Abstract
Cyclins play a central role in cell-cycle regulation; in mammals, the D family of cyclins consists of cyclin D1, D2 and D3. In Xenopus, only homologs of cyclins D1 and D2 have been reported, while a novel cyclin, cyclin Dx (ccndx), was found to be required for the maintenance of motor neuron progenitors during embryogenesis. It remains unknown whether zebrafish possess cyclin D3 or cyclin Dx. In this study, we identified a zebrafish ccndx gene encoding a protein which can form a complex with Cdk4. Through whole-mount in situ hybridization, we observed that zccndx mRNA is expressed in the motor neurons of hindbrain and spinal cord during development. Analysis of a 4-kb promoter sequence of the zccndx gene revealed the presence of HRE sites, which can be regulated by HIF2α. Morpholino knockdown of zebrafish Hif2α and cyclin Dx resulted in the abolishment of isl1 and oligo2 expression in the precursors of motor neurons and also disrupted axon growth. Overexpression of cyclin Dx mRNA in Hif2α morphants partially rescued zccndx expression. Taken together, our data indicate that zebrafish cyclin Dx plays a role in maintaining the precursors of motor neurons.
Similar content being viewed by others
Introduction
Cyclins and cyclin-dependent kinases (Cdks) were first identified in an effort to understand cell cycle control in different organisms, such as yeast, frogs and mammals1,2,3,4,5,6. All cyclins have a conserved cyclin box domain that can bind to a similar region on Cdk7. In mammalian cells, cyclin D-Cdk4/6 is the first kinase to be activated by mitogenic signals that allow cells to exit G0 phase and enter G1 phase; this is followed by sequential activation of cyclin E-Cdk2 at the G1/S phase boundary, cyclin A-Cdk2 during S phase, cyclin A-Cdk1 during G2 phase and finally, cyclin B-Cdk1 during mitosis8,9. Cyclins D1 and E are under mitogenic control and are frequently deregulated during oncogenesis8.
Besides cell cycle control, cyclins, such as A1, A2, B1, B2, B3, D2 and D3, also play crucial roles in the control of mitotic and meiotic divisions during mammalian spermatogenesis10. In addition, cyclin E plays important roles in cell fate determination in the central nervous system11, while cyclin D1 can act as a transcriptional cofactor in a CDK-independent manner12.
In mammals, the cyclin D family consists of three members, cyclin D1, D2 and D3, which are also called G1-phase regulators13,14,15,16. In Xenopus, the cyclin D1 and cyclin D2 homologs have been cloned and characterized. At stage 33/34, cyclin D1 can be detected in eye, brain, branchial arches, neural tube and tailbud, while cyclin D2 is also localized to the head region, including the brain, otic vesicle, nasal pits and branchial arches17,18,19,20. Overexpression of cyclin D2 induces cell cycle arrest during early development18. However, a homolog of the cyclin D3 gene has not been identified in Xenopus. Instead, a novel Xenopus cyclin, cyclin Dx (ccndx), was recently identified and found to be required for the maintenance of motor neuron progenitors (pMNs) during embryogenesis21.
In zebrafish, only cyclin D1 has been reported; this cyclin is expressed in G1 phase during development22 and is required for early eye development23. Down-regulation of cyclin D1 by morpholino (MO) injection was reported to cause microphthalmia and microcephaly24. Other cyclin D family members in zebrafish have not previously been reported. In this study, we identified and characterized a cyclin Dx homolog from zebrafish. The developmental and tissue-specific expression patterns of this zccndx mRNA were determined by RT-PCR and whole-mount in situ hybridization analysis. The promoter region of the zebrafish zccndx gene was also characterized, suggesting that it is regulated by Hif2α. Morpholino knockdown of zccndx or Hif2α expression demonstrated the importance of these genes in maintaining the precursors of motor neurons.
Initiation of anti-neural factor expression expands the pool of neural progenitors; subsequent cell cycle exit and differentiation results in the development of primary motor neurons. Expression of two genes, Islet1 (Isl1) and oligo2 (olig2), is required to maintain a pro-neurogenic state. Isl1 is required for motor neuron generation in vertebrates25, loss of zebrafish isl1 leads to the loss of primary motor neurons26. On the other hand, olig2 is required for primary motor neuron differentiation of neural precursor cells27; phosphorylation of olig2 maintains cells in a pro-neurogenic state28. Both genes are essential for primary motor neuron development. In this study, we identified that the zebrafish ccndx gene acts as a cell-cycle regulator to maintain the pool of motor neuron progenitors, as previously reported for Xenopus21. Furthermore, knockdown of zccndx resulted in loss of differentiated motor neurons and ultimately, the disruption of axon growth.
Experimental Procedures
Fish
The Tg (isl:GFP) transgenic line29 was obtained from Zebrafish Core Facility at NHRI (National Health Research Institutes). Zebrafish (Danio rerio) were maintained at 28 °C on a 14 h-light/10 h-dark cycle. Embryos were incubated at 28 °C and different developmental stages were determined according to the description provided in the Zebrafish Book30. All animal procedures were approved by Academia Sinica Institutional Animal Care and Utilization Committee (ASIACUC) (protocol #10-12-114). The methods were carried out in accordance with the approved guidelines.
Cell culture
Monkey kidney fibroblast COS-1 cells and human embryonic kidney HEK 293T cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% fetal bovine serum (FBS; HyClone, UT, USA) in a humidified atmosphere of 5% CO2 at 37 °C.
Cloning of the full-length cDNAs encoding zebrafish cyclin dx and cyclin d1
The cDNA encoding the complete open reading frame (ORF) of zebrafish ccndx (cyclin dx), ccnd1 (cyclind1) and smo were obtained by PCR amplification using gene-specific primers, which were designed based on the NCBI’s zebrafish EST database. DNA fragments containing full-length cDNA were obtained by PCR amplification using forward primer (ccndx-F1: 5′-GGA CAG ACT TGA GTG TTG GAG CAG-3′; ccnd1-F1: 5′-AAC ACT GAA GAT ATG GAG CAC CAG-3′) and reverse primer (ccndx-R1: 5′-TAT ACG AGT GAT GAA TGA ACC GCG-3′; ccnd1-R1: 5′-ACA CAC GAG ATG AAT AGC GAT CCC-3′). The PCR products were subcloned into the pGEM-T Easy vector (Promega, WI, USA) and the resulting clones were subjected to sequence analysis.
Luciferase assay
The 2,432bp upstream region of the zebrafish cyclin dx gene was amplified from BAC clone, CH211-152F23 (RZPD, Berlin, Germany), using forward primer (F1: 5′- GGT ATC CCT GGA CCT TGT ATA CGC AGTGGT-3′) and reverse primer (R1: 5′- CAA TGT GTC CTG TTG CAC GTG TGT CTT GTG-3′). The amplified fragments were then inserted into pGEM-T Easy cloning vector (Promega, WI) for sequencing and subsequently subcloned into the KpnI/NcoI sites of the pGL3 vector to generate pGL3-ccndx-2.5k-pro plasmid. Reduced promoter region of pGL3-ccndx-2.5 kb to 1,096bp by restriction enzyme digestion in HindIII and KpnI sites was generated pGL3-1 b-pro plasmid. The pGL3-0.7 -pro plasmid was generated by reduced promoter region to 654bp using SacI and KpnI sites. Constructs containing mutated HRE elements were generated using the following primers: pGL3-ccnx-0.7 -pro-HRE1-Mut – 0.7KPROF (5′-CAC GGT ACC CAC AGT TTT CGA GGG ATA C-3′), HRE1MF (5′-CAC GGT ACC CAC AGT TTT CGA GGG ATA C-3′) and HRE1MR (5′-AAT CTC GAG CAA ATC TGT CCT GTT GAT ATC G-3′); 0.7KPROF (5′-CAC GGT ACC CAC AGT TTT CGA GGG ATA C-3′), 0.7KPROR (5′-AAT CTC GAG CAA TGT GTC CTG TTG CAC GTG-3’), pGL3-ccnx-0.7 -pro-HRE2-Mut – HRE2MF (5′-CAA AAT ATC TTC TTT TTG ATA TCA ACA TTT-3′) and HRE2MR (5′-AAA TGT TGA TAT CAA AAA GAA GAT ATT TTG-3′). The 5 × 105 COS-1 cells were placed in each of 12-well plate overnight before transfection. Cells was transfected with 0.5 g of pGL3-2.5k-pro, pGL3-1k-pro, pGL3-0.7k-pro, pGL3-0.7k-pro-HRE1-Mut, pGL3-0.7k-pro-HRE2-Mut plasmid along or co-transfected with 1 g of pcDNA3-Hif2α-HA and pcDNA3-Arnt1α-HA using the Lipofectamine Plus reagent (Invitrogen), according to the manufacturer′s instructions. Luciferase activity were measured at 48 h post-transfection using the ONE-GLO Luciferase Assay Reagent (Promega, WI, USA).
Electrophoretic mobility shift assay
After COS-1 cells were transfected with pcDNA3-HA or pcDNA3-Hif2α-HA and pcDNA3-Arnt1α-HA plasmids for 48 h, nuclear extracts were prepared by minipreparation method as described previously31 The nuclear extracts were storage at −70 °C until use. The HRE1 and HRE2 sites of the ccndx promoter were used as probes for EMSA experiment. HRE1 (5′-CAC AAG ACA CAC GTG CAA CAG GAC ACA TTG-3′) and HRE2 (5′- CAA AAT ATC TTC TTT TTG CGT GCA ACA TTT-3′) core sequence and corresponding mutant oligonucleotides were synthesized with or without labeled at the 5′-end with biotin (Genomics, Taipei, Taiwan). For each EMSA experiment, 20 g of nuclear extracts were incubated with HRE1 or HRE2 probe and Binding Reagents (Thermo, IL, USA) at room temperature for 30 minutes. For competitive experiments, 50X normal or mutant probes were added. The DNA-protein complexes were separated on a non-denaturing 10% polyacrylamide gel. The gel was transferred to a nylon membrane (Pall, NY, USA), followed by UV cross-linking and detection with a LightShift Chemiluminescent EMSA Kit (Thermo, IL, USA).
Plasmid construction
The expression vectors, pcDNA3-ccndx-myc and pcDNA3-ccnd1-myc, were generated by inserting full-length cyclin dx and cyclin d1 cDNA between BamHI and EcoRI sites of pcDNA3-myc-HisA plasmid, respectively. The pcDNA3-cdk4-HA plasmid was constructed by inserting the full-length cdk4 cDNA between the BamHI and EcoRI sites of the pcDNA3-HA-YUN plasmid.
Immunoblotting and co-immunoprecipitation analysis
Total cell lysate was collected with IP-lysis buffer (150 mM NaCl, 20 mM NaCl, 20 mM HEPES, pH 7.2, 10 mM NaF, 1 mM EDTA, 1% NP-40, 1 mM Na3V04, 1 mM PMSF, 1 DTT and proteinase inhibitor cocktail) at 72 h and subjected to immunoblotting with anti-myc (1:3000, Santa Cruz, CA, USA) and anti-HA (1:1000, Santa Cruz, CA, USA) monoclonal antibody. Co-immunoprecipitation was performed with 25 μl Protein A/G PLUS-Agarose (Santa Cruz, CA, USA) and anti-myc monoclonal antibody (1 μg) to pull-down the complex, which was then immunoblotted with anti-HA monoclonal antibody.
Capped mRNA synthesis
Full-length cDNA fragments of zebrafish ccndx (JX099349.1), mouse ccnd1 (NM_007631.2), ccnd2 (NM_009829.3) and ccnd3 (NM_001081635.1) were amplified and inserted into the pCS2 + vector. Each of the resulting pCS2 + plasmids was linearized with NotI. Capped mRNA was transcribed from 1 μg linearized plasmid using the mMESSAGE mMACHINE SP6 Kit (Ambion, Foster City, CA, USA).
Morpholino knockdown of hif2α and ccndx
The antisense morpholino oligonucleotides (MO) used to knockdown hif2α were designed according to a previous report32. The sequences of the ccndx MOs were as follows: ccndx ATG MO- 5′ ATC ACT GCT CCA ACA CTC AAG TCT G 3′; ccndx splicing MO- 5′- TGG TTT ATG ATA TCT AAA CAC TAC C 3′. The control MO sequence was as follows: 5′- CCT CTT ACC TCA GTT ACA ATT TAT A-3′. All antisense MOs were synthesized by Gene Tools (Philomath, OR, USA). Each MO was dissolved in 1x Danieau solution (5 mM HEPES, pH 7.6, 58 mM NaCl, 0.7 mM KCl, 0.4 mM MgSO4 and 0.6 mM (NO3)2) containing 0.5% phenol red to a concentration of 0.3 mM.
Injection of morpholinos and capped mRNA
MOs and capped mRNA were injected into wild-type zebrafish embryos using a microinjection system consisting of a SZX9 stereomicroscope (Olympus, Tokyo, Japan) and an IM300 Microinjector (Narishige, Tokyo, Japan). The capped mRNA was injected into embryos at the 1-cell stage, while MOs were injected at the 2- to 4-cell stage of wild-type zebrafish embryos. The MO-mediated effects were evaluated at 24 h after injection.
Whole-mount immunostaining
Whole-mount immunostaining was performed following standard protocols as previously described33 with some modifications. The antibodies used were as follows: mouse monoclonal anti-synaptotagmin antibody against znp134 (Developmental Studies Hybridoma Bank (DSHB), Iowa City, IA) and Cy2-conjugated anti-mouse IgG (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA).
Whole-mount in situ hybridization
To synthesize digoxigenin-labeled (Roche, Penzberg, Germany) antisense RNA probes, we linearized pGEM-T easy-ccndx with PstI and transcribed it with T7 RNA polymerase. The probes for motor neuron progenitors and interneurons (isl1, olig2, nkx6.1 and vsx1) have been described previously27,35,36,37; these probes were linearized with NcoI and transcribed with SP6 RNA polymerase. To generate fluorescein-labeled (Roche, Penzberg, Germany) antisense probes, we linearized pcDNA3-CMV-GFP with HindIII and transcribed it with SP6 RNA polymerase. Whole-mount in situ hybridization was performed as previously described38,39. For double fluorescence in situ hybridization, the digoxigenin-labeled probes were detected using anti-digoxigenin POD (Roche, Penzberg, Germany) (1:500 dilution) and signals were amplified using a tyramide-FITC kit (Perkin Elmer, Boston, MA, USA). After the first staining step, the tyramide working solution was washed away and the embryos were incubated for 30 minutes with 1% H2O2, to inactivate the peroxidase activity of the first antibody. The embryos were then blocked for 1 hour and the fluorescein-labeled probes were detected with anti-fluorescein POD (Roche, Penzberg, Germany) (1:500 dilution); the signals were subsequently amplified using a tyramide-Cy2 kit (Perkin Elmer, Boston, MA, USA).
Proliferating cell labeling and quantification
Proliferating cells were labeled with rabbit anti-pH3 (Cell signaling Technology) and BrdU (Sigma) as previously described40,41; and goat anti-mouse-IgG conjugated to Cy2 and Cy3 (Invitrogen, USA) were utilized in our study. The fluorescent images were acquired on a Zeiss LSM 710 laser confocal microscope. The labeled proliferating cells within the image of the developing brain were counted through ImageJ analysis. All the statistical analysis was performed by SPSS. Significance was evaluated by one-way ANOVA while considered differences at P < 0.05.
High resolution confocal imaging
Zebrafish embryos were fixed with 4% paraformaldehyde in PBS at 4 °C for 24 and then washed 3 times in PBS (5 min/wash). Embryos were cleared as described previously42; embryos were incubated in FocusClear in a small chamber with a spacer ring for 5 min and then mounted with MountClear at room temperature for 30 min. Images were captured with a Zeiss LSM 710 confocal microscope (Carl Zeiss, Jena).
Results
Cloning and characterization of zebrafish cyclin Dx
Mammals possess three members of the cyclin D family: cyclin D1, D2 and D313,14,43,44. In Xenopus, cyclin D1 and D2 have been identified17,45, but not cyclin D3. Instead, a novel cyclin D, cyclin Dx, was recently identified in Xenopus21. From an evolutionary perspective, zebrafish may be expected to possess cyclin D family members similar to those of Xenopus. However, only cyclin D1 has been identified in zebrafish24,46. To search for zebrafish cDNAs related to Xenopus cyclin Dx and cyclin D2, we used the coding regions of Xenopus cyclin Dx (accession no. AAI58908) and cyclin D2 (P53782) to BLAST search GenBank for related expression sequence tag (EST) sequences. Sequence assembly and 5′-RACE enabled the identification of one zebrafish cyclin Dx and two cyclin D2-related cDNAs, designated zccnDx, zccnD2A and zccnD2B, respectively; the assembled sequences were deposited in GenBank with the following accession numbers: FJ648072, zccnDx; FJ648073, zccnD2A; and FJ648074, zccnD2B. The full-length coding regions of each cDNA were amplified by PCR using gene-specific primers and were confirmed by sequence analysis.
An alignment of the deduced amino acid sequences of zebrafish, Xenopus and human cyclin D-related proteins is shown in Supplemental Fig. 1. The zebrafish cyclin Dx protein contains 297 amino acid residues and displayed low identity to all examined cyclin D-related proteins (31% to 35% identity; 50% to 53% similarity). The zccnD2A and zccnD2B cDNA sequences are predicted to encode proteins of 298 and 296 amino acid residues, respectively. The zebrafish cyclin D2A protein exhibits higher identity to human and Xenopus cyclin D2 (79% and 80% identity, respectively). On the other hand, zebrafish cyclin D2B protein displays 68% identity to human and Xenopus cyclin D2. Interestingly, zebrafish cyclin D2B is also homologous to cyclin D2A with 72% identity, suggesting that they may be derived from a gene duplication event. The cyclin box domain of cyclin Dx was conserved with that of other cyclin D family members. Phylogenetic tree analysis, as shown in Fig. 1A, indicated that zebrafish cyclin Dx is grouped with Xenopus cyclin Dx, but is separate from other cyclin D family members.
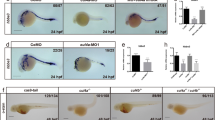
Zebrafish cyclin Dx is a member of the cyclin D family and zccndx is expressed during early development.
(A) Phylogenetic tree analysis of zccndx with other cyclin D proteins. Four groups of the cyclin D family, ccnD1, D2, D3 and Dx, exhibited perfect separation. The zccndx protein was grouped with Xenous ccndx. Analysis was performed using the parsimony method with 1000 bootstraps. (B) The zccndx protein binds to Cdk4. COS-1 cells were cotransfected with the indicated constructs and cell lysates were co-immunoprecipitated (IP) with anti-HA monoclonal antibody; the anti-Myc monoclonal Ab was used for detection. The immunoblot was probed using the indicated antibodies. (C) At 24 hpf, zccndx mRNA was observed to be expressed in the motor neurons of hindbrain and spinal cord (a and a′). At 48 hpf, zccndx mRNA was also expressed in the hindbrain, but not in the spinal cord (b and b′). At 72 hpf, zccndx mRNA was weakly expressed in the hindbrain, with no signal in the spinal cord (c and c′). Dorsal views of zccndx mRNA signal in the hindbrain are shown (a′–c′). Scale bar, 100 mm. (D) Sections were made in the hindbrain at 22 hpf. The zccndx signal is shown in green and the isl1 (panel a), olig2 (panel b) and vsx1 (panel c) and nkx6.1 (panel d) mRNA signals are shown in red. The cartoon (panel e) shows the location of zccndx (green), isl1 (purple), olig2 (orange), vsx1 (blue) and nkx6.1 (red) signals. The Shh (blue) signaling gradient has been created from the floor plate to diffuse along the ventral tube while BMP/Wnt (purple) gradient shows opposite direction. The interaction of the dorsalizing and ventralizing signals defines the proper position of motoneurons. Scale bar, 100 mm.
Xenopus cyclin Dx has been reported to bind to Cyclin-dependent kinase 4 (Cdk4) in HEK 293T cells21. Based on this report and the above finding, we hypothesized that zebrafish cyclin Dx may be a member of the cyclin D family and bind to Cdk4. Indeed, co-immunoprecipitation experiments revealed that zebrafish cyclin Dx binds to Cdk4 in COS-1 cells (Fig. 1B).
Spatial and temporal distribution of zebrafish cyclin Dx mRNA during development
Whole-mount in situ hybridization using the 3′-UTR region of zccndx gene as probe was performed to detect the expression pattern of zccndx mRNA during development. At 24 hpf, zccndx mRNA was expressed on both sides of the midline in hindbrain and spinal cord (Fig. 1C, a and a′). At 48 hpf, zccndx mRNA continued to be expressed in the hindbrain, but the signal in the spinal cord became weak (Fig. 1C,b,b′). At 72 hpf, the expression of zccndx mRNA was very weak in the hindbrain and no signal was detected in the spinal cord (Fig. 1C,c,c′). The expression pattern of zccndx mRNA is similar to that of xccndx mRNA, which has been reported to be expressed in the progenitors of motor neurons (pMNs)21. Markers for pMNs, such as isl135 and olig227, were subsequently used to determine co-localization of zccndx mRNA expression in the experiments described below.
The zebrafish cyclin Dx gene is expressed in the motor neuron progenitor region
During the early development of zebrafish, motor neuron progenitors (pMNs) are distributed in the ventral neural tube between the V2 and V3 interneuron progenitors. The isl1 and olig2 genes are significantly expressed in this neural tube layer and were thus used as markers to identify pMNs27,35. The vsx1 gene is expressed in the V2 interneuron progenitor layer in the neural tube and was thus used as the interneuron marker47. Finally, the nkx6.1 gene is expressed in V3 interneuron progenitors48.
We performed double in situ hybridization of zebrafish embryo sections to investigate whether the zccndx gene is expressed in the pMN layer. Different markers for the progenitors of motor neurons and interneurons in the ventral spinal cord, such as isl1, olig2, vsx1, nkx6.1 and endogenous zccndx, were double labeled with Cy3 and Cy5 at 22 hpf. The zccndx mRNA was observed to be expressed in the ventral spinal cord (Fig. 1D, panel a). The isl1 mRNA signal in the pMN layer of the ventral spinal cord partially overlapped with the zccndx mRNA signal in the same layer (Fig. 1D, panel a). An olig2 mRNA signal was also observed in the pMN layer of the ventral spinal cord and this signal partially overlapped with the zccndx mRNA in the same layer (Fig. 1D, panel b). A vsx1 signal was observed in the V2 layer of the ventral spinal cord; this signal was above the zccndx mRNA signals and the two signals did not overlap (Fig. 1D, panel c). Another marker of the V3 layer of interneuron progenitors, nkx6.1 mRNA, was observed to be expressed below the zccndx mRNA signal (Fig. 1D, panel d). Taken together, our data demonstrate that expression of ccndx mRNA is restricted to the pMN layer of ventral spinal cord and partially overlaps with the expression of olig2 and isl1 mRNA (Fig. 1D, panel e).
The ccndx gene is required for the maintenance of motor neuron progenitors
To explore the functions of zebrafish cyclin Dx, we performed MO knockdown of the ccndx gene; this resulted in the loss of motor neurons. To determine the specificities of the MOs used, we created pCMV-GFP reporter plasmids. The 25-bp target sequence of the ATG MO was cloned upstream of the GFP open reading frame (ORF) in the pCMV-GFP reporter plasmid. We injected zebrafish embryos with a pCMV-GFP reporter plasmid containing the MO target sequence, with or without ccndx ATG-MO (Supplementary Fig. 2A, panels a and b). The GFP signal intensity was decreased in MO-injected embryos as compared to embryos uninjected with MO, confirming the specificity of ccndx MO; in addition, levels of ccndx were shown to be reduced by the MO (Supplementary Fig. 2A,B). While 81.1% and 68.3% of ccndx morphant embryos exhibited decreased expression of olig2 and isl1 signals in motor neurons as compared to this control, these percentages could be decreased to 15% and 37.6% by co-injection of ccndx mRNA, indicative of partial rescue (Fig. 2A, panels a–h). In addition, we analyzed the effects of the loss of ccndx on motoneurons by using the Tg:(isl1:GFP) transgenic line. GFP expression was observed in all the cranial motoneurons and primary motoneurons located in the ventral region of the spinal cord within control MO-injected embryos at 72 hpf (Fig. 2B, panel a). However, ccndx morphant embryos exhibited lower numbers of GFP-labeled primary motoneurons compared to control MO-injected embryos (Fig. 2B, panels b and d). This phenotype could be partially rescued by injection of ccndx mRNA (Fig. 2B, panels c and d). To determine whether ccndx is required for normal outgrowth by primary motor axons, we injected ccndx MO into fertilized embryos and assayed primary motor axons with antibody of znp1 after early development. We observed that 62.8% of ccndx morphants exhibited shorter CaP axon branches compared with control morphants (Fig. 2C, panel b). Again, injection of ccndx mRNA could partially rescue this phenotype (Fig. 2D, panels c and d). Thus, zccndx functions in maintaining the expression of motor neuron progenitors, which are specifically required for subsequent primary motor neuron formation during development in the ventral spinal cord and is also necessary for the subsequent appropriate outgrowth of CaP axons.
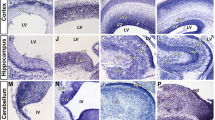
Morpholino (MO) knockdown of zccndx affects motor neuron development.
(A) MO knockdown of zccndx reduced the olig2 and isl1 signals in motor neurons (panels a, b, e and f); rescue experiments performed by co-injecting ccndx mRNA are shown in panels c and g. Panels d and h: graphs showing the qPCR analysis confirmed the results of morphants with signals lower than control MO-injected embryos at 24 hpf. **P < 0.05. Scale bar, 100 mm. (B) The ccndx morphant showed a significantly higher ratio of embryos with lower numbers of GFP-labeled primary motoneurons than the control MO-injected embryos (panels a and b) in the Tg:(isl1:GFP) transgenic line. The ccndx mRNA recue experiment is shown in panel c. Panel d: the Statistical charts represent the quantitative results of GFP-labeled primary motoneurons cells at 24 hpf. (C) Labeling of primary motor axons with Znp1 antibody revealed that CaP axon branches were shorter in ccndx morphants than in control morphants (panels a and b); the rescue experiment is shown in panel c. Panel d: graphs showing the percentages of morphants with primary motor defects as compared to control MO-injected embryos at 24 hpf in panels d and h. *P < 0.05.
We then compared the cell proliferation statuses of ccndx mutants and ccndx morphants by utilizing anti-phosphohistone 3 (pH3) antibody staining and BrdU labeling. The cell proliferation assays showed that at 24 hpf, the pH3-positive cell numbers in the brain region of ccndx morphants were significantly reduced compared to control MO-injected embryos (Fig. 3A, panels a and b). Consistent with the findings of the anti-pH3 assay, the numbers of BrdU-positive cells in brain region in ccndx morphants were reduced compared to those in control MO-injected embryos at 24 hpf (panels d and e). These data indicate that ccndx modulates cell proliferation in the brain region. To further investigate the effect of ccndx on motor neuron precursors, we injected ccndx MO into the embryos of wild-type zebrafish at 2–4 cell stages. At 24 hpf, MO-injected embryos were collected and subjected to whole-mount in situ hybridization using different markers for each progenitor domain of motor neurons and interneurons in the ventral spinal cord, namely isl1 (pMN), olig2 (pMN), vsx1 (V2) and nkx6.1 (V3). In ccndx morphants, the isl1 and olig2 signals in motor neurons were lost (Fig. 3B, panels a, b, e and f). In contrast, ccndx MO-injected embryos (panels c and g) displayed similar expression patterns of nkx6.1 and vsx1 mRNA to those observed in the control MO-injected embryos (panels c and d).
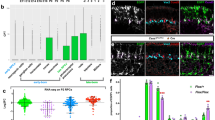
The role of zccndx in cell proliferation and regulation of the numbers of motor neuron progenitors (pMNs).
(A) Confocal images showing lateral views of the heads of control-MO-injected (panel a) or ccndx-MO-injected (panel b) embryos stained with anti-pH3 (red in panels a and b) or anti-BrdU (green in d and e). Panels c and f: graphs showing the number of proliferating cells (pH3 or BrdU positive) at 24 hpf. The asterisks indicate significant differences between the control and experimental morphants. The n value is indicated. (B) Whole–mount in situ hybridization of motor neurons and interneuron markers in control-MO and zccndx ATG-MO-injected embryos. Probes against olig2 (a and b), nkx6.1(c and d), isl1 (e and f) and vsx1 (g and h) were used for detection. control MO-injected embryos are shown in panels a, c, e and g while ccndx ATG-MO-injected embryos are shown in panels b, d, f and h. Motor neurons (MN) were lost (panels b, f, lost MNs indicated by arrowheads), while interneurons (IN) (panels d and h) were unaffected. Scale bar, 100 mm.
Expression of the zebrafish ccndx gene is regulated by HIF2α through HRE sites
In a previous report, HIF2α was found to regulate birc5a/5b gene expression by directly binding to Hypoxia-Response Elements (HREs) in their promoter region, thereby protecting neural progenitor cells32. We report here that ccndx is required for the maintenance of pMNs and that knockdown of ccndx mRNA expression causes apoptosis of pMNs. Analysis of the upstream region of the ccndx gene revealed several HRE sites. Two HRE sites are located in the upstream region of 654 bp (0.7 ) of ccndx gene. Transactivation analysis using luciferase assays in COS-1 cells revealed significant activation of this 0.7 promoter by HIF2α and ARNT1α protein (Fig. 4A). Mutation of these two HREs reduced transactivation activity (Fig. 4B). These data suggest that HIF2α regulates ccndx gene expression through HRE sites. To determine whether zebrafish HIF2α specifically binds to the HRE sites in the ccndx promoter, we performed electrophoretic mobility shift assay (EMSA) with biotin-labeled oligonucleotides for each of the two HRE core sequences (ACGTG) (Fig. 4C). When HIF2α was expressed together with ARNT1α in COS-1 cells, a mobility complex was formed with each of the two wild-type HRE probes (lanes 2 and 6). This mobility complex was successfully out-competed by a 200-fold molar excess of the unlabeled HRE (lanes 3 and 7). On contrary, the mobility complex was unaffected by a 200-fold molar excess of unlabeled mutated HRE (lanes 4 and 8). These results thus demonstrate that HIF2α binds specifically to each of the two HRE sites.
Expression of the ccndx gene is regulated by the HIF2α transcription factor.
(A) The promoter region of the ccndx gene contains several HRE sites, as indicated in the schematic diagram (top left). The 2.5-kb, 1-kb and 0.7-kb promoters are all activated by HIF2α and arnt1a transcription factors. (B) The HIF2α and ARNT1α proteins co-activate ccndx gene expression through two HRE sites in the 700bp promoter region. Data represent mean ± s.d. (n = 3). (C) The DNA binding ability of HIF2α and the ARNT1α complex were confirmed by EMSA. Nuclear extract (NE) from pcDNA3-HA (Con) or pcDNA3-Hif2α-HA and pcDNA3-Arnt1α-HA (HIF2α + ARNT1α) transfection were indicated. Two HRE oligonucleotides (as indicated) were used as probes and normal (Cold) or mutant (Mut) oligonucleotides were used for competition. The asterisks in lanes 2, 4, 6 and 8 indicate the specific binding complex.
The HIF2α transcription factor regulates ccndx gene expression, which is required for motor neuron development
To explore how HIF2α and cyclin Dx regulate the formation of motor neuron progenitors expressing olig2 and isl1, we performed rescue experiments by injecting mRNA into MO knockdown morphants. The olig2 and isl1 signals were lost from the majority of HIF2α morphants (Fig. 5A, panels b and f); motor neuron progenitor cells (expressing olig2 and isl1) were partially restored by co-injection of zccndx mRNA (Fig. 5A, panels c and g, d and h). Similar effects were observed in GFP-labeled cranial motoneurons and primary motoneurons cells of the Tg(Isl1:GFP) transgenic line; HIF2α morphants exhibited reduced GFP-labeled primary motoneurons cells number as compared to control MO-injected embryos (Fig. 5B, panels b and d), while the number of GFP-labeled motoneurons could be rescued by injection of ccndx mRNA (Fig. 5B, panels c and d). Rescue was also observed for Znp1 antibody-labeled primary motor axons during early development. We found that 53.8% of HIF2α morphants had shorter CaP axon branches than those of control morphants (Fig. 5C, panels b and d) and this could be partially reversed by injection of ccndx mRNA (Fig. 5D, panels c and d). Taken together, these results suggest that ccndx gene is regulated by HIF2α, which functions in maintaining the expression of olig2 and isl1 and the subsequent formation of primary motor axons.
The HIF2α transcription factor helps regulate ccndx gene expression and subsequent motor neuron development.
(A) MO knockdown of hif2a reduced the olig2 and isl1 signals in motor neurons (panels a, b, e and f); co-injection of ccndx mRNA partially rescued this phenotype (panels c and g). Panels d and h: graphs showing the qPCR analysis confirmed the results of morphants with signals lower than control MO-injected embryos at 24 hpf. *P < 0.05. Scale bar, 100 mm. (B) A significantly higher proportion of hif2a morphant embryos had lower numbers of GFP-labeled primary motoneurons as compared to control MO-injected embryos (panels a and b) in the Tg:(isl1:GFP) transgenic line. The effect of rescue experiments with ccndx mRNA is shown in panel c. Quantitative data at 24 hpf are shown in panel d. (C) A significantly higher proportion of hif2a morphant embryos exhibited shorter CaP axons branches as compared to control MO-injected embryos (panels a and b). The effect of rescue experiments with ccndx mRNA is shown in panel c. Quantitative data at 24 hpf are shown in panel d. *P < 0.05.
Mouse cyclin D1, D2 and D3 cannot restore the expression of isl1 and olig2 in the motor neuron progenitor region in the absence of zebrafish cyclin Dx
At the time of writing, cyclin Dx has been identified only in zebrafish and Xenopus; mammals and chicken appear to express cyclin D3 instead. To explore the functional relationships between members of the mammalian cyclin D family, we injected cyclin Dx morphants with cyclin D1, D2, or D3 mRNA. The olig2 and isl1 signals were lost from the motor neurons of ccndx morphants (Fig. 6A, panels b and h). However, the expression of isl1 and olig2 could not be restored by overexpression of mouse cyclin D1, D2, or D3 mRNA (Fig. 6A, panels c, d, e, f, h, i, j, k and l). In addition, overexpression of any one of these genes was not able to significantly restore GFP-labeled primary motoneurons of Tg:(isl1:GFP) transgenic line (Fig. 6B) and the length of primary motor axons in zccndx morphants (Fig. 6C). These data suggest that mouse cyclin D1, D2 and D3 do not have similar functions to that of zebrafish and Xenopus ccndx.
Mouse cyclin D1, D2 and D3 cannot compensate for the loss of zebrafish cyclin Dx.
(A) MO knockdown of ccndx reduced the isl1 and olig2 signals in motor neurons (panels a, b, g and h); co-injection of mouse ccnd1, ccnd2 and ccnd3 mRNA had no effect, as shown in panels c to e and i to k. Panels r and l: graphs showing the qPCR analysis confirmed the results of morphants with signals lower than control MO-injected embryos at 24 hpf. *P < 0.05. Scale bar, 100 mm. (B) A significant number of ccndx morphant embryos contained fewer GFP-labeled primary motoneurons as compared to ccndx morphants (panels a and b) in the Tg:(isl1:GFP) transgenic line and co-injection of mouse ccnd1, ccnd2, or ccnd3 mRNA was unable to rescue this defect (panels c, d and e). Panel f: graph showing the quantitative results of GFP-labeled primary motoneurons cells at 24 hpf. (C) A significant number of ccndx morphant embryos exhibited shorter primary motor axons as compared to ccndx morphants (panels a and b) and co-injection of mouse ccnd1, ccnd2, or ccnd3 mRNA was unable to rescue this defect (panels c, d and e). Panel f: graph showing the proportion of embryos with signals lower than those in control MO-injected embryos at 24 hpf.
Taken together, these results indicate that zebrafish cyclin Dx is specifically required for the formation of motor neuron progenitors during development. Its expression is regulated by HIF2a through binding to two HRE sites in the promoter region. However, we discovered that mouse cyclin D1, D2 and D3 do not share the role of zebrafish cyclin Dx in maintaining the pool of motor neuron progenitors and subsequent promotion of isl1 and olig2 expression and primary motor axon development.
Discussion
During the early stages of ventral neural tube development, three types of cells are generated: floor plate cells, motor neurons and interneurons. The differentiation of these cells is initially triggered by the notochord and later by floor plate cells through the Sonic hedgehog (Shh) signaling pathway49. Five progenitor domains in the ventral neural tube, consisting of four interneurons (V0 to V3) and one motor neuron (pMN), are defined by Shh signaling. In the present study, the zebrafish cyclin Dx transcript was observed to be localized to pMN, distal from the midline and partially overlapping with expression of oligo2 and isl1 (Fig. 1). Moreover, the zccndx gene was observed to be essential and required for the formation of motor neuron progenitors and later, expression of the olig2 and isl1 genes (Figs 2 and 3), through a process regulated by HIF2α (Figs 4 and 5). Mammals lack the ccndx gene (it either evolved after the divergence of mammals and other chordates or was lost from the mammalian lineage), but the mammalian cyclin D genes, ccnd1, d2 and d3, were unable to restore the expansion of motor neuron progenitor pools in mutants, or rescue subsequent expression of isl1 and olig2 in ccndx morphants (Fig. 6).
The cyclin Dx gene was first cloned and characterized in Xenopus21. In this study, we cloned a cyclin Dx ortholog from zebrafish. Although the amino acid sequence identity between zebrafish and Xenopus cyclin Dx is only 36%, the similar expression patterns of these genes in the motor neurons of hindbrain and spinal cord and their specific function in the maintenance of pMNs, confirm that zebrafish cyclin Dx is a homolog of Xenopus cyclin Dx. Data mining revealed that zebrafish cyclin Dx has higher identities with the predicted sequences of cyclin Dx from other bony fish, such as medaka (59%), tetraodon (57%) and fugu (60%) (Supplementary Fig. 3). These genes were placed in the same group upon phylogenetic tree analysis using MEGA software50. However, no homolog of this gene is present in the genome of human or mouse. Therefore, such a cyclin Dx family may be present only in lower vertebrates and Xenopus and not in mammals or chicken. Instead, mammals and chicken possess the cyclin D3 gene. However, based on the importance of cyclin Dx in maintaining the pool of motor neuron progenitor cells, it is possible that a cyclin Dx-like protein may be encoded by the mammalian genome, but has not yet been identified due to the great evolutionary variation between fish and mammals.
Oxygen concentration is implicated in stem cell self-renewal and differentiation51. During hypoxia, increased expression of hypoxic response factors, such as HIF2α, regulate several cellular processes and signal transduction52. The HIF2α has been reported to be stabilized by association with ARNT to form a hetero-dimer leading to activate downstream target genes53. In zebrafish, there are two Arnt1 homologues, Arnt1a and Arnt1b54. While HIF2α has been demonstrated to be required for the differentiation of neural progenitor cells in the hindbrain and spinal cord through its downstream effects on the Birc5a and Birc5b genes, by forming a hetero-dimer with ARNT1A32. In this study, we have demonstrated that ccndx is one of the downstream target genes of HIF2α/ARNT1A and such regulation may be responsible for the formation of motor neuron progenitor cells, but not interneurons, during zebrafish early development. Importantly, the expression of the ccndx gene is essential for the expression of both isl1 and olig2 genes for primary neuron development. The regulation of ccndx gene expression is thus critical for primary motor neuron development and motor neuron progenitor cell renewal. In summary, our data indicate that zebrafish cyclin Dx plays a key role in the proliferation and specification of motor neuron progenitor cells.
Additional Information
How to cite this article: Lien, H.-W. et al. Zebrafish cyclin Dx is required for development of motor neuron progenitors and its expression is regulated by hypoxia-inducible factor 2α. Sci. Rep. 6, 28297; doi: 10.1038/srep28297 (2016).
References
Hayles, J. & Nurse, P. Cell cycle regulation in yeast. J Cell Sci Suppl 4, 155–170 (1986).
Lee, M. & Nurse, P. Cell cycle control genes in fission yeast and mammalian cells. Trends Genet 4, 287–290(1988).
Pelech, S. L., Sanghera, J. S. & Daya-Makin, M. Protein kinase cascades in meiotic and mitotic cell cycle control. Biochem Cell Biol 68, 1297–1330 (1990).
Maller, J. L. Xenopus oocytes and the biochemistry of cell division. Biochemistry 29, 3157–3166 (1990).
Norbury, C. & Nurse, P. Animal cell cycles and their control. Annu Rev Biochem 61, 441–470 (1992).
Forsburg, S. L. & Nurse, P. Cell cycle regulation in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. Annu Rev Cell Biol 7, 227–256 (1991).
Morgan, D. O. Cyclin-dependent kinases: engines, clocks and microprocessors. Annu Rev Cell Dev Biol 13, 261–291 (1997).
Malumbres, M. & Barbacid, M. To cycle or not to cycle: a critical decision in cancer. Nat Rev Cancer 1, 222–231 (2001).
Sherr, C. J. & Roberts, J. M. Living with or without cyclins and cyclin-dependent kinases. Genes Dev 18, 2699–271 (2004).
Yu, Q. & Wu, J. Involvement of cyclins in mammalian spermatogenesis. Mol Cell Biochem 315, 17–24 (2008).
Berger, C., Pallavi, S. K., Prasad, M., Shashidhara, L. S. & Technau, G. M. Cyclin E acts under the control of Hox-genes as a cell fate determinant in the developing central nervous system. Cell Cycle 4, 422–425 (2005).
Coqueret, O. Linking cyclins to transcriptional control. Gene 299, 35–55 (2002).
Inaba, T. et al. Genomic organization, chromosomal localization and independent expression of human cyclin D genes. Genomics 13, 565–574 (1992).
Xiong, Y., Menninger, J., Beach, D. & Ward, D. C. Molecular cloning and chromosomal mapping of CCND genes encoding human D-type cyclins. Genomics 13, 575–584 (1992).
Ewen, M. E. et al. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell 73, 487–497 (1993).
Motokura, T., Keyomarsi, K., Kronenberg, H. M. & Arnold, A. Cloning and characterization of human cyclin D3, a cDNA closely related in sequence to the PRAD1/cyclin D1 proto-oncogene. J Biol Chem 267, 20412–20415 (1992).
Taieb, F. & Jessus, C. Xenopus cyclin D2: cloning and expression in oocytes and during early development. Biol Cell 88, 99–111 (1996).
Taieb, F., Chartrain, I., Chevalier, S., Haccard, O. & Jessus, C. Cyclin D2 arrests Xenopus early embryonic cell cycles. Exp Cell Res 237, 338–346 (1997).
Vernon, A. E. & Philpott, A. The developmental expression of cell cycle regulators in Xenopus laevis. Gene Expr Patterns 3, 179–192 (2003).
Wianny, F. et al. G1-phase regulators, cyclin D1, cyclin D2 and cyclin D3: up-regulation at gastrulation and dynamic expression during neurulation. Dev Dyn 212, 49–62 (1998).
Chen, J. A., Chu, S. T. & Amaya, E. Maintenance of motor neuron progenitors in Xenopus requires a novel localized cyclin. EMBO Rep 8, 287–292 (2007).
Yarden, A., Salomon, D. & Geiger, B. Zebrafish cyclin D1 is differentially expressed during early embryogenesis. Biochim Biophys Acta 1264, 257–260 (1995).
Bessa, J. et al. meis1 regulates cyclin D1 and c-myc expression and controls the proliferation of the multipotent cells in the early developing zebrafish eye. Development 135, 799–803 (2008).
Duffy, K. T. et al. Coordinate control of cell cycle regulatory genes in zebrafish development tested by cyclin D1 knockdown with morpholino phosphorodiamidates and hydroxyprolyl-phosphono peptide nucleic acids. Nucleic Acids Res. 33, 4914–4921 (2005).
Pfaff, S. L., Mendelsohn, M., Stewart, C. L., Edlund, T. & Jessell, T. M. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell 84, 309–320 (1996).
Hutchinson, S. A. & Eisen, J. S. Islet1 and Islet2 have equivalent abilities to promote motoneuron formation and to specify motoneuron subtype identity. Development 133, 2137–2147 (2006).
Park, H. C., Mehta, A., Richardson, J. S. & Appel, B. olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Dev Biol 248, 356–368 (2002).
Sun, Y. et al. Phosphorylation state of Olig2 regulates proliferation of neural progenitors. Neuron 69, 906–917 (2011).
Higashijima, S., Hotta, Y. & Okamoto, H. Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer. J Neurosci 20, 206–218 (2000).
Westerfield, M. The Zebrafish Book. third edn, (University of Oregon Press, Eugene, OR, USA, 1995).
Deryckere, F. & Gannon, F. A one-hour minipreparation technique for extraction of DNA-binding proteins from animal tissues. Biotechniques 16, 405 (1994).
Ko, C. Y. et al. Integration of CNS survival and differentiation by HIF2alpha. Cell death and differentiation 18, 1757–1770 (2011).
Ungos, J. M., Karlstrom, R. O. & Raible, D. W. Hedgehog signaling is directly required for the development of zebrafish dorsal root ganglia neurons. Development 130, 5351–5362 (2003).
Fox, M. A. & Sanes, J. R. Synaptotagmin I and II are present in distinct subsets of central synapses. The Journal of comparative neurology 503, 280–296 (2007).
Appel, B. et al. Motoneuron fate specification revealed by patterned LIM homeobox gene expression in embryonic zebrafish. Development 121, 4117–4125 (1995).
Cheesman, S. E., Layden, M. J., Von Ohlen, T., Doe, C. Q. & Eisen, J. S. Zebrafish and fly Nkx6 proteins have similar CNS expression patterns and regulate motoneuron formation. Development 131, 5221–5232 (2004).
Avaron, F. et al. Comparison of even-skipped related gene expression pattern in vertebrates shows an association between expression domain loss and modification of selective constraints on sequences. Evol Dev 5, 145–156 (2003).
Thisse, C., Thisse, B., Schilling, T. F. & Postlethwait, J. H. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development 119, 1203–1215 (1993).
Chu, C. Y. et al. The zebrafish erythropoietin: functional identification and biochemical characterization. FEBS Lett 581, 4265–4271 (2007).
Berghmans, S. et al. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proceedings of the National Academy of Sciences of the United States of America 102, 407–412 (2005).
Han, H. W. et al. The Nogo-C2/Nogo receptor complex regulates the morphogenesis of zebrafish lateral line primordium through modulating the expression of dkk1b, a Wnt signal inhibitor. PLoS One 9, e86345 (2014).
Liu, Y. C. & Chiang, A. S. High-resolution confocal imaging and three-dimensional rendering. Methods 30, 86–93 (2003).
Motokura, T. et al. A novel cyclin encoded by a bcl1-linked candidate oncogene. Nature 350, 512–515 (1991).
Motokura, T., Yi, H. F., Kronenberg, H. M., McBride, O. W. & Arnold, A. Assignment of the human cyclin D3 gene (CCND3) to chromosome 6p–q13. Cytogenet Cell Genet 61, 5–7 (1992).
Klein, S. L. et al. Genetic and genomic tools for Xenopus research: The NIH Xenopus initiative. Dev Dyn 225, 384–391 (2002).
Nadauld, L. D., Sandoval, I. T., Chidester, S., Yost, H. J. & Jones, D. A. Adenomatous polyposis coli control of retinoic acid biosynthesis is critical for zebrafish intestinal development and differentiation. J Biol Chem 279, 51581–51589 (2004).
Passini, M. A. et al. Cloning of zebrafish vsx1: expression of a paired-like homeobox gene during CNS development. Dev Genet 23, 128–141(1998).
Barth, K. A. & Wilson, S. W. Expression of zebrafish nk2.2 is influenced by sonic hedgehog/vertebrate hedgehog-1 and demarcates a zone of neuronal differentiation in the embryonic forebrain. Development 121, 1755–1768 (1995).
Jessell, T. M. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet 1, 20–29 (2000).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular biology and evolution 30, 2725–2729 (2013).
Grayson, W. L., Zhao, F., Bunnell, B. & Ma, T. Hypoxia enhances proliferation and tissue formation of human mesenchymal stem cells. Biochemical and biophysical research communications 358, 948–953 (2007).
Jain, S., Maltepe, E., Lu, M. M., Simon, C. & Bradfield, C. A. Expression of ARNT, ARNT2, HIF1 alpha, HIF2 alpha and Ah receptor mRNAs in the developing mouse. Mechanisms of development 73, 117–123 (1998).
Epstein, A. C. et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43–54 (2001).
Prasch, A. L., Tanguay, R. L., Mehta, V., Heideman, W. & Peterson, R. E. Identification of zebrafish ARNT1 homologs: 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity in the developing zebrafish requires ARNT1. Mol Pharmacol 69, 776–787 (2006).
Acknowledgements
We thank Dr. Yung-Shu Kuan for helpful discussions. We thank the Taiwan Zebrafish Core Facility at Academia Sinica (ZCAS) for providing AB wild type zebrafish. We are also grateful to the Zebrafish Core Facility at NHRI (National Health Research Institutes at Miaoli, Taiwan) for providing the Tg (isl:GFP) transgenic line.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: H.-W.L., C.-H.C. and C.-J.H. Performed the experiments: H.-W.L., Y.-C.C. and C.-H.C. Analyzed the data: H.-W.L., Y.-C.C., C.-C.H., C.-H.C. and C.-J.H. Contributed reagents/materials/analysis tools: R.-Y.Y., C.-M.C., C.-H.H., S.-P.H., P.-P.H., C.-N.S. and C.-L.C. Wrote the paper: H.-W.L., C.-H.C. and C.-J.H.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Lien, HW., Yuan, RY., Chou, CM. et al. Zebrafish cyclin Dx is required for development of motor neuron progenitors and its expression is regulated by hypoxia-inducible factor 2α. Sci Rep 6, 28297 (2016). https://doi.org/10.1038/srep28297
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep28297
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.