Abstract
RNA-binding motif 4 (RBM4) is a multifunctional protein that participates in regulating alternative splicing and mRNA translation. Its reduced expression has been associated with poor overall survival in lung cancer, breast cancer and ovarian cancer. We assessed RBM4 protein expression levels with immunohistochemistry in tissue microarrays containing malignant gastric cancer tissues and benign tissues from 813 patients. We also examined the expression levels of RBM4 mRNA in twenty-five paired gastric cancer samples and adjacent noncancerous tissues. Both RBM4 protein and mRNA expression levels were significantly lower in gastric cancer tissues compared with the adjacent noncancerous tissues. There was a significant association between reduced RBM4 protein expression and differentiation (P < 0.001), lymph node metastasis (P = 0.026), TNM state (P = 0.014) and distant metastasis (P = 0.036). Patients with reduced RBM4 expression (P < 0.001, CI = 0.315–0.710) and TNM stage III and IV (P < 0.001, CI = 4.757–11.166) had a poor overall survival. These findings suggest that RBM4 is a new biomarker in gastric cancer, as the reduced expression of this protein is correlated with poor differentiation, lymph node status and distant metastasis. Further, lower RBM4 expression is an independent prognostic marker for gastric cancer.
Similar content being viewed by others
Introduction
Even though its incidence has been decreasing during past decades, Gastric cancer remains as the second leading cause of cancer death worldwide1. More than 70% of the gastric cancer cases occur in developing countries and China has the highest incidence2. Gastric cancer is the third leading cause of death among cancer patients in China with an age standardized incidence of 22.7 per 100,0003. Metastatic spread of tumor cells from the stomach to other sites, such as the peritoneum and liver, is the cause of the high mortality rates in gastric cancer patients4. A variety of significant treatments have been used with early-stage gastric cancer patients. However, the overall prognosis of gastric cancer patients at advanced stages is still unfavorable in spite of aggressive treatments5,6. Therefore, it is very important to identify novel biological markers for gastric cancer that can help us develop an early diagnosis method and target therapy for extending the life of gastric cancer patients.
Initially described in Drosophila, RNA-binding motif 4 (RBM4), which is also known as Lark, is expressed ubiquitously throughout development, with relatively high abundance in heart, brain, skeletal muscle and testis7. It is particularly highly expressed in the embryonic nervous system8. In humans, the gene is located on chromosome 11q13 and encodes a protein of 364 -amino acids long9,10. The human gene has 95% homology with the mouse ortholog and 53% homology with the Xenopus homolog. Two RNA recognition motifs (RRMs) and a CCHC-type zinc finger in the N-terminal region have been found in RBM4, whereas a low-complexity region is located in the C-terminal that can interact with other proteins11.
RBM4 is a multifunctional protein that participates at least in modulating alternative splicing and mRNA translation12,13. RBM4 can regulate exon selection and alternative splicing in both in vivo and in vitro splicing models14,15. RBM4 has been shown to regulate translation, under cell stress by interacting directly with argonaute 216, inhibiting cap-dependent translation13, mediating an oxygen-regulated translation switch11 or activating internal ribosomal entry site-mediated translation17. Furthermore, RBM4 is able to selectively associate with specific microRNAs in muscle cells and repress their translation activity by promoting micro-ribonucleoprotein connection with target mRNAs18. In addition, overexpression of RBM4 promotes differentiation of pancreas and muscle cells19,20. Even though RBM4 has been reported to be a tumor suppressor that inhibits lung cancer progression in both cultured cells and in a tumor xenograft model21, little is known about RBM4 expression in cancer.
In this study, we sought to explore the role of RBM4 expression in the prognosis of gastric cancer patients. RBM4 expression was assessed by immunohistochemistry and qRT-PCR techniques. The association of RBM4 expression with clinicopathologic characteristics and overall survival (OS) was evaluated as well.
Results
Reduced RBM4 mRNA expression in cancer
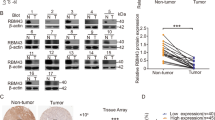
We measured RBM4 mRNA levels in 25 paired gastric cancer samples and adjacent noncancerous tissues. The results showed that 19 primary gastric cancer samples had substantially reduced RBM4 expression levels on mRNA compared with the paired adjacent noncancerous tissues, with an average downregulation fold of 0.643 (P < 0.001) (Fig. 1).
mRNA expression of RBM4 in paired gastric cancer samples and adjacent noncancerous tissue.
Quantitative polymerase chain reaction (qRT-PCR) was performed to detect the expression of RBM4 mRNA in twenty five gastric cancer compared with adjacent noncancerous tissues using β-actin, as a normalization control. RBM4 mRNA levels were significantly lower in gastric cancer compared with corresponding noncancerous tissues (P < 0.001). Each bar represents one patient on the x-axis.
Clinicopathologic features of patients
Using TMAs, we analyzed RBM4 protein expression in 813 (94.4%) of the 861 samples in the TMA, The remaining samples were lost during antigen retrieval or there were no tumor tissues found in the core. The level of core loss was similar to the loss rates described in previous TMA studies22.
Of the 813 benign and malignant gastric tissues analyzed (Table 1, Supplementary dataset 1), there were 29 chronic gastritis, 25 intestinal metaplasia, 24 low-grade intraepithelial neoplasia, 27 high-grade intraepithelial neoplasia, 103 matched adjacent noncancerous and 605 cancer. The baseline characteristics of the 605 gastric cancer were shown in Table 2 (Supplementary dataset 2). There were 458 male and 157 female patients. Their median age was 59.5 years (range, 33–86). The distribution of TNM stage was as follows: 352 patients at stage 0, I and II, 253 at stage III and IV. For the histological type, most patients initially presented as the tubular (528/605) type. For the differentiation status, 37 tumors were well-differentiated, 224 were moderately- differentiated, 278 were poorly- differentiated and 66 were others (not of the tubular or papillary adenocarcinoma type).
RBM4 protein expression in benign and malignant gastric tissues by IHC
RBM4 protein expression mostly presented in the cytoplasm and nucleus, especially in nuclear speckles (Fig. 2). Using the X-tile software program for TMA data analysis (http://www.tissuearray.org/rimmlab), we first identified the significant cutoff point in terms of OS in gastric cancer. We found the appropriate cutoff point to be 100: Score 0–100 was considered no or low expression while 101–300 was considered high expression. For the subsequent analyses, RBM4 protein expression levels were considered either as “Low or no” or “High” using these cutoff values.
Representation of RBM4 protein expression in gastric benign and malignant tissues on TMA sections.
Column A: normal surgical margin of gastric cancer with high RBM4 expression (IHC score, 270); column B: chronic gastritis with high RBM4 expression (IHC score, 140); column C: intestinal metaplasia with high RBM4 expression (IHC score, 160); column D: low-grade intraepithelial neoplasia with high RBM4 expression (IHC score, 110); column E: high-grade intraepithelial neoplasia with low RBM4 expression (IHC score, 90); column F: well differentiated gastric cancer with high RBM4 expression (IHC score, 180); column G: moderately- differentiated gastric cancer with low RBM4 expression (IHC score, 20); column H: poorly differentiated gastric cancer with low RBM4 expression (IHC score, 0). Row 1 and 3 are RBM4 staining with ×40 magnification and row 2 and 4 are RBM4 staining with ×400 magnification.
High RBM4 protein expression was recorded in 58.6%, 56.0%, 54.2%, 51.9% and 63.1% of the stomach benign tissues in chronic gastritis, intestinal metaplasia, low-grade intraepithelial neoplasia, high-grade intraepithelial neoplasia and adjacent noncancerous tissues, respectively (Table 1). In gastric cancer, high RBM4 protein expression was only 43.8%, significantly lower than in the benign tissues (P = 0.006).
Association of RBM4 expression with clinicopathologic characteristics in gastric cancer
Table 2 shows a summary of the correlations between RBM4 protein expression levels and clinicopathologic variables in the gastric cancer patients. Reduced RBM4 expression was significantly associated with poor differentiation (P < 0.001), lymph node metastasis (P = 0.026), distant metastasis (P = 0.036) and advanced TNM stage (P = 0.014). However, we did not find a significant correlation between RBM4 expression with depth of invasion and other clinicopathologic variables, including age, sex and histological type.
Reduced RBM4 expression correlates with poor OS
To evaluate the predictive value of RBM4 expression in gastric cancer, Kaplan-Meier survival curves were performed to compare the patients with high RBM4 expression and those with low or no RBM4 expression using overall cumulative survival. Data showed reduced RBM4 expression to be correlated with worse OS (P < 0.001, Fig. 3A) and worse disease-free survival (DFS) (P < 0.001, Fig. 3B). For OS, the overall cumulative survival rate was 31.4% in the low or no RBM4 expression groups and 52.4% in the high RBM4 expression group. DFS was 20.9% and 54.7%, respectively. We also found poorer OS in TNM stages III and IV compared with that in TNM stages 0, I and II (P < 0.001, Fig. 3C).
Correlation between RBM4 expression and clinicopathologic parameters and patients survival in human gastric cancer.
(A) Low or no RBM4 expression correlates with a poorer overall cumulative survival for 605 gastric cancer patients (P < 0.001, log-rank test). (B) Low or no RBM4 expression was found to be correlated with a poorer disease-free survival for 605 gastric cancer patients (P < 0.001, log-rank test). (C) TNM III + IV correlates with a poorer overall cumulative survival for 605 gastric cancer patients (P < 0.001, log-rank test).
The univariate and multivariate analyses results for prognostic markers in gastric cancer are shown in Table 3. Reduced RBM4 expression (HR = 0.449, 95% CI = 0.324–0.624; P < 0.001) was significantly associated with a shorter survival in univariate analysis, along with other prognostic factors, including age (HR = 0.711, 95% CI = 0.512–0.987; P = 0.042), differentiation (HR = 1.607, 95% CI = 1.290–2.002; P < 0.001) and TNM stage (HR = 8.415, 95% CI = 5.691–12.444; P < 0.001). In multivariate analysis, both reduced RBM4 expression (HR = 0.470, 95% CI = 0.315–0.710; P < 0.001) and TNM stage (HR = 7.288, 95% CI = 4.757–11.166; P < 0.001) worsened the prognosis independently (Table 3).
Discussion
The two isoforms of RBM4 that have been reported in mammals (i.e.,—RBM4a and RBM4b) have a very similar structure and sequence. Interestingly, the entire RBM4a gene is situated within intron 2 of RBM4b19. Since RBM4a is the only isoform that has been studied, the present review will only refer to this isoform. Not much is known about the role of RBM4 in cancer. To date, only two studies have investigated the potential role of RBM4 in tumors: Lin et al. found that the SRPK1-RMB4 network may contribute to tumorigenesis through altered sensitivity to apoptotic signals in breast cancer cells23. Wang et al. have shown that only in lung, breast and ovarian cancer, RBM4 expression is decreased dramatically in cancer patients and a reduced RBM4 level is correlated with poor survival21. The function of RBM4 in gastric cancer has not been clearly studied.
In this study, we first found that RBM4 staining was localized in the nucleus and cytoplasm and this observation was in line with previous findings14. According to immunohistochemistry and qRT-PCR analyses, the expression of RBM4 protein and mRNA is downregulated in gastric cancer relative to levels in human chronic gastritis, intestinal metaplasia, low-grade intraepithelial neoplasia, high-grade intraepithelial neoplasia and adjacent noncancerous tissues. These results were similar to Wang et al.21, indicating that inactivation of RBM4 may play an important role in tumorigenesis of gastric cancer. However, our study showed that RBM4 protein expression levels were generally higher (43.8%) than Wang’s study (19.0%) in cancers and the reasons may be as follows: 1) different secondary antibodies were used: Abcam company (Wang study) vs Dako company (our study); 2) different research objects: ovarian cancer, non small cell lung cancer and pancreatic cancer (Wang study) vs gastric cancer (our study), as the expression of RBM4 protein may not be the same in different tumors.
Subsequently, we studied the correlation between RBM4 protein expression levels and clinicopathologic variables in gastric cancer patients. We found that reduced RBM4 protein expression was associated with lymph node metastasis, TNM stage and distant metastasis. There was an especially obvious correlation with differentiation (P < 0.001). RBM4 protein expression is only in 34.5% of poorly-differentiated cancers, while we found it in 54.5% of moderately-differentiated and 67.6% of well-differentiated cancers. RBM4 promotes myoblast differentiation by selectively binding to muscle-specific miR-1 and miR-20619 and elevates pancreas cell differentiation via alternative splicing regulation20. However, it is not known how RBM4 mediates gastric cancer cell differentiation. A prospective study is needed to clarify this. Although this study did not indicate a significant correlation between RBM4 expression and histological type, RBM4 protein expression levels were found to be only 17.4% in signet ring cell carcinoma (SRCC), much lower than in any other form of gastric cancer. SRCC is a poorly differentiated cancer, which has been thought to have a worse prognosis in advanced gastric cancer24,25,26, For this reason, identifying the various types and stages of gastric cancer could potentially increase curative effect of targeted therapies. The current study is limited by its small sample size and retrospective design. Future studies with larger samples of SRCC could help clarify the role of RBM4 in gastric cancer.
In addition, we showed that lower expression levels of RBM4 were closely associated with reduced OS and DFS in patients with gastric cancers. Also, RBM4 was an independent gastric cancer prognostic factor according to multivariate Cox regression analysis. These results are consistent with what was found by Wang et al.21, implying RBM4 may be used as a new therapeutic target for gastric cancer. Of course, it is not only a single gene that is involved in each step of the metastasis process in the clinic of gastric cancer. It has been reported previously that some known factors are associated with RBM4 in cell and mouse models, including SR protein kinase 1(SRPK1)23, Bcl-x21, CD4427 and SRSF121. Future studies are needed to directly compare these factors of protein expression with RMB4 protein expression by IHC in gastric cancer and their association with OS.
It has been reported that RBM4 affects cell growth by controlling both splicing and translation. Splicing dysregulation has been recently shown to be a major molecular hallmark of human cancer28,29. Our study shows that RBM4 expression is reduced in gastric cancer and RBM4 may serve as a tumor suppressor. Oncogene activation and tumor suppressor gene inactivation are studied most often in tumorigenesis, cancer development and cancer progression30. Tumor suppressor genes most often activate antiproliferative and pro-apoptotic pathways and they, therefore, protect cells from advancing to cancer. However, it is not known yet how RBM4 inhibits tumor growth in gastric cancer. Wang et al. showed that RBM4 shifts splicing of Bcl-x to suppress cancer progression by controlling the balance between pro- and anti-apoptotic pathways and RBM4 counteracts SRSF1 to inhibit tumor progression by mediating the activation of the mTORC1 pathway21. Further experiments are needed to investigate whether RBM4 interacts directly with Bcl-x and SRSF1 or binds to other splicing factors to suppress tumor progression in vitro and in mouse models of gastric cancer.
In conclusion, our study found that RBM4 is a new biomarker in gastric cancer. The reduced expression of RBM4 is correlated with poor differentiation, lymph node status and distant metastasis and is an independent prognostic marker for poor outcome in patient with gastric cancer.
Materials and Methods
Patients and tissue specimens
Stomach tissue samples (n = 861) that were formalin-fixed paraffin-embedded (FFPE) and processed in tissue microarray (TMA) were obtained from the Department of Pathology, Affiliated Hospital of Nantong University. The samples included 33 intestinal metaplasia, 27 chronic gastritis, 29 low-grade intraepithelial neoplasia, 31 high-grade intraepithelial neoplasia, 621 gastric cancer tissues and 120 matched adjacent noncancerous tissues. Clinical data included age, sex, histological type, differentiation grade and TNM stage et al. Clinical follow-up results were available from 741 patients (range, 2 to 11 years) and at the last follow-up date patients who were alive were censored from the analysis. The date of surgery to death or last follow-up served as the value for OS and the period from surgery to recurrence was defined as the DFS value. No cancer patients received radiation therapy, chemotherapy or immunotherapy before surgery. To perform qRT-PCR test for mRNA expression level, an additional twenty-five freshly gastric cancer tissues and matching adjacent noncancerous tissues obtained from Huai’an Second People’s Hospital, Zhang Jia Gang Ao Yang Hospital, Second Affiliated Hospital of Nanjing Medical University, Drum Tower Hospital of Nanjing Medical University and the First Affiliated Hospital of Nanjing Medical University were also included in this study. This study was approved by the Human Research Ethics Committee of the abovenamed hospitals and carried out in accordance with the approved guidelines. All patients provided written informed consent for their stomach tissue samples to be used for research.
TMA construction and immunohistochemistry analysis (IHC)
A total 13 gastric TMAs were manufactured by Shanghai Outdo Biotech (China). Core tissue biopsies that were 2 mm in diameter were obtained from ~70 individual FFPE blocks. They were sequentially arranged in one prepared blank paraffin block. To generate TMA slides, four-micron thick sections were cut and mounted on super frost-charged glass microscope slides.
IHC was carried out as previously reported31. In brief, we deparaffinized and rehydrated the tissue sections with graded alcohol. The tissue sections were boiled with 0.01 M citrate buffer (pH 6.0) to achieve antigen retrieval. After pre-incubation with 10% FCS (fetal calf serum) for 20 min, the TMA slide was incubated with rabbit anti-human RBM4 antibody (polyclonal, dilution 1:100, Proteintech, Chicago, IL) and subsequently with Envision HRP-congjugated goat anti-rabbit-IgG antibody (monoclonal, dilution 1:100, Dako). A sample in which diluted PBS replaced primary antibody during incubation served as a negative control. All slides were performed at the same time under the same conditions.
RBM4 expression was evaluated by the percentage of cells and the staining intensity with the semi-quantitative H-score method32. To evaluate the staining intensity four different groups were defined: 0 = no staining, 1+ = weak staining, 2+ = moderate staining, or 3+ = intense staining. To obtain a final staining score, the intensity score was multiplied by the percentage of cells stained at the respective intensity. Thus the staining score had a minimum value of 0 (no staining) and a maximum of 300 (100% of cells with 3+ staining intensity). Immunostained sections were evaluated by two experienced pathologists under blinded experimental conditions.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Trizol reagent (Invitrogen, Carlsbad, CA, USA) was used to extract total RNA. RNA quantity and quality were checked by agarose gel electrophoresis and spectrophotometry. Total RNA was reverse transcribed to cDNA using a PrimeScriptTM RT reagent kit (Takara, Glen Burnie, MD). An ABI PRISM 7500HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA) was used for the qPT-PCR reactions. PCR primers were designed with Primer Express Software and 96-well plates were used. Following primers were used for RBM4: forward, 5′-GCAGACTTGACCGAGCAATA-3′ and reverse, 5′- TCCGTACGCGTTGTTGTAAT-3′ (Synthesized by Genescript. Nanjing, China). Human β-actin was used as a control: forward, 5′- TGGAGAAAATCTGGCACCAC-3′ and reverse, 5′- GAGGCGTACAGGGATAGCAC-3′ (Genescript). The reaction system (20 μL) contained 2 μL of cDNA template; each primer, 20 nmol/L and 10 μL of 2 × SYBR Green PCR Master Mixtures (Applied Biosystems). qRT-PCR conditions were as follows: after an initial annealing temperature of 50 °C for 2 min to allow AmpErase-UNG activity and 10 min at 95 °C, the samples were cycled 40 times at 95 °C for 15 sec and 56 °C for 1 min. The Ct-value for each sample was calculated with the ΔΔCt method33,34,35 and results were expressed as 2-ΔΔCt. clinicopathological parameters of the 25 cancer patients and primitive Ct-value of qRT-PCR are shown in Supplementary dataset 3 and 4.
Statistical analysis
SPSS 20.0 statistical software package (SPSS Inc, Chicago, IL) was used for all statistical analyses. Continuous RBM4 expression data from IHC were initially converted into dichotic data (low or no vs high) using specific cutoff values, The cutoff values were selected to be significant regarding OS using the X-tile software program for TMA data analysis (The Rimm Lab at Yale University; http://www.tissuearray.org/rimmlab)35,36,37. To assess the relationship between RBM4 expression and clinicopathological parameters, we used χ2 test. Survival curves were made with the Kaplan–Meier method. The significance of differences between the curves was analyzed with a log-rank test. The Cox proportional hazards regression model was used to perform univariate and multivariate analyses. For qRT-PCR, paired-samples t test was used. Levels of statistical significance were set at P < 0.05.
Additional Information
How to cite this article: Yong, H. et al. Prognostic value of decreased expression of RBM4 in human gastric cancer. Sci. Rep. 6, 28222; doi: 10.1038/srep28222 (2016).
References
Ferro, A. et al. Worldwide trends in gastric cancer mortality (1980–2011), with predictions to 2015 and incidence by subtype. Eur J Cancer 50, 1330–1344 (2014).
Lin, Y. et al. Comparative epidemiology of gastric cancer between japan and china. World J Gastroenterol 17, 4421–4428 (2011).
Zhu, X. et al. Gastric carcinoma in china: Current status and future perspectives (review). Oncol Lett 1, 407–412 (2010).
Yoo, C. H. et al. Recurrence following curative resection for gastric carcinoma. Br J Surg 87, 236–242 (2000).
Degiuli, M. et al. Survival results of a multicentre phase ii study to evaluate d2 gastrectomy for gastric cancer. Br J Cancer 90, 1727–1732 (2004).
Sano, T. et al. Gastric cancer surgery: Morbidity and mortality results from a prospective randomized controlled trial comparing d2 and extended para-aortic lymphadenectomy–japan clinical oncology group study 9501. J Clin Oncol 22, 2767–2773 (2004).
Lin, J. C. et al. Exon selection in alpha-tropomyosin mrna is regulated by the antagonistic action of rbm4 and ptb. Mol Cell Biol 25, 10111–10121 (2005).
Newby, L. M. et al. A new biological rhythm mutant of drosophila melanogaster that identifies a gene with an essential embryonic function. Genetics 135, 1077–1090 (1993).
Jackson, F. R. et al. A novel zinc finger-containing rna-binding protein conserved from fruitflies to humans. Genomics 41, 444–452 (1997).
Markus, M. A. et al. Rbm4: A multifunctional rna-binding protein. Int J Biochem Cell Biol 41, 740–743 (2009).
Lin, J. C. et al. Rna-binding motif protein 4 translocates to cytoplasmic granules and suppresses translation via argonaute2 during muscle cell differentiation. J Biol Chem 284, 34658–34665 (2009).
Lin, J. C. et al. Rbm4 down-regulates ptb and antagonizes its activity in muscle cell-specific alternative splicing. J Cell Biol 193, 509–520 (2011).
Lin, J. C. et al. Emerging role for rna binding motif protein 4 in the development of brown adipocytes. Biochim Biophys Acta 1843, 769–79 (2014).
Lai, M. C. et al. A novel splicing regulator shares a nuclear import pathway with sr proteins. EMBO J 22, 1359–1369 (2003).
Markus, M. A. et al. Wt1 interacts with the splicing protein rbm4 and regulates its ability to modulate alternative splicing in vivo. Exp Cell Res 312, 3379–3388 (2006).
Uniacke, J. et al. An oxygen-regulated switch in the protein synthesis machinery. Nature 486, 126–129 (2012).
Hock, J. et al. Proteomic and functional analysis of argonaute-containing mrna-protein complexes in human cells. EMBO Rep 8, 1052–1060 (2007).
Lin, J. C. et al. Cell stress modulates the function of splicing regulatory protein rbm4 in translation control. Proc Natl Acad Sci USA 104, 2235–2240 (2007).
Lin, J. C. et al. Multiple roles of rbm4 in muscle cell differentiation. Front Biosci (Schol Ed). 4, 181–189 (2012).
Lin, J. C. et al. Rbm4 promotes pancreas cell differentiation and insulin expression. Mol Cell Biol 33, 319–327 (2013).
Wang, Y. et al. The splicing factor rbm4 controls apoptosis, proliferation and migration to suppress tumor progression. Cancer Cell 26, 374–389 (2014).
Bai, J. et al. Overexpression of cullin1 is associated with poor prognosis of patients with gastric cancer. Hum Pathol 42, 375–383 (2011).
Lin, J. C. et al. Elevated srpk1 lessens apoptosis in breast cancer cells through rbm4-regulated splicing events. RNA 20, 1621–1631 (2014).
Taghavi, S. et al. Prognostic significance of signet ring gastric cancer. J Clin Oncol 30, 3493–3498 (2012).
Phalanusitthepha, C. et al. Endoscopic features of early-stage signet-ring-cell carcinoma of the stomach. World J Gastrointest Endosc 7, 741–746 (2015).
Pernot, S. et al. Signet-ring cell carcinoma of the stomach: Impact on prognosis and specific therapeutic challenge. World J Gastroenterol 21, 11428–11438 (2015).
Williams, K. et al. Cd44 integrates signaling in normal stem cell, cancer stem cell and (pre)metastatic niches. Exp Biol Med (Maywood) 238, 324–38 (2013).
Oltean, S. et al. Hallmarks of alternative splicing in cancer. Oncogene 33, 5311–5318 (2014).
David, C. J. et al. Alternative pre-mrna splicing regulation in cancer: Pathways and programs unhinged. Genes Dev 24, 2343–2364 (2010).
Nag, S. et al. The mdm2-p53 pathway revisited. J Biomed Res 27, 254–271 (2013).
Bai, J. et al. Cullin1 is a novel marker of poor prognosis and a potential therapeutic target in human breast cancer. Ann Oncol 24, 2016–2022 (2013).
Detre, S. et al. A “quickscore” method for immunohistochemical semiquantitation: Validation for oestrogen receptor in breast carcinomas. J Clin Pathol 48, 876–878 (1995).
Tang, Q. et al. Expression and prognostic significance of the alpha b-crystallin gene in human hepatocellular carcinoma. Hum Pathol 40, 300–305 (2009).
Han, L. et al. High expression of cxcr2 is associated with tumorigenesis, progression and prognosis of laryngeal squamous cell carcinoma. Med Oncol 29, 2466–2472 (2012).
Ni, S. et al. Increased zo-1 expression predicts valuable prognosis in non-small cell lung cancer. Int J Clin Exp Pathol 6, 2887–2895 (2013).
Sun, R. et al. Prognostic value of lamp3 and tp53 overexpression in benign and malignant gastrointestinal tissues. Oncotarget 5, 12398–12409 (2014).
Zhai, X. et al. Abnormal expression of emt-related proteins, s100a4, vimentin and e-cadherin, is correlated with clinicopathological features and prognosis in hcc. Med Oncol 31, 970 (2014).
Acknowledgements
This project was supported by the National Natural Science Foundation of China (Grant number 81201596), Special Fund of Clinical Medicine in Jiangsu Province (BL2013038), Science and Technology Program of Huaian (HAS2015009-2) and the Priority Academic Program Development of Jiangsu Higher Education Institutions. We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
Z.F. and B.L. conceived the idea and designed the experiments; S.Z. and W.W. performed the immunohistochemistry and qRT-PCR experiments; W.Z., C.C., G.D., L.Z., Z.Z., H.L., Y.Z. and J.W. collected and analyzed data; X.K. and J.Z. provided fresh stomach tissues from their surgical practices; H.Y. and H.Z. wrote the manuscript. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Yong, H., Zhu, H., Zhang, S. et al. Prognostic value of decreased expression of RBM4 in human gastric cancer. Sci Rep 6, 28222 (2016). https://doi.org/10.1038/srep28222
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep28222
This article is cited by
-
RBM4 dictates ESCC cell fate switch from cellular senescence to glutamine-addiction survival through inhibiting LKB1-AMPK-axis
Signal Transduction and Targeted Therapy (2023)
-
RBM4 modulates the proliferation and expression of inflammatory factors via the alternative splicing of regulatory factors in HeLa cells
Molecular Genetics and Genomics (2020)
-
The impact of the RBM4-initiated splicing cascade on modulating the carcinogenic signature of colorectal cancer cells
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.