA-responsive gene transcription36. Some evidence suggested that over-expression of TaSnRK2S could enhance the tolerance of abiotic stressors, such as drought and salt stress, in A. thaliana37. In addition, protein phosphatase 2C (PP2C) was another key regulator and got involved in inhibiting SnRK’s substrate phosphorylation which caused signaling cascade in the ABA signaling pathway38. PP2C-47 (gi|357147223) was found and down regulated at 6 h, which was in agreement with Sheard’s study38: PP2C acted as a constitutive negative regulaor of SnRKs whose autophosphorylation is required for kinase activity towards downstream targets. Thus, it indicated that Bd21 responded positively to defense the drought stress with phosphorylation at 6 h (Supplemental Table S2a)
In plants, the signals from the upstream elicitor receptors/sensors to the downstream mitogen-activated protein kinase (MAPK) substrates largely occur in three steps: the MAP kinase kinase kinase (MAPKKK), MAP kinase kinase (MAPKK), and MAPK cascade39. Some experiments have shown that various MAPK cascade members are involved in mediating signal transduction by transcription regulating P and N deprivation40. The activation of these MAP kinases is a sequential process through the phosphorylation of some conserved residues within an activation loop in their kinase domains41. In our study, three phosphorylated proteins were identified: MPK17 (gi|405778409), MPK7-1 (gi|405778403), and MPK3 (gi|405778399) (Table S1). MPK17 had two phosphorylation sites (Ser465, Ser491) and the phosphorylation level at Ser465 had significant up-regulation at 24 h. A recent study showed that MAP kinases could serve as upstream activators of the stress marker genes under saline conditions41. A. thaliana MPK20 and proline dehydrogenase might interact with each other and be part of a functional hypoosmotic stress-signaling pathway42. Sequence alignment and conservation analyses demonstrated that the expression of MPK17 was high, similar to Arabidopsis thaliana (Fig. 5; Table S5c). In addition, we identified some important CDPKs, such as CDPK26-like (gi|357125710; Table S2a) phosphorylated at Ser18, which supports the view that many of the autophosphorylation sites in CDPKs are localized at the N terminus.
Transcription factors involved in gene expression
Under abiotic stress, many defense-related genes are regulated by transcription factors (TFs). In the present study, many TFs were phosphorylated (Table S2; Table 2). For example, nuclear TFs Y subunit A-7-like isoforms X1 (NF-Y A, gi|721672572), NF-Y B-1 (gi|357122032), and NF-Y beta (gi|721661240) were phosphorylated at Ser202 (_GAS(ph)PANQTGNRE_), Ser4 (_PDS(ph)DNEDS GGGGGIGGGGNNK_), and Ser105 (_LDTLDS(ph)PK_), respectively. Both NF-Y B and NF-Y A proteins were up-regulated, whereas the NF-Y beta protein was down-regulated. Some studies involving maize, Arabidopsis, and wheat have proven that NF-Y B subunits may respond to drought stress43,44. Nelson et al.43 reported tolerance to drought stress in transgenic maize, based on increasing ZmNFYB2 expression. The Bd21 phosphorylation levels were conserved (Fig. 5; Table S5c). Thus, these TFs responded positively to abiotic stresses.
Heat shock proteins (Hsps) and heat shock TFs (Hsfs) are involved in cellular responses to various sources of stress beyond heat stress45. The conformation of Hsps is affected by phosphorylation modification, such as the formation of active sites, flexibility, and inter-domain communication46. In other researches, HsfA1, as a main regulator, could enhance thermo tolerance and other stresses tolerance47,48. In our study, HsfA-2e-like (gi|357111341) was identified in Table S2a and HsfA-2e-like and Hsp81-1 (gi|357148345) have a complex relationship with other SCPL phosphoproteins (Figs 6 and S8). Hsfs recognized and combined with the Hsps gene promoter region of the heat shock element conservation motif. Thus, Hsfs may regulate Hsps transcription and gene expression to improve the stress tolerance of plants.
Phosphoproteins involved in water and ion transport
In the face of various biotic and abiotic stresses, particularly drought stress, plants gradually adjust water and ion transport mechanisms. AQPs belong to a membrane protein family that rapidly transports water under osmotic pressure49. In our study, three AQPs were identified as belonging to the plasma membrane intrinsic protein (PIP) and nodulin 26-like intrinsic protein (NIP) subfamilies (Table S2a). All were up-regulated at 6 h under drought stress, which indicates that they had the same function in this process. NIP2-2 and PIP2-7 were phosphorylated at Ser271 and Ser282, respectively, in agreement with Guenther et al.50. Nodulin 26 phosphorylation at the C terminal tail (at Ser262) enhances water transport and is involved in the Ca2+ signal pathway to accelerate regulation of the water channel switch. Thus, phosphorylation could influence the activity of these AQPs.
There are three types of H+-ATPase: F-type, V-type, and P-type. The P-type is a large family of molecular pumps that act as intermediates in ATP hydrolysis and ion transport or ion counter-transport51,52. Moreover, activation of the plasma membrane H+-ATPase is associated with phosphorylation of a penultimate Thr and binding to the phosphorylated C-terminus of 14-3-3 protein to control the stomatal aperture53. A recent study has showed that the stomatal aperture is a limiting factor in photosynthesis and plant growth, and the overexpression of H+-ATPase in guard cells is useful for promoting plant growth54. We also identified some proteins related to H+-ATPase. Plasma membrane ATPase (gi|357166497) and V-type proton ATPase subunit a1 (gi|357125740) were up-regulated at 6 h and phosphorylated at Thr881 and Ser688 (Table S2a), which enhanced enzyme activity and related to osmotic regulation.
ROS detoxification and stress defense
Many biotic and abiotic stresses might induce plant cells to produce ROS, leading to the acceleration of lipid peroxidation and leaf damage55,56 or beyond the antioxidant system57, particularly when subjected to drought stress. ROS scavenging or detoxification is a defense strategy in plants, involving a series of enzymes and proteins that reduce oxidative damage under drought stress58,59. In this work, one of the probable phospholipid hydroperoxide glutathione peroxidases (PHGPx, gi|357165189) was identified as an SCPL protein (Table S2b), which displayed up-regulated at 24 h. Phosphorylated PHGPx could become active to eliminate ROS and play a key role in the detoxification response to drought stress.
In addition, to sustain normal morphological structure and physiological function, plant cells must complete normal homeostasis, including electron transfer and oxidation reduction, and activate oxygen-scavenging to resist stress. In this study, we identified photosynthesis- and electron transport-related phosphorylated proteins that alleviate stress from drought. Chlorophyll a-b binding protein (chloroplast; LHCB, gi|357121229) was identified to be a specific SCPL protein at 6 h, and had significant up-regulation, as identified in salt stress (Table S2a; Table 2). LHCB was regulated by the redox state of the plastoquinone mediating electron transfer between photosystems I and II, which involve reversible phosphorylation. Moreover, LHCB occurred redox dependent phosphorylation by specific thylakoid-bound kinases to affect the reconstruction of light harvesting complexes60. Furthermore, the changes in physiological and ultrastructure (Fig. 1) also implied that chloroplast structure was injured under drought stress and LHCB phosphorylation activity was weakened, which influenced photosynthesis and energy metabolism.
LEA proteins are made up of a group of dehydration-inducible protein families and their function is to help maintain the minimum cellular water requirement. Comparison of dehydrin DHN3-like (gi|357149091, Table S1) with a LEA protein (RAB-17) in maize61, showed the same sequence motif of –SSSSSSSEDD– (Table S1). Two LEAs were identified in SCPL proteins (LEA1, gi|721622359; LWAB19.3, gi|721639026) and up-regulated at Ser81 and Ser91. In maize cells, phosphorylation of LEA could facilitate binding to the targeted proteins and involve nuclear location62. Moreover, LEA is involved in ABA induction under drought stress.
In addition, molecular chaperones were involved in regulating plant growth processes and the chaperone complexes of Hsp90 could respond to ABA stimulation activitily63. Hsp81-1 (gi|357148345), a molecular chaperone member of the Hsp90 family, has the same motif with some identified sequence phosphopeptides, such as -TTEKEIS(ph)DDEDEEEK- in A. thaliana64, rice65, and wheat63 (Table S5a). Hsp90 plays a key role in protein folding66 and participates in signal-transduction networks and protein degradation and trafficking67,68,69. Moreover, in the present study, Hsp81-1 was up-regulated at 6 h (Table S2a), suggesting that it could play an active role in response to drought stress.
A putative pathway of drought stress responses in Bd21 leaves through phosphorylation modification
Based on the results from this study and previous studies, we propose a putative pathway of drought stress response in Bd21 seedling leaves through phosphorylation modification (Fig. 6). When plants were subjected to drought stress, membrane sensors sensed external environment changes, and transferred the chemical signals to the cell, which involved ABA, Ca2+, and MAPK signal pathways. Phosphate kinases/phosphatases were activated by the corresponding signal molecules. Then downstream TFs and translation factors were regulated by these phosphate kinases/phosphatases and they induced the production of drought stress response proteins such as LEA, Hsps, and LHCB. Massive accumulation of free radicals and ROS caused membrane lipid peroxidation, damaging the membrane systems, which caused the up-regulation of ROS scavenging-related phosphoproteins to defend against drought stress. At the same time, ABA and Ca2+ concentrations triggered stomata closure to defend against water stress. The AQPs and transporters on membranes also regulated the balance of osmosis by phosphorylation/dephosphorylation.
Materials and Methods
Plant materials and drought treatment
Bd21 kindly provided by Dr. John Vogel et al.24 at USDA-ARS, was used as plant material in this study. Bd21 seeds were surface sterilized by 15% sodium hypochlorite for 3 min, and rinsed 4 times in sterile distilled water. Seeds were submerged in water for 12 h at room temperature, and then transferred to wet filter paper to germinate at room temperature (22–25 °C) for 24 h. The uniformly germinated seeds were selected to grow in plastic pots containing Hoagland solution, in which was changed every two days. In a greenhouse, the experimental conditions included daily photo cycle of 16 h light/8 h dark (26 °C/18 °C) and 65–75% air humidity. Three biological replicates were performed under the same conditions. At the three leaf stage, the seedlings for different drought treatment times (0, 6, 12, 24 and 48 h) were cultivated in Hoagland solution containing 20% PEG600070 (ψs = −0.75) measured by VAPRO pressure osmometer (Wescor 5520, USA), whereas the seedlings for the control were only cultivated in Hoagland solution (ψs = −0.045). After treatment for different times, some sampled leaves for each biological replicates were used for physiological indicator measurement and ultrastructure observation, and the remaining leaves were kept frozen in −80 °C for later protein extraction.
Seedling morphology, physiological parameter measurement and leaf ultrastructure observation
Phenotypic, physiological characterization and ultrastructure of Bd21 seedling leaves under the control and treatment conditions were assessed. RWC measurements were conducted according to Larbi & Mekliche71 with minor modifications. Proline content and MDA content were measured, according to Bates et al.72 and Zhao et al.73 with minor modifications, respectively. Chlorophyll (chlorophyll a and b) were determined according to Zhang at al.19. Statistical analyses of the physiological measurements were conducted using an analysis of independent Student’s T-tests by SPSS statistics software (17.0). For ultrastructural observation by TEM, conventional chemical fixation, dehydration and embedding of 2 mm × 2 mm leaves segments were performed, according to the procedure described by Thiel et al.74. For electron microscopic analysis with a H-7500 transmission electron microscope (HITACHI Company, Japan).
Protein extraction
Leaf total proteins were extracted according to the method of Lv et al.21 with minor modifications. Approximately 500 mg of fresh leaves from each biological replicate was ground into a fine powder in liquid nitrogen. The ground powder was suspended in 4 mL SDS buffer (30% sucrose, 2% SDS, 100 mM Tris-HCl, pH 8.0, 50 mM EDTA-Na2, 20 mM DTT) and 4 mL phenol (Tris-buffered, pH 8.0) in a 10 mL tube, followed by the addition of 1 mM phenylmethanesulfonyl fluoride (PMSF) and 20 μL protease inhibitor cocktail (Merck KGaA, Germany) per 1 g of fresh sample to inhibit the protease activities. Samples were mixed vigorously for 15 min at room temperature, centrifuged twice at 15700× g for 15 min each, and the supernatants were precipitated with 100 mM cold ethanolamine-methanol solution at −20 °C over night. After centrifuging at 15700× g for 15 min, the pellets were rinsed with cold acetone (−20 °C) and further centrifuged three times. After freeze-drying, the pellets were added to 300 μL of solubilization buffer (7 M urea, 2 M Thiourea, 1% DTT (w/v) and 4% CHAPS) at room temperature for 2 h. Insoluble materials were removed by centrifugation at 15700× g for 15 min, and protein samples were measured using a 2-D Quant Kit (Amersham Bioscience, USA). The final protein solution was stored at −80 °C for later use.
Phosphopeptide enrichment using TiO2 microcolumns
Extracted protein mixtures were directly reduced with dithiothreitol (DTT), alkylated with iodoacetamide, and subsequently digested with endoproteinase Lys-C and trypsin as described previously75. The enrichment for phosphopeptides procedure was performed as reported by Wu et al.76 with some modifications. In detail, the TiO2 beads (GL Sciences, Tokyo, Japan) were incubated in 400 μL loading buffer containing 65% Acetonitrile (ACN)/2% trifluoroacetic acid (TFA)/saturated by glutamic acid. A total of 2 mg tryptic peptide were resolved in 600 μL loading buffer, and then incubated with appropriate amount of the TiO2 beads. After washing by 600 μL wash buffer (65% ACN/0.1% TFA), phosphopeptides were eluted twice with 300 μL elution buffer (500 mM NH4OH/60% ACN). The eluates were dried down and reconstituted in 0.1% formic acid (FA)/H2O for MS analysis.
Phosphopeptide identification using LC-MS/MS
Enriched phosphopeptides were separated on a self-packed C18 reversed phase column (75 μm I. D., 150 mm length) (Column Technology Inc.), which directly connected on the nano electrospray ion source on a LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific). Pump flow was split to achieve a flow rate at 1 μL/min for sample loading and 300 nL/min for MS analysis. The mobile phases consisted of 0.1% FA (A) and 0.1% FA and 80% ACN (B). A five-step linear gradient of 5% to 30% B in 105 min, 35% to 90% B in 16 min, 90% B in 4 min, 90% to 2% B for 0.5 min and 2% B for 14.5 min was employed. The spray voltage was set to 2.0 kV and the temperature of heated capillary was 240 °C. For each sample, triplicate LC-MS/MS experiments were performed.
For data acquisition, each MS scan was acquired at a resolution of 60,000 (at 400 m/z) with lock mass option enabled and was followed by a data-dependent top 10 MS/MS scans using collision induced dissociation (CID). The threshold for precursor ion selection was 500. Mass window for precursor ion selection was 2.0 Da. The dynamic exclusion duration was 120 s, repeat count was 1 and repeat duration was 30 s. The analyzer for the MS scans was Orbitrap and for the MS/MS scans was LTQ (37% relative collision energy). Three biological replicates were performed from sample collection to the phosphopeptide identification using LC-MS/MS.
The raw files were processed with Maxquant (version 1.2.2.25)77 and searched against own Brachypodium distachyon L. Database (NCBI_B. distachyon_25824 entries_20150320. Fasta). Up to two missing cleavage points were allowed. The precursor ion mass tolerances were 7 ppm, and fragment ion mass tolerance was 0.5 Da for MS/MS spectra. The false discovery rate (FDR) was set to < 1.0% for both peptide and protein identification, the minimum peptide length was set to 6.
Label free quantification and phosphorylation residue localization
Label free quantification was similar to the qualitative analysis on the experimental operation, but it needed more complex data requirements in the data extraction and processing methods. Each sample should be measured at least three times in parallel, in order to eliminate accidental errors. These datas of each phosphopeptide were normalized in calculated. MaxQuant software was used to calculated the integrating the ion intensities over its chromatographic elution profile coupled26,77,78. Phosphorylation residue localization was evaluated based on PTM scores that assign probabilities for each of the possible residues according to their residue-determining ions. In this study, Maxquant (version 1.2.2.25) was used to calculate PTM scores and PTM localization probabilities. Then, potential phosphorylation residues were grouped into three categories depending on their PTM localization probabilities75 namely class I (localization probability, p ≥ 0.9), class II (0.9 > p ≥ 0.75), class III (0.75 > p ≥ 0.5) and class IV (p < 0.5). The FDR of 1% was used for phosphorylation residues identification. Spectra without residue-determining ions resulted in the identification of phosphopeptides with undetermined residues.
Verification of identified phosphoproteins by Pro-Q diamond gel staining
Extracted leaf total proteins were separated by 2-DE and then 2D gels were stained with Pro-Q Diamond (Invitrogen, USA) according to the manufacturer’s instructions. Briefly, the gels were fixed twice for 30 min/each time, and washed three times with ddH2O for 10 min/each time. The gels were incubated in Pro-Q Diamond stain in darkness for 2 h. The gels were destained with 20% acetonitrile in 50 mM sodium acetate (pH 4.0) for four times (30 min/each time), and then thrice washed for 5 min/each time. Finally, the gels were scanned on a TyphoonTM 9400 scanner (GE Healthcare, USA) with a 532 nm excitation laser and a 610 nm long pass filter with a resolution of 100 microns. After fluorescent image acquisition, the gels were stained with CBB to visualize total proteins. In addition, the same 2D gels were stained with CBB to visualize total proteins and used as control. The protein spots stained by Pro-Q Diamond were further identified by matrix-assisted laser desorption/ionization time-of-flight/time-of-flight mass spectrometry (MALDI-TOF/TOF MS) according to Lv et al.21.
Bioinformatics
The significantly enriched phosphorylation motifs were extracted from the Motif-X algorithm79 (http://motif-x.med.harvard.edu/). Search Tool for the Retrieval of Interacting Genes/Proteins (STRING version 9.1) database of physical and functional interactions was used to analyze the Protein-Protein Interaction (PPI) of all the phosphoproteins identified in the current study80 (http://string-db.org). The sequences of all phosphoproteins were blasted in NCBI. Function annotations were based on Blast2Go81,82 (http://www.blast2go.com/b2ghome). Using EuKaryotic Orthologous Groups database (http://eggnog.embl.de/version_3.0/), KOG search and numbers were obtained. Then a dataset containing all the KOG numbers was used to do PPI analysis by using STRING and the interested network was displayed by Cytoscape (version 3.1.0) software (http://www.cytoscape.org/). Secondary structure of phosphoproteins was predicted via SABLE server version 2 (http://sable.cchmc.org/). Phyre2 online server (/redict the 3D structure of some interested phosphoproteins. Then the phosphorylated residues were displayed by SPDBV (version 4.1) software83 (http://spdbv.vital-it.ch/).
Additional Information
How to cite this article: Yuan, L.-L. et al. Dynamic Phosphoproteome Analysis of Seedling Leaves in Brachypodium distachyon L. Reveals Central Phosphorylated Proteins Involved in the Drought Stress Response. Sci. Rep. 6, 35280; doi: 10.1038/srep35280 (2016).
References
Barnabás, B., Jäger, K. & Fehér, A. The effect of drought and heat stress on reproductive processes in cereals. Plant Cell Environ. 31, 11–38 (2008).
Harb, A., Krishnan, A., Ambavaram, M. M. & Pereira, A. Molecular and physiological analysis of drought stress in Arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiol. 154, 1254–1271(2010).
Zhu, J. K. Salt and drought stress signal transduction in plants. Ann Rev Plant Biol. 53, 247 (2002).
Wilkinson, S. & Davies W. J., Drought, ozone, ABA and ethylene: new insights from cell to plant to community. Plant, Cell Environ. 33, 510–525 (2010).
Lee, S. C. & Luan, S. ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant, Cell Environ. 35, 53–60 (2012).
Berridge, M. J., Lipp, P. & Bootman, M. D. The versatility and universality of calcium signaling. Nat Rev Mol Cell Biol. 1, 11–21 (2000).
D’Autréaux, B. & Toledano, M. B. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 8, 813–824 (2007).
Finkel, T. Signal transduction by reactive oxygen species. J Cell Biol. 194, 7–15 (2011).
Thingholm, T. E., Jensen, O. N. & Larsen, M. R. Analytical strategies for phosphoproteomics. Proteomics. 9, 1451–1468 (2009).
Melo-Braga, M. N. et al. Modulation of protein phosphorylation, N-glycosylation and Lys-acetylation in grape (Vitisvinifera) mesocarp and exocarp owing to lobesiabotrana infection. Mol Cell Proteomics. 1, 945–956 (2012).
Tanou, G. et al. Oxidative and nitrosative‐based signaling and associated post‐translational modifications orchestrate the acclimation of citrus plants to salinity stress. Plant J. 72, 585–599 (2012).
Guo, Y. et al. Casein kinase1-like protein2 regulates actin filament stability and stomatal closure via phosphorylation of actin depolymerizing factor. Plant Cell. 28, 1422–1439 (2016).
Silva-Sanchez, C., Li, H. & Chen, S. Recent advances and challenges in plant phosphoproteomics. Proteomics. 15, 1127–1141 (2015).
Khan, M., Takasaki, H. & Komatsu, S. Comprehensive phosphoproteome analysis in rice and identification of phosphoproteins responsive to different hormones/stresses. J Proteome Res. 4, 1592–1599 (2005).
Kline, K. G., Barrett-Wilt, G. A. & Sussman, M. R. In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc Natl Acad Sci. USA 107, 15986–15991 (2010).
Nguyen, T. H. N. et al. Quantitative phosphoproteomic analysis of soybean root hairs inoculated with Bradyrhizobium japonicum. Mol Cell Proteomic. 11, 1140–1155 (2012).
Bonhomme, L., Valot, B., Tardieu, F. & Zivy, M. Phosphoproteome dynamics upon changes in plant water status reveal early events associated with rapid growth adjustment in maize leaves. Mol Cell Proteomics. 11, 957–972 (2012).
Zhang, M. et al. Comparative phosphoproteome analysis of the developing grains in bread wheat (Triticum aestivum L.) under well-watered and water-deficit conditions. J Proteome Res. 13, 4281–4297 (2014).
Zhang, M. et al. Phosphoproteome analysis reveals new drought response and defense mechanisms of seedling leaves in bread wheat (Triticum aestivum L.). J Proteomics. 109, 290–308 (2014).
Lv, D. W. et al. Proteomic and phosphoproteomic analysis reveals the response and defense mechanism in leaves of diploid wheat T. monococcum under salt stress and recovery. J Proteomics. 143, 93–105 (2016).
Lv, D. W. et al. Large-scale phosphoproteome analysis in seedling leaves of Brachypodium distachyon L. BMC Genomics. 15, 1 (2014).
Draper, J. et al. Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol. 127, 1539–1555 (2001).
Douché, T. Brachypodium distachyon as a model plant toward improved biofuel crops: Search for secreted proteins involved in biogenesis and disassembly of cell wall polymers. Proteomics. 13, 2438–2454 (2013).
Vogel, J. P. et al. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 463, 763–768 (2010).
Kosová, K. et al. Proteome analysis of cold response in spring and winter wheat (Triticum aestivum) crowns reveals similarities in stress adaptation and differences in regulatory processes between the growth habits. J Proteome Res. 12, 4830–4845 (2013).
Lv, D. W. et al. Proteome and phosphoproteome characterization reveals new response and defense mechanisms of Brachypodium distachyon leaves under salt stress. Mol Cell Proteomics. 13, 632–652 (2014).
Sharma, P. et al. Phosphorylation of mek1 by cdk5/p35 down-regulates the mitogen-activated protein kinase pathway. J Biol Chem. 277(1), 528–534 (2002)
Oesch-Bartlomowicz, B. & Oesch, F. Cytochrome-p450 phosphorylation as a functional switch. Arch Biochem Biophy. 409(1), 228–234 (2003).
Lv, D. W. et al. Integrative network analysis of the signaling cascades in seedling leaves of bread wheat by large-scale phosphoproteomic profiling. J Proteome Res. 13(5), 2381–2395 (2014).
Yeong, S. S. et al. The last 10 amino acid residues beyond the hydrophobic motif are critical for the catalytic competence and function of protein kinase Cα. J Biol Chem. 281(41), 30768–30781 (2006).
Yao, Q. et al. P3DB: An integrated database for plant protein phosphorylation. Front Plant Sci. 3, 206 (2012).
Rose, C. M. et al. Rapid phosphoproteomic and transcriptomics changes in the rhizobia-legume symbiosis. Mol Cell Proteomics. 11(9), 724−744 (2012).
Heazlewood, J. L. et al. PhosPhAt: a database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Res. 36(D), D1015−D1021 (2008).
Guo, M. & Huang, B. X. Integration of phosphoproteomic, chemical, and biological strategies for the functional analysis of targeted protein phosphorylation. Proteomics. 13, 424–437 (2013).
Park, S. Y. et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 324, 1068–1071 (2009).
Gómez-Cadenas, A. et al. An abscisic acid-induced protein kinase, PKABA1, mediates abscisic acid-suppressed gene expression in barley aleurone layers. Proc Natl Acad Sci. USA. 96, 1767–1772 (1999).
Zhang, H., Mao, X., Wang, C. & Jing, R. Overexpression of a common wheat gene TaSnRK2. 8enhances tolerance to drought, salt and low temperature in Arabidopsis. PLoS One. 5, e16041 (2010).
Sheard, L. B. & Zheng, N. Plant biology: signal advance for abscisic acid. Nature. 462(7273), 575–576 (2009).
Meng, X. & Zhang, S. MAPK cascades in plant disease resistance signaling. Ann Rev Phytopath. 51, 245–266 (2013).
Wen, Y. et al. Characterization and expression analysis of mitogen-activated protein kinase cascade genes in wheat subjected to phosphorus and nitrogen deprivation, high salinity, and drought. J Plant Biochem Biotechnol. 24, 184–196 (2015).
Moustafa, K., AbuQamar, S., Jarrar, M., Al-Rajab, A. J. & Trémouillaux-Guiller, J. MAPK cascades and major abiotic stresses. Plant Cell Rep. 33, 1217–1225 (2014).
Moustafa, K., De Vos, D. L., Leprince, A. S., Savouré, A. & Lauriere, C. Analysis of the Arabidopsis mitogen-activated protein kinase families: organ specificity and transcriptional regulation upon water stresses. Sch Res Exch. 225, 196–214 (2008).
Nelson, D. E. et al. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc Natl Acad Sci. USA 104, 16450–16455 (2007).
Petroni, K. et al. The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell. 24, 4777–4792 (2012).
Swindell, W. R., Huebner, M. & Weber, A. P. Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics. 8, 1 (2007).
Li, J. & Buchner, J. Structure, function and regulation of the hsp90 machinery. Biomed J 36, 106 (2013).
Mishra, S. K., Tripp, J., Winkelhaus, S., Tschiersch, B. & Theres, K. In the complex family of heat stress transcription factors, HSFA1 has a unique role as master regulator of thermo tolerance in tomato. Gene Dev. 16, 1555–1567 (2002)
Liu, H. C., Liao, H. T. & Charng, Y. Y. The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 34. 738–751 (2011)
Gomes, D. et al. Aquaporins are multifunctional water and solute transporters highly divergent in living organisms. BBA-Biomembranes. 1788, 1213–1228 (2009).
Guenther, J. F. et al. Phosphorylation of soybean nodulin 26 on serine 262 enhances water permeability and is regulated developmentally and by osmotic signals. Plant Cell. 15, 981–991 (2003).
Morth, J. P. et al. A structural overview of the plasma membrane Na+, K+-ATPase and H+-ATPase ion pumps. Nat Rev Mol Cell Bio. 12, 60–70 (2011).
Wang, H., Wu, K., Liu, Y., Wu, Y. & Wang, X. Integrative proteomics to understand the transmission mechanism of Barley yellow dwarf virus-GPV by its insect vector Rhopalosiphum padi. Sci Rep. 5. 10971 (2015)
Hayashi, M., Inoue, S. I., Takahashi, K. & Kinoshita, T. Immunohistochemical detection of blue light-induced phosphorylation of the plasma membrane H+-ATPase in stomatal guard cells. Plant Cell Physiol. 52, 1238–1248 (2011).
Wang, Y. et al. Overexpression of plasma membrane H+-ATPase in guard cells promotes light-induced stomatal opening and enhances plant growth. Proc Natl Acad Sci. USA 111, 533–538(2014).
Upadhyaya, H., Khan, M. H. & Panda, S. K. Hydrogen peroxide induces oxidative stress in detached leaves of Oryza sativa L. Plant Physiol. 33, 83–95 (2007).
Liu, Y. & He, C. Regulation of plant reactive oxygen species (ROS) in stress responses: learning from atrbohd. Plant Cell Rep. 35(5), 1–13 (2016).
Liu, B., Qin, F., Liu, W. & Wang, X. Differential proteomics profiling of the ova between healthy and rice stripe virus-infected female insects of Laodelphax striatellus. Sci Rep. 6, 27216 (2016).
Asada, K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 141, 391–396 (2006).
Bailey-Serres, J. & Mittler, R. The roles of reactive oxygen species in plant cells. Plant Physiol. 141, 311–311 (2006).
Kargul, J. & Barber, J. Photosynthetic acclimation: Structural reorganisation of light harvesting antenna–role of redox‐dependent phosphorylation of major and minor chlorophyll a/b binding proteins. FRBS J. 275, 1056–1068 (2008).
Vilardell, J. et al. Gene sequence, developmental expression, and protein phosphorylation ofRAB-17 in maize. Plant Mol Biol. 14, 423–432 (1990).
Jensen, A. B., Goday, A., Figueras, M. & Jessop, A. C. Phosphorylation mediates the nuclear targeting of the maize Rab17 protein. Plant J. 13, 691–697 (1998).
Hao, P. C. et al. An integrative proteome analysis of different seedling organs in tolerant and sensitive wheat cultivars under drought stress and recovery. Proteomics. 15, 1544–1563 (2015).
Ito, J. et al. A survey of the Arabidopsis thaliana mitochondrial phosphoproteome. Proteomics. 9, 4229–4240 (2009).
Nakagami, H. et al. Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 153, 1161–1174 (2010).
Frydman, J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Ann Rev Biochem. 70, 603–647 (2001).
Richter, K. & Buchner, J. Hsp90: chaperoning signal transduction. J Cell Physiol. 188, 281–290 (2001).
Pratt, W. B., Krishna, P. & Olsen, L. J. Hsp90-binding immunophilins in plants: the protein movers. Trends Plant Sci. 6, 54–58 (2001).
Young, J. C., Moarefi, I. & Hartl, F. U. Hsp90: a specialized but essential proteinfolding tool. J Cell Biol. 154, 267–273 (2001).
Li, Z. et al. The physiological and itraq-based proteomic analyses reveal the function of spermidine on improving drought tolerance in white clover. J Proteome Res. 15, 1563−1579 (2016).
Larbi, A. & Mekliche, A. Relative water content (RWC) and leaf senescence as screening tools for drought tolerance in wheat. Options Mediterr. Série ASém. Méditerr. 60, 193–196 (2004).
Bates, L. S., Waldren, R. P. & Teare, I. D. Rapid determination of free proline for water-stress studies. Plant Soil. 39, 205–207 (1973).
Zhao, S., Xu, C. C., Zou, Q. & Meng, Q. W. Improvements of method for measurement of malondialdehyde in plant tissues. Physiol. Commun. 30, 207–210 (1994).
Thiel, J. et al. Differentiation of endosperm transfer cells of barley: a comprehensive analysis at the micro-scale. Plant J. 71, 639–655 (2012).
Olsen, J. V. et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 127, 635–648 (2006).
Wu, J. et al. Global profiling of phosphopeptides by titania affinity enrichment. J Proteome Res. 6, 4684–4689 (2007).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 26, 1367–1372 (2008).
Sandin, M., Krogh, M., Hansson, K. & Levander, F. Generic workflow for quality assessment of quantitative label-free LC–MS analysis. Proteomics. 11, 1114–1124 (2011).
Schwartz, D. & Gygi, S. P. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol. 23, 1391–1398 (2005).
Franceschini, A. et al. STRING v9. 1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815 (2013).
Conesa, A. & Götz, S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Intl J Plant Genomics. 2008, 619832 (2008).
Liu, W., Gray, S., Huo, Y., Li, L., Wei, T. & Wang, X. Proteomic analysis of interaction between a plant virus and its vector insect reveals new functions of hemipteran cuticular protein. Mol Cell Proteomics. 14(8), 2229–2242 (2015).
Guex, N. & Peitsch, M. C. SWISS‐MODEL and the Swiss‐Pdb Viewer: an environment for comparative protein modeling. Electrophoresis. 18, 2714–2723 (1997).
Acknowledgements
This research was financially supported by the grants from the Ministry of Science and Technology China (2016YFD0100500) and the International Science & Technology Cooperation Program of China (2013DFG30530).
Author information
Authors and Affiliations
Contributions
L.Y., M.Z. and X.Y. performed the experiment, data analysis, draft the manuscript. Y.B. and S.Z. contributed to analyze and interpret experimental data. Y.Y. designed and supervised the project. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Yuan, LL., Zhang, M., Yan, X. et al. Dynamic Phosphoproteome Analysis of Seedling Leaves in Brachypodium distachyon L. Reveals Central Phosphorylated Proteins Involved in the Drought Stress Response. Sci Rep 6, 35280 (2016). https://doi.org/10.1038/srep35280
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep35280
This article is cited by
-
Protein phosphorylation associated with drought priming-enhanced heat tolerance in a temperate grass species
Horticulture Research (2020)
-
DynaVenn: web-based computation of the most significant overlap between ordered sets
BMC Bioinformatics (2019)
-
Identification of phosphorylation proteins in response to water deficit during wheat flag leaf and grain development
Botanical Studies (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.