Abstract
The heart-failure relevant Potassium Channel Interacting Protein 2 (KChIP2) augments CaV1.2 and KV4.3. KChIP3 represses CaV1.2 transcription in cardiomyocytes via interaction with regulatory DNA elements. Hence, we tested nuclear presence of KChIP2 and if KChIP2 translocates into the nucleus in a Ca2+ dependent manner. Cardiac biopsies from human heart-failure patients and healthy donor controls showed that nuclear KChIP2 abundance was significantly increased in heart failure; however, this was secondary to a large variation of total KChIP2 content. Administration of ouabain did not increase KChIP2 content in nuclear protein fractions in anesthetized mice. KChIP2 was expressed in cell lines and Ca2+ ionophores were applied in a concentration- and time-dependent manner. The cell lines had KChIP2-immunoreactive protein in the nucleus in the absence of treatments to modulate intracellular Ca2+ concentration. Neither increasing nor decreasing intracellular Ca2+ concentrations caused translocation of KChIP2. Microarray analysis did not identify relief of transcriptional repression in murine KChIP2−/− heart samples. We conclude that although there is a baseline presence of KChIP2 in the nucleus both in vivo and in vitro, KChIP2 does not directly regulate transcriptional activity. Moreover, the nuclear transport of KChIP2 is not dependent on Ca2+. Thus, KChIP2 does not function as a conventional transcription factor in the heart.
Similar content being viewed by others
Introduction
Heart failure is a disease of the elderly1 and the prevalence is rising both in Western and developing countries following the general longer lifespan2. Heart failure is characterized by decreased pump efficiency leading to symptoms like shortness of breath, edema and fatigue. Besides the left ventricular structural remodeling seen in HF2, electrophysiological modifications are taking place as well. The most dominant electrophysiological changes in heart failure are modulations of repolarizing ion currents prolonging the cardiac action potential3,4,5,6 and changes in the intracellular Ca2+ handling7,8 aiming to improve cardiac pump function, but as a byproduct the electrical remodeling creates a substrate for ventricular arrhythmias. Underlying these structural and electrophysiological remodeling processes lie a range of transcriptional alterations where the regulating mechanisms are largely unknown9.
Potassium Channel Interacting Proteins (KChIPs) are accessory β-subunit proteins belonging to a family of small Ca2+-binding cytosolic proteins consisting of four isoforms, KChIP1-4. KChIP1-3 are structurally related as they all contain a highly conserved C-terminal core region of approximately 180 amino acids that house the 4 EF-hand domains capable of binding Ca2+ ions which induces a conformational change in the protein10. These 3 isoforms share a common mode of action on potassium channel kinetics: They all increase peak KV4 current, slow channel inactivation and quickens the recovery from inactivation. All isoforms are expressed in brain, but KChIP2 is the only isoform expressed in the heart, where it regulates both the repolarizing KV4.3 current4,11,12 and the depolarizing CaV1.2 current8,13,14,15. The heightened interest of KChIP2 comes from its reported downregulation in HF16,17, which is thought to contribute to the modified repolarization and altered Ca2+ handling in the disease.
It has been shown that KChIP3 besides its electrophysiological effects also holds properties that affect gene transcription. KChIP3, primarily expressed in brain tissue11,18, has also independently been identified as Downstream Regulatory Element Agonist Modulator (DREAM)19 and as calsenilin20. KChIP3 translocates from the cytosol to the nucleus upon a raise in cytosolic Ca2+ concentration following activation of a calmodulin kinase II (CaMKII) mediated pathway21. In the nucleus, KChIP3 only binds DNA in a Ca2+-free state, whereas binding of Ca2+ to KChIP3 leads to conformational changes preventing binding to DNA19,22. KChIP3 binds Downstream Regulatory Elements (DRE) on the DNA and represses transcription of several genes, including Na+/Ca2+ exchanger 3 in cerebellar neurons22, prodynorphin, involved in pain sensation19,23 and CaV1.2 channel in neonatal rat cardiomyocytes21.
It has been shown that in addition to KChIP3, the other KChIP isoforms bind DRE sites24, so based on this and the structural familiarity between KChIP2 and KChIP3, we investigated whether KChIP2 have transcriptional regulatory functions. If KChIP2 proves to be transcriptionally active, the repression of KChIP2-controlled genes would potentially be lost in HF as a result of the downregulation of the protein. At the same time, an increase in the intracellular Ca2+ concentration seen in HF would promote translocation of KChIP2 from the cytosol to the nucleus. In the present study, we investigated whether KChIP2 can translocate from the cytosol to the nucleus and tested whether KChIP2 binds DNA and alters gene expression.
Results
KChIP2 expression and localization in human failing and healthy hearts
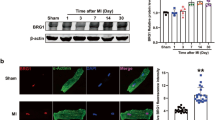
Heart failure is a disease characterized by an elevated adrenergic state, resulting in an chronic increased intracellular Ca2+ concentration25, suggesting that the nuclear fraction of KChIP2 could be increased in this disease. Tissue samples were obtained from hearts from heart-failure patients with an ejection fraction of 22 ± 9% (n = 5) and from healthy hearts (ejection fraction 66 ± 2%; n = 5, p < 0.05; Table 1). We found that total protein levels were comparable in human samples from failing and non-failing cardiac tissue. KChIP2 migrated as a single band to the expected size around 32 kDa on immunoblots (Fig. 1A). We observed comparable total KChIP2 levels in human non-failing and failing heart samples; although there was a trend towards increased total KChIP2 levels in samples from failing hearts (P = 0.064; Fig. 1B). Successful separation of nuclear and cytosolic fractions was confirmed by the presence of Lamin A/C in nuclear fractions only (Fig. 1C). KChIP2 was present in both cytosolic and nuclear fractions, but the nuclear KChIP2 content was significantly larger in failing hearts relative to non-failing hearts (P = 0.025; Fig. 1D). Notwithstanding, the difference in nuclear KChIP2 content seemed to be dependent on the non-significant change in total KChIP2, since the nuclear-to-total KChIP2 ratio was comparable in failing and non-failing hearts (P = 0.6). Hence, in human hearts, KChIP2 is present in the nucleus; however, the nuclear-to-cytosolic ratio is not altered in end-stage heart failure.
KChIP2 expression and subcellular localization in human hearts.
(A) Representative immunoblots showing detection of KChIP2 in cytosolic (C) and nuclear (N) protein fractions from heart samples from heart-failure (HF; n = 3) and non-failing (NF; n = 2) patients. Lanes with molecular weight marker are indicated with M. Arrowheads indicate KChIP2 and β-tubulin bands in the upper and lower panel, respectively. The double band of lamin A/C is indicated with a “{”. Detecting β-tubulin and Lamin A/C confirms equal loading of cytosolic protein and successful fractionation, respectively. Each sample is analyzed in duplicate. (B) Quantification of the total amount of KChIP2 in the two experimental groups. (C) Quantification of Lamin A/C in nuclear and cytosolic fractions. (D) Ratio of the nuclear KChIP2 and nuclear Lamin A/C in NF and HF samples (*P = 0.025). (E) Ratio of the nuclear KChIP2 to total KChIP2. P values from Student’s t test.
Nuclear translocation in vivo
Next, we investigated whether it is possible to induce a KChIP2 translocation process into the nucleus in cardiomyocytes in vivo. Mice were injected with a range of pharmacological compounds increasing the heart rate and/or inotropic state and thereby the intracellular Ca2+ concentration26, with the aim of initiating an acute KChIP2 translocation to the nucleus (Fig. 2A). Mice treated with adrenaline and caffeine showed an increase of nuclear KChIP2 in cardiomyocytes compared to controls in most experiments, e.g., Fig. 2B; however not consistently. Ouabain, isoprenaline and dobutamine had no consistent effect on KChIP2 localization, e.g. the nuclear-to-cytosolic ratios of KChIP2 after administration of saline or ouabain were comparable (Fig. 2C,D). Generally, the variation between the translocation responses in the treated mice was large and the outcomes did not relate to the chronotropic or inotropic effects of the pharmacological compounds. Overall, we did not identify statistically significant KChIP2 translocation based on the pharmacologically induced acute elevation of intracellular Ca2+ in vivo.
KChIP2 expression and subcellular localization in mouse hearts.
(A) Representative two lead surface ECG from anesthetized mice at baseline and after treatment with 1 mg/g ouabain IP. (B) Exemplary immunoblots showing an increase of KChIP2 in the nuclear fraction when the mouse was treated with adrenaline and caffeine (A + C). Saline (S) treated mice served as controls. (C) Representative immunoblots showing the cytosolic (C) and nuclear (N) content of KChIP2 after administration of ouabain, isoprenaline and dobutamine, all increasing heart rate and inotropy and saline as control. (D) Quantification of the nuclear-to-cytosolic ratio of KChIP2 in hearts from 5 ouabain-treated and 4 saline-treated mice and compared with Student’s t test.
Nuclear translocation in vitro
To directly visualize the subcellular localization of KChIP2 and determining if an increase in the intracellular Ca2+ concentration is a triggering factor for translocation from the cytosol to the nucleus, KChIP2-transfected cells were treated in a time- and concentration dependent manner with Ca2+ ionophores (Fig. 3). The transfected cells, in absence of treatments to elevate the intracellular Ca2+ concentration, had already significant levels of KChIP2-immunoreactive protein in the nucleus (Fig. 3A). Increasing the intracellular Ca2+ concentration caused no change in nuclear levels of KChIP2 (Fig. 3C). Comparable results were obtained when treating the cells with another Ca2+ ionophore (A23187, data not shown).
KChIP2 localization in heterologous expression systems.
(A) COS-1 cells transfected with KChIP2 and treated with 0 μM and 10 μM ionomycin for 40 min. The extracellular [Ca2+] was 1.8 mM. Pictures showing representative cells stained using a KChIP2 antibody (green). The nucleus is stained blue using DAPI. (B) Graph showing the intensity of the KChIP2 and DAPI signals along the red line in panel A for the cell treated with 10 μM ionomycin. One hundred consecutive values were chosen within both the cytosol and nucleus for quantification of protein localization (indicated by red lines). (C) Quantitative analysis of the time- and concentration-dependency of KChIP2 translocation from the cytosol to the nucleus using ionomycin as a trigger. Five cells from each transfection (n = 4–5) were chosen for quantification and compared using a 1-way ANOVA.
We transfected cells with the Ca2+-insensitive KChIP2-∆EF mutant to determine if KChIP2’s Ca2+ sensitivity is responsible for translocation. There was no difference in KChIP2 localization between control cells and cells treated with Ca2+ ionophores in medium containing a physiological level of Ca2+ (Fig. 4). These experiments corroborate the in-vivo studies and show that KChIP2 is present in the nucleus of transfected, un-stimulated cells and they suggest that increases in intracellular Ca2+ concentrations do not trigger a translocation of KChIP2 from the cytosol into the nucleus in heterologous expression systems.
Regular KChIP2 (WT) and KChIP2-∆EF (∆EF) expression and localization in COS-1 cells.
COS-1 cells transfected with WT KChIP2 and KChIP2-∆EF and treated with 10 μM ionomycin for 40 min. Pictures showing representative cells stained using a KChIP2 antibody (green). The nucleus is stained blue using DAPI. Both KChIP2 and KChIP2-∆EF are present in the nucleus.
Localization of KChIP2 in Ca2+-depleted cells
KChIP2 localization in the nucleus in absence of stimulation indicates that the Ca2+ concentration during normal conditions is already elevated to a level that allows translocation, or that KChIP2 is independent of a raise in the intracellular Ca2+ concentration in order to translocate to the nucleus. To test this, we observed protein localization in a Ca2+-free environment. HL-1 cells overexpressing KChIP2 were incubated for 40 min in a Ca2+ free medium and treated with A23187 in order to deplete intracellular Ca2+ (Fig. 5). The variations within both groups were large, but on average the nucleus/cytosol ratio did not differ in Ca2+-depleted and control cells. Thus, the presence of KChIP2 in the nucleus is not regulated by Ca2+.
Ca2+ depletion in HL-1 cells.
(A) HL-1 cells transfected with KChIP2 cDNA and treated with 0 μM A23187 in a Ca2+-containing medium or 10 μM A23187 in a Ca2+-free medium for 40 min. Pictures showing representative cells stained using a KChIP2 antibody (green). The nucleus is stained blue using DAPI. (B) Quantitative analysis of KChIP2 localization after Ca2+ depletion and the control. Five cells from each transfection (n = 2–3) were chosen for quantification and compared using a Student’s t test.
KChIP2 DNA binding studies using the Cacna1c promoter region
To test if KChIP2 regulates gene expression in general, we performed expression studies. If KChIP2 is an important transcript factor in vivo, we expect that transcription of a large number of genes would be altered. Microarray data detected the expression of 14,905 genes in the WT mouse hearts; however, only 37 genes were expressed with a statistically significant difference in KChIP2−/− hearts that exceeded a 1.4 fold cut-off (Table 2). Figure 6 depicts the expression levels of membrane and ion-channel proteins in WT and KChIP2−/− hearts. Most changes in gene expression are very minor and involved mostly genes which were detected near background levels (log2 values below 5), indicating that KChIP2 is not a transcriptional modifier. Finally, the microarray confirmed that KChIP2 is the only KChIP isoform expressed in the heart and showed that no other KChIP isoform is expressed in KChIP2−/− hearts (Fig. 6).
Expression of genes encoding membrane and ion-channel proteins in WT (n = 3) and KChIP2−/− (n = 3) hearts using the Affymetrix Mouse Gene 482 1.0 array.
Colours indicate log2-transformed and mean normalized expression levels from high (red) to low (blue) shown in pseudo colours of the z-score (standard deviations from the mean). *P < 0.05 (Student’s t test). †Gene not expressed above the level of background noise (indicated for genes with statistically significant differences only). Integrin α-9 (Itga9) and the β2 subunit of the L-type Ca2+ channel (Cacn2b) were expressed at 82% and 135%, respectively, in KChIP2−/− hearts, relative to WT. KChIP2 was not detected in KChIP2−/− hearts.
The Cacna1c promoter region contains a Downstream Regulatory Element, a reported KChIP3 binding site21, whereby KChIP3 can repress transcriptional activity. In the microarray analysis, there was no difference in expression levels of Cacna1c in WT and KChIP2−/− hearts. To confirm the finding that KChIP2 does now affect transcription of this gene, we used a chromatin immunoprecipitation assay on neonatal mouse cardiomyocytes treated with Ca2+ ionophores and used the obtained DNA as a template for quantitative PCR. Despite using 2 different KChIP2 antibodies, the amount of precipitated input DNA was low relative to control, indicating that KChIP2 does not bind to DNA under the present conditions. Thus, the present experiments show that KChIP2 does not function as a repressor of gene expression in vivo in the mouse and that KChIP2 does not bind DNA in vitro.
Discussion
Three proteins were described practically simultaneously and we now know that these proteins are identical: Calsenilin was initially described as a calcium-binding protein that binds presenilin, which is critically involved in early-onset familial Alzheimer disease20. Downstream regulatory element antagonist modulator (DREAM) was described as a calcium-regulated transcriptional repressor of key genes involved in memory and pain19. Finally, the potassium-channel interacting proteins (KChIP) were described as important modulators of the neuronal A-type potassium current, which is comparable to the cardiac transient outward potassium current (Ito)11. Later, it was clear that calsenilin, DREAM and KChIP3 are the same.
In the heart, only KChIP2 is expressed. It significantly modulates Ito11,4,27,28 and recently we have identified direct functional interaction between KChIP2 and the cardiac calcium channel: KChIP2 binding to the cytosolic N-terminal domain of CaV1.2 causes a 40% increase in current amplitude8,13,15. KChIP1, 2 and 3 share 70% amino acid identity with an identical core domain containing 4 EF hands, of which 3 can bind Ca2+11. In neurons, KChIP3 is upregulated in KChIP2−/− mice and KChIP2 is upregulated in KChIP3−/− mice29, suggesting a compensatory expression profile displayed by the two proteins and potentially a complementary function. In the present study, none of the KChIPs are expressed in hearts from KChIP2−/− mice (Fig. 6).
In 2011, Ronkainen and colleagues21 identified a mechanism by which the calcium concentration of the cell could transcriptionally feedback on the calcium channel itself: A downstream regulatory element (DRE) in the promoter region of Cacna1c was sensitive to CaMKII-mediated repression of transcription. When CaMKII was activated by rising cytosolic [Ca2+], KChIP3 would translocate from the cytosol to the nucleus, bind to the DRE and repress transcription of Cacna1c, the gene encoding CaV1.2, the cardiac calcium channel. Based on the overall similarity between KChIP2 and -3 and given the physiological expression of KChIP2 in the heart, we decided to test the hypothesis that KChIP2 was transcriptionally active.
KChIP2 is palmitoylated at the N-terminal which causes effective localization of KChIP2 to the cytoplasmic side of the plasma membrane and is required for achieving the full effect on Ito in heterologous expression systems30. Palmitoylation is the addition of palmitic acid derivatives to cysteine residues and it has been suggested that it may not serve as a targeting signal per se, but rather stabilize the ion channel complex at the cell surface30. Both KChIP2 and -3 has a comparable cellular distribution, with clear staining in both cytoplasm and in nuclei in primary neuronal cultures18. In the present study, we show that KChIP2 can be identified in nuclear protein fractions from cardiac tissue from humans and from mice and we show by immunofluorescence that cell lines transiently overexpressing KChIP2 have immunoreactive protein in the nucleus.
From the present study, it is clear that KChIP2 is present in the nucleus; however, it is unclear what function the protein has there, far from its described interaction partners at the cell surface. Small molecules and proteins up to a weight of ~40 kDa can passively diffuse through the nuclear pore complexes31, which could suggest that KChIP2 (32 kDa) is merely present in the nucleus as a consequence of non-regulation. Nevertheless, the documented regulation of the nuclear import of KChIP3 (28 kDa) is a clear example of a small, transcriptionally active protein that is actively transported to the nucleus when cytosolic calcium increases.
Using a host of stimulation protocols in vivo and in vitro, we were unable to unequivocally initiate a translocation process stimulating the transport of KChIP2 into the nucleus. Moreover, microarray data from KChIP2−/− hearts suggested that KChIP2 was transcriptionally inactive and the ChIP assay failed to show specific KChIP2-DNA interaction. Hence, based on these experiments, we conclude that unlike KChIP3, the nuclear presence of KChIP2 is not regulated by cytosolic calcium concentrations and KChIP2 does not interact with the DRE’s to repress transcriptional activity. One important limitation of the ChIP assay is that it is very useful for detecting proteins that binds DNA; however, it is not very specific in discriminating false negative and true negative outcomes.
The data shows that KChIP2 does not directly regulate transcription; however KChIP2 augments both sarcolemmal K+ currents4,11,12 and Ca2+ currents8,13,15,32 with complex impact on action potential morphology8,32. This may set the stage for an electrical-transcriptional feedback loop, where current amplitudes and membrane potentials signals to the nucleus, which have been described previously, e.g., during atrial arrhythmias33. Thus, changes in KChIP2 levels may cause cardiac remodelling via this electrical-transcriptional coupling; however, a direct effect of KChIP2 on transcription is not likely.
Soltysinska and colleagues16 have previously shown that KChIP2 mRNA and protein levels are significantly reduced in ventricular tissue samples from end-stage heart-failure patients (New York Heart Association class IV). In the present study, we found a surprising trend (P = 0.064) towards upregulated total KChIP2 protein in samples from failing hearts (Fig. 1C). Heart samples and sources differed in the two studies; however, the primary antibody was identical. The age of the healthy donors may have contributed to the difference in results, as our donors were older (59 years of age versus 42 years of age in the Soltysinska study) and thereby potentially had lower KChIP2 expression. It is clear that we need larger and better controlled studies of the proteomic changes that occur in the heart with disease and with age. Notwithstanding, KChIP2 may not be unequivocally downregulated in heart disease.
In conclusion, we show that KChIP2 does not translocate from the cytosol to the nucleus in a regulated manner and that KChIP2 cannot bind DNA or modulate gene expression as a conventional transcription factor. The potential function of nuclear KChIP2 is presently unknown. Moreover, our data suggest that the general assumption that KChIP2 reduction in heart failure is the primary electrophysiological hallmark of the disease may not be universal.
Methods
Ethical considerations and experimental animals
Human left ventricular tissue was harvested from end-stage failing hearts removed during heart transplantation or from donor hearts not suitable for transplantation as approved by the Local Ethics Committee (Ref No. 20–277 ex 08/09) of Medical University of Graz, Austria. All procedures were carried out in accordance with the approved guideline and the patients had all provided written informed consent. None of the donors had a clinical history of heart failure and were classified as non-failing with preserved ejection fraction according to echocardiographic assessment before explant.
Animal experiments were performed using male C57BL6 (wild type) and KChIP2−/− mice. Mice were genotyped (GeneTyper, NY, USA) using DNA isolated from tail samples. Animals had access to water and food ad libitum and were housed in a room with a temperature of 22 °C and a 12 h light/dark schedule. Body temperature was kept at 37 °C during surgical procedures. Euthanasia was done by cervical dislocation at the end of the experiments. The experiments were approved by the national ethics committee (The Ministry of Food, Agriculture and Fisheries, Denmark) and were carried out in accordance with the approved guidelines. Further, the experiments conform to The Council Directive of the European Communities on The Protection of Animals used for Scientific Purposes (2010/63/EU) and the declaration of Helsinki.
KChIP2 localization in vivo
Mice were anesthetized with 1.5% isoflurane and surface ECG and core temperature was monitored throughout the experiment. Adrenaline (2 μg/g) and caffeine (120 μg/g) was co-administered intraperitoneally (IP) in order to raise the intracellular Ca2+ concentration in the cardiomyocytes. In addition, dobutamine (2 μg/g IP), ouabain (1 mg/g IP or 60 μg/min intravenously (IV)) or isoprenaline (2 μg/g IP) were tested. Mice injected with saline served as control. The hearts were explanted after 10 minutes treatment and snap frozen in liquid N2. Tissue from human and mouse hearts were stored at −80 °C until use.
The tissues were homogenized using Precellys system (Bertin Technologies, France) and the proteins were isolated and divided into cytosolic and nuclear fractions using NE-PER fractionation kit following the manufacturer’s instructions (Thermo Scientific, MA, USA). Total protein concentration in the samples was determined using a Lowry protein assay (BioRad, CA, USA). Human and murine samples were separated on 4–15% TGX SDS-PAGE or 4–20% Tris-HCl SDS-PAGE (BioRad), transferred to immobilon hybond-p polyvinylidene flouride (PVDF) transfer membranes (Millipore, MA, USA) and blocked in Odyssey blocking buffer (LI-COR, NE, USA). The membranes were blocked in 4% milk in PBS. Primary antibody incubation was done overnight at 4 °C using a KChIP2 antibody (3.47 μg/ml, NeuroMab, UC Davis, CA, USA). Lamin A/C antibody (Cell Signaling Technology, MA, USA) was used to test if the fractionation protocol was successful, as lamin A/C is confined to the nucleus. GAPDH (0.2 μg/ml, Sigma-Aldrich, MO, USA) and β-tubulin antibodies (1 μg/ml, Millipore, MA, USA) served as loading control: Total (nuclear + cytosolic) content should be comparable between subjects; however, it is not possible to compare protein loading between cytosolic and nuclear lanes. Immunoreactive proteins were detected using fluorescent donkey anti-mouse and donkey anti-rabbit antibodies (0.1 μg/ml, LI-COR, NE, USA), or with peroxidase conjungated donkey anti-mouse and donkey anti-rabbit antibodies (0.08 μg/ml, Jackson Immunoresearch, PA, USA). Visualization of the proteins was done using the Odyssey system (LI-COR, NE, USA) and enhanced chemiluminescence for human and murine samples, respectively. Each sample was tested in duplicate. Densitometric quantification of the bands was performed with the Image Studio (LI-COR, NE, USA) and ImageJ software.
KChIP2 translocation in vitro
Experiments were performed using COS-1 cells cultured in Dulbecco’s Modified Eagles Medium (DMEM) containing 10% Fetal Bovine Serum (FBS) and 1% penicillin-streptomyocin and HL-1 cells cultured in Claycomb medium (Sigma-Aldrich, MO, USA) supplemented with 2 mM L-glutamine, 100 μM noradrenaline, 10% modified FBS and 1% penicillin-streptomyocin. COS-1 and HL-1 cells were plated in 35 mm dishes in DMEM and Claycomb medium, respectively, for 24 hours and transiently transfected using SilentFect (Invitrogen, CA, USA) according to the manufacturer’s instructions. Human KChIP2.1 in a pXOOM vector was used for transfections. In some experiments a Ca2+-insensitive KChIP2 mutant with asparagine-to-alanine substitutions in the Ca2+-binding EF hands (KChIP2-∆EF) was used to probe calcium-dependency of translocation15.
Transfected COS-1 cells were treated with either 0, 1 or 10 μM ionomycin for 0, 10, 20 or 40 min or with 0 or 10 μM A23187 (Sigma-Aldrich) for 0 or 40 min, in order to elevate the intracellular Ca2+ concentration. The Ca2+ ionophores were dissolved in DMSO and diluted in DMEM to reach final concentrations. Moreover, HL-1 cells were treated with 10 μM A23187 in Ca2+ free DMEM for 40 min. Cells treated with DMSO in Ca2+-containing DMEM served as control. In both experiments the cells were fixed in 4% paraformaldehyde for 20 min. KChIP2 was stained using a KChIP2 antibody (10.4 μg/ml, NeuroMab) and fluorescent secondary antibody (4 μg/ml, Alexa Flour 488 donkey anti-mouse, Invitrogen) and the nuclei were stained using 4′,6-diamidino-2-phenylindole (DAPI, 16.7 μg/ml). Protein localization was visualized using confocal microscopy (LSM710, Zeiss) and ImageJ software was used to quantify the fluorescent signal across the cell. This was done by drawing a line through both the nucleus and cytosol, using the fluorescent signal intensities as a measurement for protein density. One hundred consecutive values were chosen within both nucleus and cytosol, the values were averaged and a nucleus/cytosol ratio was calculated.
Microarray analysis
Ventricular heart tissue from 3 wild type and 3 KChIP2−/− mice was used for the analysis. The tissue was homogenized using Precellys system and from the homogenates the RNA was isolated. NanoDrop Spectrophotometer and Flourospectrometer (both Thermo Scientific) were used for RNA concentration determination. RNA was amplified and labelled using a pico-amplification kit according to manufactures instructions. In short, 50 ng total RNA was amplified using the Ovation Pico WTA v.2 RNA Amplification System from (NuGEN, San Carlos, CA, USA) and biotin labelling was performed with the Encore Biotin Module (NuGEN). The labelled samples were hybridized to the Mouse Gene 1.0 ST GeneChip array (Affymetrix, Santa Clara, CA, USA). The arrays were washed and stained with phycoerytrin conjugated streptavidin (SAPE) using the Affymetrix Fluidics Station 450 and the arrays were scanned in the Affymetrix GeneArray 3000 7G scanner to generate fluorescent images. Cell intensity files (CEL files) were generated in the GeneChip Command Console Software (AGCC) (Affymetrix, Santa Clara, CA, USA) and imported into the software Partek Genomics Suite. The data was pre-filtered to exclude genes with an average expression level within each group (WT and KO) below noise or background level of log2 values of 3. Class comparison between the WT and KO was performed and a gene was defined to be differentially expressed between the two groups if p-value was below 0.05 in t-test and fold change above 1.4. For hierarchical cluster visualization of membrane proteins and ion channels, the average expression value for each of the two groups was imported into Qlucore Omics explorer v. 3.2 (Qlucore, NY, USA) and genes were normalized to have mean equal to zero.
Chromatin ImmunoPrecipitation (ChIP)
Neonatal ventricular mouse cardiomyocytes were isolated 1–2 days postpartum and cultured as described previously34. Cardiomyocytes were treated with 1 μM A23187 (Sigma-Aldrich) or DMSO as control for 1 hour. Cells were fixed in 1% formaldehyde for 15 min at room temperature, washed in ice-cold PBS, scraped in Farnham lysis buffer containing Protease Inhibitor Cocktail (PIC, Roche, Switzerland), centrifuged for 5 min at 700 g and pellets were snap frozen. Pellets were then resuspended in RIPA buffer containing PIC and sonicated to an averaged chromatin size of 200–300 base pairs using Bioruptor sonicator 4 × 10 min, followed by centrifugation for 15 min at 15.000 rpm. Both monoclonal and polyclonal KChIP2 antibodies (13 μg/ml for NeuroMab and 2.5 μg/ml for Santa Cruz Biotechnology, TX, USA) were used to immunoprecipitate protein-DNA complexes. Rabbit IgG was used as negative control while KChIP3 antibodies were used for positive control. The DNA was purified using QIAquick PCR Purification Kit (Qiagen, Germany) following the manufacturer’s instructions and the DNA fragments used as template for quantitative PCR (qPCR). KChIP2 binding was examined using primers specific for the Cacna1c promoter region containing a Downstream Regulatory Element (DRE) sequence, which is a putative KChIP2 binding site, or for a negative control region about 1000 base pairs downstream from the binding site.
Statistical analysis
All data are presented as mean ± SEM. Statistical analysis were done with Student’s t-test or 1-way ANOVA for comparison of two groups or more underlying two variable conditions. P-values < 0.05 were considered statistical significant.
Additional Information
How to cite this article: Winther, S. V. et al. Potassium Channel Interacting Protein 2 (KChIP2) is not a transcriptional regulator of cardiac electrical remodeling. Sci. Rep. 6, 28760; doi: 10.1038/srep28760 (2016).
References
Stewart, S., MacIntyre, K., Hole, D. J., Capewell, S. & McMurray, J. J. V. More ‘malignant’ than cancer? Five-year survival following a first admission for heart failure. European Journal of Heart Failure 3, 315–322 (2001).
Braunwald, E. Heart Failure. JACC: Heart Failure 1, 1–20 (2013).
Beuckelmann, D. J., Näbauer, M. & Erdmann, E. Alterations of K+ currents in isolated human ventricular myocytes from patients with terminal heart failure. Circ. Res. 73, 379–385 (1993).
Grubb, S. et al. Loss of K+ Currents in Heart Failure Is Accentuated in KChIP2 Deficient Mice. J Cardiovasc Electrophysiol 25, 896–904 (2014).
Näbauer, M., Beuckelmann, D. J. & Erdmann, E. Characteristics of transient outward current in human ventricular myocytes from patients with terminal heart failure. Circ. Res. 73, 386–394 (1993).
Wickenden, A. D. et al. The role of action potential prolongation and altered intracellular calcium handling in the pathogenesis of heart failure. Cardiovascular Research 37, 312–323 (1998).
Gómez, A. M. et al. Defective excitation-contraction coupling in experimental cardiac hypertrophy and heart failure. Science 276, 800–806 (1997).
Grubb, S. et al. Preservation of cardiac function by prolonged action potentials in mice deficient of KChIP2. Am. J. Physiol. Heart Circ. Physiol. 309, H481–489 (2015).
Braunwald, E. Research Advances in Heart Failure A Compendium. Circulation Research 113, 633–645 (2013).
Pongs, O. & Schwarz, J. R. Ancillary Subunits Associated With Voltage-Dependent K+ Channels. Physiological Reviews 90, 755–796 (2010).
An, W. F. et al. Modulation of A-type potassium channels by a family of calcium sensors. Nature 403, 553–556 (2000).
Kuo, H.-C. et al. A Defect in the Kv Channel-Interacting Protein 2 (KChIP2) Gene Leads to a Complete Loss of Ito and Confers Susceptibility to Ventricular Tachycardia. Cell 107, 801–813 (2001).
Foeger, N. C., Wang, W., Mellor, R. L. & Nerbonne, J. M. Stabilization of Kv4 protein by the accessory K(+) channel interacting protein 2 (KChIP2) subunit is required for the generation of native myocardial fast transient outward K(+) currents. J. Physiol. (Lond.) 591, 4149–4166 (2013).
Thomsen, M. B., Foster, E., Nguyen, K. H. & Sosunov, E. A. Transcriptional and electrophysiological consequences of KChIP2-mediated regulation of CaV1.2. Channels 3, 308–310 (2009).
Thomsen, M. B. et al. Accessory Subunit KChIP2 Modulates the Cardiac L-Type Calcium Current. Cir. Res. 104, 1382–1389 (2009).
Soltysinska, E. et al. Transmural expression of ion channels and transporters in human nondiseased and end-stage failing hearts. Pflugers Arch. 459, 11–23 (2009).
Ambrosi, C. M., Yamada, K. A., Nerbonne, J. M. & Efimov, I. R. Gender Differences in Electrophysiological Gene Expression in Failing and Non-Failing Human Hearts. PLoS ONE 8, e54635 (2013).
Pruunsild, P. & Timmusk, T. Subcellular localization and transcription regulatory potency of KCNIP/Calsenilin/DREAM/KChIP proteins in cultured primary cortical neurons do not provide support for their role in CRE-dependent gene expression. J. Neurochem. 123, 29–43 (2012).
Carrión, A. M., Link, W. A., Ledo, F., Mellström, B. & Naranjo, J. R. DREAM is a Ca2+-regulated transcriptional repressor. Nature 398, 80–84 (1999).
Buxbaum, J. D. et al. Calsenilin: a calcium-binding protein that interacts with the presenilins and regulates the levels of a presenilin fragment. Nat. Med. 4, 1177–1181 (1998).
Ronkainen, J. J. et al. Ca2+-calmodulin-dependent protein kinase II represses cardiac transcription of the L-type calcium channel alpha(1C)-subunit gene (Cacna1c) by DREAM translocation. J. Physiol. (Lond.) 589, 2669–2686 (2011).
Gomez-Villafuertes, R. et al. Downstream Regulatory Element Antagonist Modulator Regulates Ca2+ Homeostasis and Viability in Cerebellar Neurons. J. Neurosci. 25, 10822–10830 (2005).
Ledo, F. et al. The DREAM–DRE interaction: key nucleotides and dominant negative mutants. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1498, 162–168 (2000).
Link, W. A. et al. Day-Night Changes in Downstream Regulatory Element Antagonist Modulator/Potassium Channel Interacting Protein Activity Contribute to Circadian Gene Expression in Pineal Gland. J. Neurosci. 24, 5346–5355 (2004).
Hasenfuss, G. & Pieske, B. Calcium cycling in congestive heart failure. J. Mol. Cell. Cardiol. 34, 951–969 (2002).
Lundby, A. et al. In vivo phosphoproteomics analysis reveals the cardiac targets of β-adrenergic receptor signaling. Sci Signal 6, rs11 (2013).
Lundby, A. et al. Effect of the Ito activator NS5806 on cloned Kv4 channels depends on the accessory protein KChIP2. Br J Pharmacol 160, 2028–2044 (2010).
Thomsen, M. B. et al. Deleting the accessory subunit KChIP2 results in loss of I(to, f) and increased I(K, slow) that maintains normal action potential configuration. Heart Rhythm 6, 370–377 (2009).
Norris, A. J., Foeger, N. C. & Nerbonne, J. M. Interdependent roles for accessory KChIP2, KChIP3 and KChIP4 subunits in the generation of Kv4-encoded IA channels in cortical pyramidal neurons. J. Neurosci. 30, 13644–13655 (2010).
Takimoto, K., Yang, E.-K. & Conforti, L. Palmitoylation of KChIP Splicing Variants Is Required for Efficient Cell Surface Expression of Kv4.3 Channels. J. Biol. Chem. 277, 26904–26911 (2002).
Bauer, N. C., Doetsch, P. W. & Corbett, A. H. Mechanisms Regulating Protein Localization. Traffic 16, 1039–1061 (2015).
Nassal, D. M., Wan, X., Liu, H. & Deschênes, I. Myocardial KChIP2 Expression in Guinea Pig Resolves an Expanded Electrophysiologic Role. PLOS ONE 11, e0146561 (2016).
Qi, X. Y. et al. Cellular signaling underlying atrial tachycardia remodeling of L-type calcium current. Circ. Res. 103, 845–854 (2008).
Ronkainen, V.-P. et al. Hypoxia inducible factor regulates the cardiac expression and secretion of apelin. FASEB J. 21, 1821–1830 (2007).
Acknowledgements
The authors greatly appreciate the technical assistance and intellectual input from Artina Metoska, Amer Mujezinovic, Camilla Stampe Jensen, Tobias Speerschneider and Nancy Mutsaers. The present study was financially funded by the Novo Nordisk Foundation (to M. B. Thomsen) and The Danish Agency for Science, Technology and Innovation, Medical Research Council (grant 12-132164 to M. B. Thomsen).
Author information
Authors and Affiliations
Contributions
S.V.W. contributed to the conception and design of the experiments, the collection, analysis and interpretation of data and to writing the manuscript. T.T. and RBH-L contributed to the collection, analysis and interpretation of data. P.T. and G.A. contributed to the conception and design of the experiments and interpretation of data. M.B.T. contributed to the conception and design of the experiments, the collection, analysis and interpretation of data and to writing the manuscript. All authors have contributed through critical review of the intellectual content of the manuscript and all have approved the submitted version.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Winther, S., Tuomainen, T., Borup, R. et al. Potassium Channel Interacting Protein 2 (KChIP2) is not a transcriptional regulator of cardiac electrical remodeling. Sci Rep 6, 28760 (2016). https://doi.org/10.1038/srep28760
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep28760
This article is cited by
-
Modulation of human Kv4.3/KChIP2 channel inactivation kinetics by cytoplasmic Ca2+
Pflügers Archiv - European Journal of Physiology (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.