Abstract
In 2011, a severe outbreak of hemolytic-uremic syndrome was caused by an unusual, highly virulent enterohemorrhagic E. coli (EHEC) O104:H4 strain, which possessed EHEC virulence traits in the genetic background of human-adapted enteroaggregative E. coli. To determine magnitude of fecal shedding and site of colonization of EHEC O104:H4 in a livestock host, 30 (ten/strain) weaned calves were inoculated with 1010 CFU of EHEC O104:H4, EHEC O157:H7 (positive control) or E. coli strain 123 (negative control) and necropsied (4 or 28 d.p.i.). E. coli O157:H7 was recovered until 28 d.p.i. and O104:H4 until 24 d.p.i. At 4 d.p.i., EHEC O104:H4 was isolated from intestinal content and detected associated with the intestinal mucosa. These results are the first evidence that cattle, the most important EHEC reservoir, can also carry unusual EHEC strains at least transiently, questioning our current understanding of the molecular basis of host adaptation of this important E. coli pathovar.
Similar content being viewed by others
Introduction
In early May 2011, an unprecedented large outbreak of hemolytic-uremic syndrome (HUS) and hemorrhagic colitis (HC) started in Germany and subsequently spread throughout Europe and North America. Nearly 4,000 individuals were affected, including almost 900 cases of HUS and 54 deaths1. The outbreak was linked to the consumption of contaminated sprouted fenugreek seeds2. An unusual enterohemorrhagic Escherichia coli (EHEC) strain of serotype O104:H4 was identified as the causative agent1. The strain possesses a combination of virulence factors from both Shiga toxin (Stx)-producing E. coli (STEC) and enteroaggregative E. coli (EAEC) strains3,4 and represents a novel hybrid pathotype of enterohemorrhagic E. coli (EHEC).
EHEC, the human pathogenic subset of STEC, has emerged as an important cause of human disease in developed countries5. The pathogens are transmitted to humans through consumption of raw or undercooked food or water contaminated by ruminant feces6 or via direct contact with infected persons or animals7. Clinical symptoms of EHEC infections range from mild diarrhea to HC8 and may be complicated by the potentially fatal HUS9. Characterization of the epidemic EHEC O104:H4 strain showed that it carries stx2 encoding for the eponymous virulence factor of STEC but, in contrast to typical EHEC, lacks the locus of enterocyte effacement (LEE). EHEC O104:H4 instead harbors genetic markers of EAEC (aatA, aggA, aggR, aap) located on the EAEC pAA virulence plasmid and expresses the corresponding phenotype of aggregative adherence to intestinal epithelial cells3,10. Typical EAEC are commonly associated with acute and persistent (>14 days) diarrhea in children11, immunosuppressed persons12, and travelers to developing countries13, and with some cases of diarrhea in developed countries14.
The principal reservoir of STEC strains associated with human disease are ruminants. Several bacterial factors (e.g., intimin15, other products of the LEE16, and Efa-117) promote bacterial colonization of the intestinal mucosa resulting in attaching and effacing lesions of E. coli O157:H7 on epithelial cells. Stxs primarily delay the host’s cellular immune response, thereby generating an opportunity for STEC bacterial colonization18. Rectum and ileocecal valve (ICV) are sites most likely to contain STEC O157:H7 bacteria19,20,21. STEC can be regularly detected in cattle herds over long periods, mainly in clinically healthy animals22. The relatively efficient transmission of STEC O157:H7 from calf-to-calf plays an important role in increasing the prevalence of cattle excreting this agent on farms23.
Except stx2, the EHEC O104:H4 outbreak strain lacks other putative STEC-associated virulence and colonization factors3. Cattle have not been reported to carry E. coli strains belonging to the EAEC pathovar, which is suspected to be adapted to humans24,25,26. Currently available data also suggest that cattle are not the reservoir of the E. coli O104:H4 outbreak strain24,27,28. It remains to be determined whether this results from an inability of the strain to colonize cattle, from insufficient exposure of the cattle population to (sufficient numbers of) the bacteria, from insufficiently low frequencies of transmission events, or combinations thereof. It is tempting to speculate that the large 2011 outbreak in the human population affecting several thousand individuals and with an undisclosed number of asymptomatic shedders29 could have led to a spillover of the strain to wild-life or livestock populations including cattle. Indeed, we detected genes characteristic of the EHEC O104:H4 strain (stx2, aggR, wzxO104, fliCH4) in fecal pools sampled in a German abattoir processing animals from farms located near the outbreak epicenter two years after the outbreak onset, while contemporarily obtained samples from an unrelated geographical area (Spain) were negative for this combination of genes30. The epidemiological evidence to suggest that the outbreak strain has already conquered an animal reservoir is weak. Currently a sound basis for a risk-assessment justifying the establishment or abandoning of targeted preventive measures by veterinary and public health authorities can only come from experimental data generated by infection studies. This prompted the current investigation assessing for the first time if EHEC strains as EHEC O104:H4 can utilize ruminants as reservoir. To this end, we determined the clinical appearance, the pattern and magnitude of fecal shedding and the site of colonization in a well described bovine infection model31,32.
Results
Duration and magnitude of fecal shedding of inoculum strains
Fecal samples from all calves inoculated with 1010 CFU were culture positive for the respective inoculum strain at several sampling points after inoculation but the magnitude and duration of detectable shedding varied between individuals (Fig. 1).
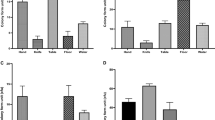
Fecal shedding of E. coli by calves inoculated with different strains.
Numbers of inoculum bacteria recovered from feces of calves inoculated with EHEC O157:H7, EHEC O104:H4 or E. coli strain 123 and followed up for four (Trial 1, N = 5 calves/group; (A)) and 28 days post inoculation (Trial 2, N = 5 calves/group; (B)) are shown. Data is presented as mean numbers ( ± standard errors of the means) of CFU/g for the indicated groups. Significant differences (Mann-Whitney U-test, p < 0.05) in mean fecal shedding between a: E. coli strain 123 and EHEC O157:H7, b: E. coli strain 123 and EHEC O104:H4, c: EHEC O157:H7 and EHEC O104:H4. All values below the dashed line represent enrichment levels. The arrow indicates the date of challenge. a.m. = ante meridiem, p.m. = post meridiem.
In Trial 1, fecal shedding of EHEC O104:H4 decreased after an initial peak of >105 CFU/g with a time slope similar to that of the EHEC O157:H7. Both strains were shed by the respective animal groups at approximately 105 CFU/g feces by day 3 pi. EHEC O104:H4 were recovered in higher numbers and for a longer time than the E. coli strain 123, which were only detectable in 2 calves at 4 dpi. There is a significant difference (p < 0.05) in shedding between E. coli strain 123 and EHEC O157:H7 on days 3 and 4 pi. No significant differences were seen between the numbers of EHEC O104:H4 CFU/g shed in the feces and the respective numbers for E. coli strain 123 or EHEC O157:H7.
A second trial conducted for a longer period reproduced the shedding pattern during the first 4 days. Later on, numbers of shed EHEC O104:H4 bacteria decreased dramatically beginning at 4 dpi. Whereas the E. coli strain 123 was not detected from 12 dpi onwards, EHEC O104:H4 were detected intermittently until 24 dpi in one calf, on days 16 and 18 pi only by enrichment. Another calf was positive for EHEC O104:H4 by enrichment culture until day 18 pi. EHEC O157:H7 were detected in all 5 calves (2 calves by enrichment culture) until day 12 pi and in 4 calves (2 calves by enrichment culture) until 28 dpi. The remaining calf was positive for EHEC O157:H7 bacteria until day 26 pi by enrichment culture. All 5 calves were negative for EHEC O157:H7 at least at one intermediate sampling date. The differences in fecal shedding between E. coli strain 123 and EHEC O157:H7 were statistically significant (p < 0.05, Mann-Whitney test) between days 1 and 28, except on days 14, 24 and 26. There was also a significant difference in shedding between E. coli strain 123 and EHEC O104:H4 on day 2 and between EHEC O157:H7 and EHEC O104:H4 on day 6, 8, 18 and 28.
Distribution of the inoculum strains at necropsy
No gross abnormalities of the mucosa of the forestomaches, the small or large intestines were observed in any of the calves during necropsy at 4 dpi. In Trial 1 EHEC O104:H4 were recovered in similar numbers as EHEC O157:H7 from feces and contents of proximal colon and rectum (RAJ contents) collected from calves inoculated with the respective inoculum strains (Table 1). Significantly lower numbers of the E. coli strain 123 were recovered from calves inoculated with E. coli 123. EHEC O104:H4 also were recovered from the contents of rumen, abomasum, and gall bladder and from most of the intestinal tissue samples. The numbers of EHEC O104:H4 and EHEC O157:H7 isolated and the number of calves positive for these strains were low within the proximal small intestine and increased in sites distal to the ileum. Distal sites most commonly containing EHEC O104:H4 and EHEC O157:H7 included the ICV, cecum, proximal colon, spiral colon, distal colon, and recto-anal junction (RAJ). EHEC O104:H4 were recovered in equal numbers as EHEC O157:H7. Numbers of EHEC O157:H7 and EHEC O104:H4 significantly differed to bacterial numbers of E. coli strain 123 at proximal colon and RAJ. Neither EHEC O104:H4, nor EHEC O157:H7 or E. coli 123 were isolated from intestinal lymph nodes, gall bladder (tissue-associated), liver or abomasum (tissue-associated).
Only EHEC O157:H7 were detectable in feces, intestinal content, and associated with intestinal tissue at 28 dpi in Trial 2. EHEC O157:H7 was sporadically recovered from the jejunum (1.6 × 103 CFU/g tissue) and content of proximal colon (102 CFU/g sample) of one calf (#24). EHEC O157:H7 bacteria were also isolated from Peyer´s patches of the ileum (IPP; 4.0 × 102 CFU/g tissue) of one calf (#25), but detected in higher numbers at the RAJ (4.0 × 102 to 4.8 × 104 CFU/g tissue; calf #21, #22, and #25), corroborating the findings at 4 dpi. Duodenum, Peyer´s patches of the jejunum, ICV, caecum, proximal colon, spiral colon, distal colon, content of the rectum, and bile were additionally examined, but negative for the inoculum strains.
Detection of adherent bacteria by microscopy
Adherent O157-positive bacteria were observed in defined mucosal sites of all 5 O157-inoculated animals. Single adherent O157-positive bacteria were seen in one calf (#13) at the IPP and in 3 calves (#4, #12 and #13) at the ICV (Supplementary Table 1). In 3 calves (#5, #6, and #13) O157-positive bacteria formed microcolonies on the surface epithelium of the rectal mucosa and single adherent bacteria were present on the squamous epithelium of the RAJ (Fig. 2).
Differences in the localization and pattern of adherence of anti-O157 and anti-O104 positive bacteria at the RAJ from calves necropsied 4 dpi.
(A) Calf #13, inoculation strain: EHEC O157:H7; O157-positive bacteria form micro-colonies on the epithelium of the rectal mucosa (arrow) and single bacteria are attached to squamous epithelial cells of the RAJ (arrowheads); (B) Calf #14, inoculation strain: EHEC O104:H4; numerous O104-positive bacteria surround detached squamous epithelial cells and single O104-positive bacteria are attached to squamous epithelial cells of the RAJ (arrows). Indirect immunoperoxidase for O157 (A) and O104 (B), bar = 100 μm.
Adherent O104-positive bacteria were seen on the squamous epithelium of the RAJ (Fig. 2) in one (calf #14) of the 5 O104-inoculated calves. The majority of bacteria were associated with detached squamous epithelial cells and single cells were present on the luminal surface of the squamous epithelium. There was no formation of microcolonies and no aggregative adherence. O104-positive bacteria were also found in dilated crypts of lymphoglandular complexes at the ICV that were filled with neutrophils and necrotic debris. Multiple groups of bacteria were seen between the neutrophils and large numbers were attached to the necrotic debris (Fig. 3). In addition, O104-positive bacteria were observed in the neutrophilic exudate of a dilated tonsillar crypt of this calf.
Localization of EHEC O104:H4 at the ICV of calf #14.
(A) Overview of the dilated crypt of a lymphoglandular complex filled with neutrophils and cellular debris. Multiple groups of O104-positive bacteria are interspersed between neutrophils. Numerous bacteria are also associated with necrotic debris. (B) Higher magnification of O104-positive bacteria between neutrophils and attached to necrotic debris. Indirect immunoperoxidase for O104; (A) bar = 500 μm, (B) bar = 50 μm.
O104- and O157-positive bacterial cells were found in the gut lumen and the intestinal content in every calf inoculated with the respective strain. Large numbers of bacteria were observed on the luminal surface of epithelial cells in some sections of the gastrointestinal tract, but they were not labelled by the primary antibodies used.
Further characterization of single isolates
Selected colonies recovered from fecal samples, intestinal content, and tissues were verified to be inoculum-type bacteria by multiplex PCR. In total, 315 randomly selected colonies from fecal samples and 189 colonies from intestinal tissues or contents were verified as EHEC O104:H4 inoculum strain by HUSEC041/EAEC multiplex PCR (Supplementary Fig. 1). Ten isolates possessed an additional astA gene, which was absent from the strain pre-inoculation. Seven of these colonies were obtained from one calf (#14). All examined isolates of EHEC O104:H4 contained the ESBL gene loci and the pESBL (data not shown). In contrast, 116 of 315 examined EHEC O104:H4 isolates had lost aggR (Supplementary Fig. 1) and the corresponding pAA plasmid (data not shown) during passage. The percentage of aggR-negative colonies steadily increased during the course of the experiment (Fig. 4). Colony morphology did not differ between aggR-positive and -negative colonies. By PFGE analysis of XbaI-restricted DNA, two differing DNA fragments of <48.5 kbp length were visible (Fig. 5). The presence of the DNA fragments corresponded to the presence of aggR as tested by multiplex PCR in the respective re-isolates (Supplementary Fig. 1).
Genetic relatedness of the inoculation strain EHEC O104:H4 and coliform bacteria from fecal samples of calf #30 (Trial 2) taken on days 3 and 8.
Overall, XbaI-digested DNA of 15 ESBL-positive colonies were analyzed by PFGE that were either putative EHEC O104:H4 re-isolates (stx2- and aggR-positive [n = 5; 3 dpi colonies no. 1, 3, 4 and 5 and 8 dpi colony no. 2] and stx2-positive but aggR-negative [n = 5] as determined by multiplex PCR [Supplementary Fig. 1]) or other coliform bacteria (stx2-negative, n = 5). 1% TBE agarose gel stained with ethidium bromide. Ctrl: EHEC O104:H4 inoculation strain, M: Lambda Ladder PFG Marker, kbp: kilobases.
Discussion
In 2011, a novel hybrid EHEC strain of serotype O104:H4, possessing a combination of virulence factors of both STEC and EAEC, caused an unprecedented, food-borne outbreak in humans. E. coli strains belonging to the EAEC pathovar have not been detected in cattle, the primary STEC/EHEC reservoir thus far. The same holds for hybrid strains like the EHEC O104:H4 outbreak strain even though cattle populations in the outbreak area have intensively been monitored during the outbreak24,27,28. However, as exposure of the cattle population to hybrid EHEC strains like EHEC O104:H4 might occur, the current study aimed at assessing the possibility that strains with this genetic make-up can cross the interspecies barrier from humans to livestock.
The study clearly demonstrated that weaned calves can be colonized by EHEC O104:H4. All calves in these trials shed EHEC O104:H4 following oral administration in numbers indicative of proliferation. Calves had higher fecal levels of inoculated bacteria than had calves inoculated with a non-pathogenic E. coli control strain, which is taken as one line of evidence for colonization16,19. After an initial period of shedding at levels equivalent to the high level shedding of EHEC O157:H7, numbers of shed EHEC O104:H4 declined below the detection limit within 28 days. Calves showed large variation in duration of EHEC O104:H4 shedding, but all calves were culture positive for a minimum of three days. EHEC O104:H4 were detectable for longer periods of time than non-pathogenic E. coli 123. One of the experimentally-infected calves even shed EHEC O104:H4 until 24 dpi. These results indicate that a niche in the bovine intestinal tract exists that promotes the replication of EHEC O104:H4 significantly better than that of non-pathogenic E. coli, i.e. a stable association between the bacteria and the mucosa.
Accordingly, inoculum-type bacteria were recovered from multiple intestinal sites where E. coli strains considered capable of intestinal colonization are found32. EHEC O104:H4 bacteria were most frequently detected in the large intestine (e.g. ICV, cecum, proximal colon, spiral colon, distal colon, RAJ). Intestinal levels of inoculum-type bacteria and number of animals positive were similar in EHEC O104:H4 inoculated and EHEC O157:H7 inoculated calves and much higher than in calves inoculated with non-pathogenic E. coli. In contrast to EHEC O157:H7, EHEC O104:H4 were not detected at any intestinal site 28 days post inoculation. However, colonization is also defined by the presence of adherent bacteria at 4 dpi31,32,33. We detected adherent bacteria in the RAJ of one calf necropsied 4 days after inoculation with EHEC O104:H4. Bacteria were attached to the surface of squamous epithelial cells. It is conceivable that numbers of EHEC O104:H4 in the other calves of this group were too low or focally distributed to be detected. Adherent bacteria were detected in all calves inoculated with EHEC O157:H7. We confirmed the mucosa-associated lymphoid tissue at the ICV and the rectal mucosa as sites in calves most likely to harbor EHEC O157:H7 A/E-inducing bacteria at 4 dpi19,20,21,32. EHEC O157:H7 bacteria were forming microcolonies on enterocytes of the rectal mucosa. The rectum and the RAJ appear to be the principal site of EHEC O104:H4 and EHEC O157:H7 colonization, although there are differences in the mode of attachment and cell types targeted. EHEC O104:H4 were bacteriologically detectable in the contents of the rumen and abomasum. This could be explained by re-infection through general exposures such as contaminated feeds or water sources or by calf-to-calf transmission. It may also be that EHEC O104:H4 is able to adhere to the squamous epithelium of the rumen. We identified the gall bladder as another site where EHEC O104:H4 settled in two calves. Thus, the gall bladder must be considered a possible niche for EHEC O104:H4, similar as for EHEC O157:H732, and may be an additional source of intestinal EHEC O104:H4. At necropsy, the O104-bacteria were also isolated and histologically detected in the tonsils of one calf. This finding may be due to the application of the inoculum (e.g., while retracting the tube after the application) or regurgitation of stomach content and it is not known if occurrence of O104:H4 at this site was only transient. However, EHEC O104:H4 showed an affinity to neutrophils and necrotic debris and because of the structure of the bovine tonsil there may be no need for bacterial attachment at epithelial cells for colonization34, introducing tonsils as another biological niche for EHEC O104:H4. Because there was no recovery from lymph nodes (Lnn. jejunalis, Lnn. cecalis) and liver, EHEC O104:H4 were not invasive in our model.
EHEC O104:H4 bacteria did not cause A/E lesions, as was expected since it lacks the eae gene3. Surprisingly, this enteroaggregative E. coli strain did not show aggregative adherence to bovine intestinal epithelial cells. Bacteria adhered usually as sparse single or few bacilli. Therefore, it is difficult to differentiate them from intestinal material that is labelled non-specifically. The lack of aggregative adherence gives evidence that colonization of the bovine intestine by EHEC O104:H4 does not rely on pAA-encoded adhesion factors. Similar findings were gained in an infant rabbit-based model, where pAA is dispensable for intestinal colonization and development of intestinal lesions35. Adherence of EHEC O104:H4 to bovine epithelial cells is perhaps mediated by putative adhesins as the iron‐regulated gene A homologue adhesin (Iha), which is responsible for adherence to epithelial cells in other eae-negative STEC36. Adherent EHEC O104:H4 were primarily found on squamous epithelial cells of the RAJ. EHEC O157:H7 bacteria have been reported to colonize both glandular and squamous epithelial cells20,32,37, but in vitro examination revealed that the respective underlying mechanisms differ37. Whether LEE-independent mechanisms to colonize the squamous epithelial cells also differ between EHEC O104:H4 and EHEC O157:H7 remains a hypothetical consideration at present. It also needs to be elucidated, whether EHEC O104:H4 has a general preference for squamous epithelium or if it is limited to lower sections of the bovine gastrointestinal tract. In this study, EHEC O104:H4 were also found in dilated crypts of lymphoglandular complexes and dilated tonsillar crypts that were filled with neutrophils and necrotic debris. EHEC O104:H4 may use aggrieved parts of the mucosa as another biological niche.
Analyses of sequential fecal samples demonstrated that EHEC O104:H4 are subject to a steady change of variable genetic elements. The virulence plasmid of enteroaggregative E. coli (pAA) is a relatively unstable genetic element as demonstrated by its intra-host loss during course of human infection38. A role of the pAA for adherence of EHEC O104:H4 to the bovine intestine remains obscure, because aggR-positive EAEC have rarely been detected in cattle30,39. pAA-negative derivatives isolated from humans lost their ability to efficiently colonize the intestinal epithelium, which results in lack of systemic absorption of Stx2 and diminished the ability of EHEC O104:H4 to cause HUS38. In this study, the adherence patterns of EHEC O104:H4 suggest that pAA is not necessary for colonization of bovine epithelium. Moreover, the selection of pAA-negative EHEC O104:H4 in bovines may lead to an enrichment of strains with reduced colonization efficiencies in humans. In contrast to the lability of pAA, the stx2a gene of EHEC O104:H4, which is encoded on an inducible bacteriophage40, and the ESBL plasmid were stable during bovine infection as demonstrated by their presence in all isolates and as reported for human isolates38. Some isolates even acquired the astA gene encoding EAST1 (enteroaggregative Escherichia coli heat-stable enterotoxin 1), which is a small protein first detected in EAEC, but is present also in other pathovars of intestinal E. coli41. Strains expressing EAST1 have been shown to induce diarrhea principally in humans, but have also been found in piglets and calves42. Beside the loss of the pAA plasmid no other genetic modifications were detectable in the re-isolates. By PFGE analysis only small DNA fragments differed between aggR-positive and -negative re-isolates corresponding in their molecular size to XbaI-fragments of the pAA plasmid as calculated by the plasmid DNA sequence (PubMed accession no. CP011332).
Inoculation of calves with 1010 CFU of EHEC O104:H4 did not induce significant clinical disease. The transient non-bloody diarrhea noted in some calves may have been due to fasting before inoculation or endotoxin absorption associated with the administration of massive numbers of gram-negative bacteria33. Spontaneous, transient diarrhea occurs occasionally in otherwise healthy experimental calves43. Although being fairly non-pathogenic for cattle similar to classical STEC strains, EHEC O104:H4 appear to be less well adapted to bovine intestine than classical STEC strains. EHEC O104:H4 was not yet found in cattle herds27,28,44 but the present experimental infection study, as well as epidemiological evidence gathered in the outbreak region30, support the hypothesis that EHEC O104:H4 and probably other EAEC may be or become part of the E. coli microbiome in this livestock species and contribute to and participate in the genome plasticity through uptake and loss of virulence genes45. Future risk assessments must therefore take cattle as an animal reservoir for EHEC/EAEC hybrid strains and a potential source of transmission to humans into account.
Methods
Bacterial strains and inocula
EHEC O104:H4 strain LB226692 (Table 2) was isolated during the outbreak 2011 in Germany3 and fully characterized by whole genome sequencing4. The strain has an ESBL phenotype3. EHEC O157:H7 strain 86–24 is a nalidixic acid-resistant mutant of an Stx2-producing strain isolated from an outbreak in Washington state46 and was used as a positive control. A nalidixic acid resistant mutant of E. coli strain 123 (O43:H28), a porcine isolate which does not produce Stx and is not pathogenic in cattle47, was used as a negative control strain. For ease of reading, strain 123 is referred to as “E. coli strain 123”, strain 86–24 Nal as “EHEC O157:H7” and strain LB226692 as “EHEC O104:H4” throughout the manuscript. Stock inocula containing approximately 1010 CFU/ml were prepared as previously described31 and stored in 1 ml aliquots at −80 °C until used. Actual numbers of viable EHEC O104:H4 bacteria in each thawed batch were confirmed by bacterial colony counts on Brilliance ESBL agar plates (Oxoid, Basingstoke, United Kingdom), whereas EHEC O157:H7 and the E. coli strain 123 were quantitated on MacConkey agar containing 50 mg of nalidixic acid per ml.
Animals and animal treatments
A detailed description of the animal model is given in the Supplementary Methods. In brief, weaned female Holstein calves were housed in an environmentally controlled animal facility at the Friedrich-Loeffler-Institut (FLI) on the Isle of Riems, Greifswald. The experimental protocol was reviewed by an independent animal welfare and ethics committee, approved by the local authority (State Office for Agriculture, Food Safety and Fisheries of Mecklenburg-Western Pomerania, Rostock, Germany, reference no. 7221.3-1.1-093/12), and carried out in accordance with the approved guidelines.
All calves were clinically normal at the time of inoculation. Calves (five/group) were inoculated intra-rumenally with1010 CFU of the particular strain.
In the short-term experiment (Trial 1), calves were observed twice a day when fecal samples were taken (0 to 4 days post inoculation [dpi]). Peripheral venous blood samples were collected pre-inoculation and every second day. Necropsy and sampling were performed at 4 dpi as previously described48. In the long-term experiment (Trial 2) calves were observed twice a day and fecal samples were collected daily in the morning for the first four days (0 to 4 dpi) and then every second day (6 to 28 dpi). Peripheral venous blood samples were collected pre-inoculation and once a week. Necropsy and sampling were performed at 28 dpi.
Bacteriologic examination
A comprehensive description is given in the Supplementary Methods. Briefly, fecal samples were obtained from all calves and screened for inoculum-type bacteria and antibiotic-resistant normal flora by plating on selective media used for quantitating the inoculum strains. Selected colonies were tested for O43, O104, and O157 antigens by slide agglutination using appropriate antigen-specific sera (E. coli O43 antiserum, E. coli K9 serum49, E. coli OK O157 antiserum; Serum Statens Institut, Denmark). For isolation of single stx2-positive colonies, colony hybridization was done as previously described22. Up to 5 stx2-positive and 5 stx2-negative colonies per sample were sub-cultured and stored with 30% glycerine.
Histologic studies
Tissues were fixed in NBF for 24 to 48 h, embedded in paraffin, sectioned, and stained with hematoxylin and eosin for routine histology. Bacteria were identified in formalin-fixed intestinal tissues by indirect immunoperoxidase method using rabbit E. coli OK O157 antiserum and rabbit E. coli K9 serum49 (Serum Statens Institut, Denmark) as the primary antibodies and peroxidase-conjugated anti-rabbit IgG antiserum (Jackson ImmunoResearch Laboratories Inc., West Grove, USA) as secondary antibody. Antigens were retrieved by trypsin digestion50. Sections were counterstained with Mayer´s hemalum.
Further characterization of single isolates
Multiplex PCR targeting typical molecular features of EHEC O104:H4 (rfbO1043, fliCH43, stx251, terD52, aggR53, pic54, and astA55) and EHEC O157:H7 (rfbO15756, fliCH757, stx258, and terD52) was used for further characterization of the stored cultures. Presence of the ESBL gene loci (blaTEM59, blaCTX-M60, and blaCTX-M-1561) was also examined by multiplex PCR. Multiplex PCR was done with the PANScript DNA-Polymerase (PAN-Biotech GmbH, Aidenbach, Germany) in a total volume of 27 μL containing 3 μL of bacterial DNA. PCR conditions included initial denaturation at 95 °C for 1 min, 35 cycles of denaturation (95 °C, 30 s), annealing (55 °C, 60 s), and extension (72 °C, 60 s), and final extension at 72 °C for 5 min. Contour-clamped homogeneous electric field-pulsed-field gel electrophoresis (CHEF PFGE) of XbaI-digested DNA was performed as previously described62.
Statistical analysis
Statistical analysis was done with “SPSS for Windows” (Version 23, SPSS Inc., Chicago, Illinois, U.S.A.). The Mann-Whitney test was used to determine the significance of differences between the numbers of bacteria recovered from the three different inoculation groups. Two-tailed p- values with p ≤ 0.05 were considered significant.
Additional Information
How to cite this article: Hamm, K. et al. Experimental Infection of Calves with Escherichia coli O104:H4 outbreak strain. Sci. Rep. 6, 32812; doi: 10.1038/srep32812 (2016).
References
Robert Koch-Institut. Abschließende Darstellung und Bewertung der epidemiologischen Erkenntnisse im EHEC O104:H4 Ausbruch (2011).
European Food Safety Authority. Tracing seeds, in particular fenugreek (Trigonella foenum-graecum) seeds, in relation to the Shiga toxin-producing E. coli (STEC) O104:H4 2011 Outbreaks in Germany and France. Technical Report of EFSA (2011).
Bielaszewska, M. et al. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect Dis 11, 671–676 (2011).
Mellmann, A. et al. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One 6, e22751 (2011).
European Food Safety Authority. The Community Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and food-borne outbreaks in the European Union in 2008. The EFSA Journal (2010).
Borczyk, A. A., Karmali, M. A., Lior, H. & Duncan, L. M. Bovine reservoir for verotoxin-producing Escherichia coli O157:H7. Lancet 1, 98 (1987).
Louie, M. et al. Molecular typing methods to investigate transmission of Escherichia coli O157:H7 from cattle to humans. Epidemiol Infect 123, 17–24 (1999).
O’Brien, A. O., Lively, T. A., Chen, M. E., Rothman, S. W. & Formal, S. B. Escherichia coli O157:H7 strains associated with haemorrhagic colitis in the United States produce a Shigella dysenteriae 1 (SHIGA) like cytotoxin. Lancet 1, 702 (1983).
Karmali, M. A., Steele, B. T., Petric, M. & Lim, C. Sporadic cases of haemolytic-uraemic syndrome associated with faecal cytotoxin and cytotoxin-producing Escherichia coli in stools. Lancet 1, 619–620 (1983).
Frank, C. et al. Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N Engl J Med 365, 1771–1780 (2011).
Cravioto, A. et al. Association of Escherichia coli HEp-2 adherence patterns with type and duration of diarrhoea. Lancet 337, 262–264 (1991).
Wanke, C. A. Enteropathogenic and enteroaggregative strains of Escherichia coli: clinical features of infection, epidemiology, and pathogenesis. Curr Clin Top Infect Dis 15, 230–252 (1995).
Mathewson, J. J. et al. A newly recognized cause of travelers’ diarrhea: enteroadherent Escherichia coli. J Infect Dis 151, 471–475 (1985).
Smith, H. R., Cheasty, T. & Rowe, B. Enteroaggregative Escherichia coli and outbreaks of gastroenteritis in UK. Lancet 350, 814–815 (1997).
Dean-Nystrom, E. A., Bosworth, B. T., Moon, H. W. & O’Brien, A. D. Escherichia coli O157:H7 requires intimin for enteropathogenicity in calves. Infect Immun 66, 4560–4563 (1998).
van Diemen, P. M., Dziva, F., Stevens, M. P. & Wallis, T. S. Identification of enterohemorrhagic Escherichia coli O26:H- genes required for intestinal colonization in calves. Infect Immun 73, 1735–1743 (2005).
Stevens, M. P., van Diemen, P. M., Frankel, G., Phillips, A. D. & Wallis, T. S. Efa1 influences colonization of the bovine intestine by shiga toxin-producing Escherichia coli serotypes O5 and O111. Infect Immun 70, 5158–5166 (2002).
Hoffman, M. A., Menge, C., Casey, T. A., Laegreid, W., Bosworth, B. T. & Dean-Nystrom, E. A. Bovine immune response to shiga-toxigenic Escherichia coli O157:H7. Clin Vaccine Immunol 13, 1322–1327 (2006).
Dean-Nystrom, E. A., Bosworth, B. T. & Moon, H. W. Pathogenesis of Escherichia coli O157:H7 in weaned calves. Adv Exp Med Biol 473, 173–177 (1999).
Naylor, S. W. et al. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect Immun 71, 1505–1512 (2003).
Naylor, S. W. et al. Escherichia coli O157:H7 forms attaching and effacing lesions at the terminal rectum of cattle and colonization requires the LEE4 operon. Microbiology 151, 2773–2781 (2005).
Geue, L. et al. A long-term study on the prevalence of Shiga toxin-producing Escherichia coli (STEC) on four German cattle farms. Epidemiol Infect 129, 173–185 (2002).
Besser, T. E., Richards, B. L., Rice, D. H. & Hancock, D. D. Escherichia coli O157:H7 infection of calves: infectious dose and direct contact transmission. Epidemiol Infect 127, 555–560 (2001).
Wieler, L. H. et al. No evidence of the Shiga toxin-producing E. coli O104:H4 outbreak strain or enteroaggregative E. coli (EAEC) found in cattle faeces in northern Germany, the hotspot of the 2011 HUS outbreak area. Gut pathogens 3, 17 (2011).
Cassar, C. A., Ottaway, M., Paiba, G. A., Futter, R., Newbould, S. & Woodward, M. J. Absence of enteroaggregative Escherichia coli in farmed animals in Great Britain. Vet Rec 154, 237–239 (2004).
Uber, A. P. et al. Enteroaggregative Escherichia coli from humans and animals differ in major phenotypical traits and virulence genes. FEMS Microbiol Lett 256, 251–257 (2006).
Auvray, F., Dilasser, F., Bibbal, D., Kerouredan, M., Oswald, E. & Brugere, H. French cattle is not a reservoir of the highly virulent enteroaggregative Shiga toxin-producing Escherichia coli of serotype O104:H4. Vet Microbiol 158, 443–445 (2012).
Paddock, Z. D., Bai, J., Shi, X., Renter, D. G. & Nagaraja, T. G. Detection of Escherichia coli O104 in the feces of feedlot cattle by a multiplex PCR assay designed to target major genetic traits of the virulent hybrid strain responsible for the 2011 German outbreak. Appl Environ Microbiol 79, 3522–3525 (2013).
Balabanova, Y. et al. Serological evidence of asymptomatic infections during Escherichia coli O104:H4 outbreak in Germany in 2011. Plos One 8, e73052 (2013).
Cabal, A. et al. Detection of virulence-associated genes characteristic of intestinal Escherichia coli pathotypes, including the Enterohemorrhagic/Enteroaggregative O104:H4 in bovines from Germany and Spain. Microbiol Immunol 59, 433–442 (2015).
Dean-Nystrom, E. A. Bovine Escherichia coli O157:H7 infection model. Methods Mol Med 73, 329–338 (2003).
Dean-Nystrom, E. A., Stoffregen, W. C., Bosworth, B. T., Moon, H. W. & Pohlenz, J. F. Early attachment sites for Shiga-Toxigenic Escherichia coli O157:H7 in experimentally inoculated weaned calves. Appl Environ Microbiol 74, 6378–6384 (2008).
Brown, C. A., Harmon, B. G., Zhao, T. & Doyle, M. P. Experimental Escherichia coli O157:H7 carriage in calves. Appl Environ Microbiol 63, 27–32 (1997).
Frank, G. H., Briggs, R. E. & Schneider, R. A. Characterization of Escherichia coli isolated from the tonsils of cattle. J Clin Microbiol 32, 256–258 (1994).
Munera, D. et al. Autotransporters but not pAA are critical for rabbit colonization by Shiga toxin-producing Escherichia coli O104:H4. Nature communications 5, 3080 (2014).
Tarr, P. I. et al. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect Immun 68, 1400–1407 (2000).
Kudva, I. T. & Dean-Nystrom, E. A. Bovine recto-anal junction squamous epithelial (RSE) cell adhesion assay for studying Escherichia coli O157 adherence. J Appl Microbiol 111, 1283–1294 (2011).
Zhang, W. et al. Lability of the pAA Virulence Plasmid in O104:H4: Implications for Virulence in Humans. Plos One 8, e66717 (2013).
Kagambega, A., Martikainen, O., Siitonen, A., Traore, A. S., Barro, N. & Haukka, K. Prevalence of diarrheagenic Escherichia coli virulence genes in the feces of slaughtered cattle, chickens, and pigs in Burkina Faso. MicrobiologyOpen 1, 276–284 (2012).
Bielaszewska, M. et al. Effects of antibiotics on Shiga toxin 2 production and bacteriophage induction by epidemic Escherichia coli O104:H4 strain. Antimicrob Agents Chemother 56, 3277–3282 (2012).
Savarino, S. J. et al. Enteroaggregative Escherichia coli heat-stable enterotoxin is not restricted to enteroaggregative E. coli. J Infect Dis 173, 1019–1022 (1996).
Menard, L. P. & Dubreuil, J. D. Enteroaggregative Escherichia coli heat-stable enterotoxin 1 (EAST1): a new toxin with an old twist. Crit Rev Microbiol 28, 43–60 (2002).
Cray, W. C. Jr. & Moon, H. W. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Appl Environ Microbiol 61, 1586–1590 (1995).
Shridhar, P. B. et al. Escherichia coli O104 in Feedlot Cattle Feces: Prevalence, Isolation and Characterization. PLoS One 11, e0152101 (2016).
Tozzoli, R. et al. Shiga toxin-converting phages and the emergence of new pathogenic Escherichia coli: a world in motion. Frontiers in cellular and infection microbiology 4, 80 (2014).
Tarr, P. I., Neill, M. A., Clausen, C. R., Newland, J. W., Neill, R. J. & Moseley, S. L. Genotypic variation in pathogenic Escherichia coli O157:H7 isolated from patients in Washington, 1984-1987. J Infect Dis 159, 344–347 (1989).
Dean-Nystrom, E. A., Bosworth, B. T., Cray, W. C. & Moon, H. W. Pathogenicity of Escherichia coli O157:H7 in the intestines of neonatal calves. Infect Immun 65, 1842–1848 (1997).
Otto, P. H., Clarke, I. N., Lambden, P. R., Salim, O., Reetz, J. & Liebler-Tenorio, E. M. Infection of calves with bovine norovirus GIII.1 strain Jena virus: an experimental model to study the pathogenesis of norovirus infection. J Virol 85, 12013–12021 (2011).
Kogan, G., Jann, B. & Jann, K. Structure of the Escherichia coli O104 polysaccharide and its identity with the capsular K9 polysaccharide. FEMS Microbiol Lett 91, 135–140 (1992).
Hautzer, N. W., Wittkuhn, J. F. & McCaughey, W. T. Trypsin digestion in immunoperoxidase staining. J Histochem Cytochem 28, 52–53 (1980).
Cebula, T. A., Payne, W. L. & Feng, P. Simultaneous identification of strains of Escherichia coli serotype O157:H7 and their Shiga-like toxin type by mismatch amplification mutation assay-multiplex PCR. J Clin Microbiol 33, 248–250 (1995).
Taylor, D. E. et al. Genomic variability of O islands encoding tellurite resistance in enterohemorrhagic Escherichia coli O157:H7 isolates. J Bacteriol 184, 4690–4698 (2002).
Yatsuyanagi, J., Saito, S., Sato, H., Miyajima, Y., Amano, K. & Enomoto, K. Characterization of enteropathogenic and enteroaggregative Escherichia coli isolated from diarrheal outbreaks. J Clin Microbiol 40, 294–297 (2002).
Al Safadi, R. et al. Correlation between in vivo biofilm formation and virulence gene expression in Escherichia coli O104:H4. PLoS One 7, e41628 (2012).
Yamamoto, T. & Echeverria, P. Detection of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 gene sequences in enterotoxigenic E. coli strains pathogenic for humans. Infect Immun 64, 1441–1445 (1996).
Paton, A. W. & Paton, J. C. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J Clin Microbiol 36, 598–602 (1998).
Gannon, V. P., D’Souza, S., Graham, T., King, R. K., Rahn, K. & Read, S. Use of the flagellar H7 gene as a target in multiplex PCR assays and improved specificity in identification of enterohemorrhagic Escherichia coli strains. J Clin Microbiol 35, 656–662 (1997).
Müller, D. et al. Identification of unconventional intestinal pathogenic Escherichia coli isolates expressing intermediate virulence factor profiles by using a novel single-step multiplex PCR. Appl Environ Microbiol 73, 3380–3390 (2007).
Grimm, V., Ezaki, S., Susa, M., Knabbe, C., Schmid, R. D. & Bachmann, T. T. Use of DNA microarrays for rapid genotyping of TEM beta-lactamases that confer resistance. J Clin Microbiol 42, 3766–3774 (2004).
Paterson, D. L. et al. Extended-spectrum beta-lactamases in Klebsiella pneumoniae bloodstream isolates from seven countries: dominance and widespread prevalence of SHV- and CTX-M-type beta-lactamases. Antimicrob Agents Chemother 47, 3554–3560 (2003).
Leflon-Guibout, V. et al. Emergence and spread of three clonally related virulent isolates of CTX-M-15-producing Escherichia coli with variable resistance to aminoglycosides and tetracycline in a French geriatric hospital. Antimicrob Agents Chemother 48, 3736–3742 (2004).
Barth, S. et al. Experimental Evaluation of Faecal Escherichia coli and Hepatitis E Virus as Biological Indicators of Contacts Between Domestic Pigs and Eurasian Wild Boar. Transbound Emerg Dis, doi: 10.1111/tbed.12389 (2015).
Acknowledgements
The study was financially supported by the European Commission (FP7 programme under project number 278976). The authors would like to thank Anja Müller, Anke Hinsching, Birgit Mintel, Sabine Lied and Lisa Wirker for her excellent technical assistance and the staff of the experimental animal facility (Friedrich-Loeffler-Institut, Isle of Riems) for their support during the animal trial. We thank Indira Kudva (National Animal Disease Center, Ames, Iowa, USA) for providing bacterial strains and Evelyn A. Dean-Nystrom for critical reading of the manuscript.
Author information
Authors and Affiliations
Contributions
K.H., J.P.T. and C.M. designed research; K.H., S.A.B., S.S., L.G., E.L., E.L.-T., K.T. and G.K. performed research; M.B. and H.K. contributed new reagents/analytic tools; K.H., S.A.B., L.G., E.L.-T. and C.M. analyzed data; K.H. and C.M. drafted the manuscript. All authors discussed the results and commented on the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Hamm, K., Barth, S., Stalb, S. et al. Experimental Infection of Calves with Escherichia coli O104:H4 outbreak strain. Sci Rep 6, 32812 (2016). https://doi.org/10.1038/srep32812
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep32812
This article is cited by
-
Using unique ORFan genes as strain-specific identifiers for Escherichia coli
BMC Microbiology (2022)
-
Shiga Toxin-Producing and Enteroaggregative Escherichia coli in Animal, Foods, and Humans: Pathogenicity Mechanisms, Detection Methods, and Epidemiology
Current Microbiology (2020)
-
Decreased STEC shedding by cattle following passive and active vaccination based on recombinant Escherichia coli Shiga toxoids
Veterinary Research (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.