Abstract
In microfluidic filtration systems, one of the leading obstacles to efficient, continuous operation is clogging of the filters. Here, we introduce a lateral flow microfluidic sieving (μ-sieving) technique to overcome clogging and to allow continuous operation of filter based microfluidic separation. A low frequency mechanical oscillation was added to the fluid flow, which made possible the release of aggregated unwanted polystyrene (PS) particles trapped between the larger target PS particles in the filter demonstrating continuous μ-sieving operation. We achieved collection of the target PS particles with 100% separation efficiency. Also, on average, more than 98% of the filtered target particles were retrieved after the filtration showing high retrieval rates. Since the oscillation was applied to the fluid but not to the microfluidic filter system, mechanical stresses to the system was minimized and no additional fabrication procedures were necessary. We also applied the μ-sieving technique to the separation of cancer cells (MDA-MB-231) from whole blood and showed that the fluidic oscillations prevented the filters from being blocked by the filtered cancer cells allowing continuous microfluidic separation with high efficiency.
Similar content being viewed by others
Introduction
Separation of micro-scale particles has been an important issue in industrial, biochemical and clinical applications for identification and analysis of specific particles1,2,3. To achieve the goal, microfluidics has been actively adopted because it is able to manipulate micro-particles precisely. Microfluidic techniques for separating micro-particles can be divided into two categories: active and passive methods. Active methods use external forces such as electric field, magnetic field, acoustic wave and optical interaction, utilizing the particle’s specific dielectric, magnetic or optical properties4,5,6,7,8,9. Although these techniques produce systems with high separation efficiency and selectivity, they generally lack high throughput due to weak target interaction that limits specimen flow rate, or they require target labeling which are time consuming and may reduce specimen viability. Passive methods utilize distinctive physical characteristics of the particles such as, size, density, and, especially for cells, deformability10,11,12,13,14,15,16,17,18,19,20. These methods have simple design, relatively simple fabrication, and high throughput. However, they usually suffer from lower selectivity compared to the active methods.
In biomedical analysis, separation of living cells is required as a powerful diagnostic and prognostic tool. Active methods based on antibodies could be a good choice because they show high efficiency in selective separation and detection of target cells. However with limited processing speed and cell viability, they are generally inappropriate for clinical applications that require high throughput and intact cells. Passive separation techniques utilizing the cell size can be a better choice for cell separation21,22,23. For example, separating circulating tumor cells (CTCs) that propagate through the peripheral circulatory system are known to be larger and more rigid than hemocytes, which enables size and deformability based separation18,21,22,24,25.
A typical approach among passive separation methods is to use microfluidic filters containing micro-fabricated pillars of specific interval or a sieve having micro-pores10,12,13,18,20. The microfluidic filters let particles with diameter smaller than the open pores pass through, while trapping targeted larger particles. However, as the trapped target particles accumulate, parts of the filters become blocked, reducing the number of open pores and they change the hydrodynamic resistance unpredictably. This results in aggregation of the smaller non-target particles around the constricted pores leading to contamination of the filtered target particles and lowering their purity15,18,26. It also induces higher shear stress, resulting in the trapped particles squeezing through the filters, or in the case of cells, the shear may induce rupture lowering the efficiency. Furthermore, hydraulic pressure applied to the filtered particles causes them to be immobile. Especially for live biological cells including non-targeted ones, pressurized contact to the filter surface gives high chances for non-specific adhesion27. This immobilization of targets hinders downstream analysis where retrieval of filtered particles is required. Even though the microfluidic filtration devices are simple in working principle and easy to fabricate, such critical issues have kept them from being widely utilized. To avoid these issues, systems incorporating actuated membranes or alternate fluid flow have been developed to prevent device clogging28,29,30,31,32,33,34. Those systems showed improved selection efficiency, but they required constant monitoring for the transition between static phase and dynamic phase, which limits processing speed and automation.
Here, we report on a lateral flow microfluidic sieving (μ-sieving) device that has overcome the limitation by adopting sieving which can be observed in our daily lives: for example, separation of grains and gravels from sand using a sieve. By adding a low frequency oscillation to the fluid flow, it was possible to purify the trapped target particles from smaller non-target particles. With the oscillated flow, the device successfully filtered polystyrene (PS) particles continuously without any contamination by smaller non-target particles. We also applied the device to the separation of a breast cancer cell line (MDA-MB-231) from whole blood without filter clogging. The sieved particles (or cells) could also be retrieved after filtration providing the potential for downstream analysis and further post-processing. Our μ-sieving device achieved high specificity and throughput. Furthermore, as we demonstrated the separation of cancer cells from blood, μ-sieving may be applied in future liquid biopsy.
Concept of μ-sieving
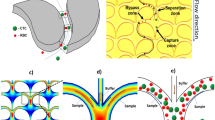
To realize the action of sieving within a microfluidic filter environment, we applied oscillatory vibrations to the fluid, rather than to the filter as compared to previous reports28,30,32. This allowed the microfluidic chip and its filter to remain standing still, reducing mechanical stresses between different components in the microfluidic system, and keeping stable separation environment. Without fluid oscillation, the microfluidic filters cannot sustain extended operation due to eventual clogging (Fig. 1a). When the fluid oscillates, all the particles in the fluid normally oscillate collectively in phase, because the small variation in their microscopic size does not create enough of a difference in their inertial moment (Fig. 1b). However, when the oscillating particles come in contact with a filter, larger particles are trapped while smaller particles pass through (Fig. 1c).
System Design and Simulation
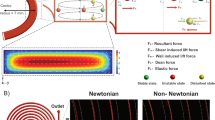
Figure 2 shows the schematic illustrations of the μ-sieving system. The figure visualizes the μ-sieving process with laminar flows. As shown in Fig. 2a, the device has an intersection of fluidic channels with a line of micro-pillars aligned diagonally at the intersection, in order to sieve particles longitudinally (Fig. 2b) and to retrieve transversely (Fig. 2c). To sieve micro-particles, the sample solution was injected from the left channel and the buffer solution was injected from the upper and lower channels, leaving the right channel as an outlet (Fig. 2b). For μ-sieving, a piezoelectric actuator was attached to the connection tube of the left channel inducing oscillation which caused the sample solution to move back and forth while advancing to the outlet. To retrieve the filtered particles, the buffer solution was injected from the upper, left, and right channel, leading the particles to the lower channel (Fig. 2c).
(a) Microfluidic chip with four channels and a piezoelectric actuator attached to the inlet tube. Red box shows the magnified view of the filter area. (b) Streamlines in the device during filtration and (c) retrieval of sieved particles. Red and blue indicate the sample and the buffer, respectively.
The fluid flow of the designed microfluidic geometry was analyzed using COMSOL multiphysics 5.1 to study the hydrodynamic conditions for μ-sieving and retrieving the target particles or cells. The simulation was performed under laminar condition, and the flow condition was controlled to properly trap and retrieve the target particles within our device geometry. The flow rate for the sample was set to be 200 μl/h, while sheath flows from the top and the bottom were 100 μl/h, respectively. For the retrieving mode, the bottom channel became the outlet, and the flow rates for the other three were set to be 1000 μl/h. With the streamline analysis, we were able to observe that the sample did not drain to the bottom channel, and the retrieving flow did not drain to the left channel as intended (Fig. 2b,c).
We also analyzed the principle of μ-sieving; how the smaller particles trapped around the larger particles could be released. We assumed that the larger particles (numbered particles in Fig. 3a) were stacked densely in front of the pillars during the forward flow, and the gaps between the stacked particles are small enough to block the passing of the smaller particles (red particles in Fig. 3a). During the backward flow by the oscillation, the stacked particles would recede with the fluid starting with the outermost particle (Fig. 3b). When the flow switches to the forward flow again, both of the smaller and larger particles start flowing at the same speed. However, as the larger particles start to impact on the filter, the gaps between the larger particles becomes narrower while the flow speed between them increases. Although the smaller particles were initially immobilized between the larger particles, now they will be swept along with the high speed inter-particle fluid, consequently escaping the stacks of larger particles and the filter (Fig. 3c).
Device Fabrication
The microfluidic device was realized on 2 × 2 cm2 surface oxidized silicon chips. Surpass 3000 (Dischem), an adhesion promoter, was spin coated on the SiO2 to enhance the adhesion of photoresist during the following processes. Photolithography was used to pattern the microfluidic channel in the SU-8 (2050, Microchem) photoresist which was spin-coated to be 30 μm thick. For encapsulation, 5 mm thick poly(dimethylsiloxane) (PDMS) was used with inlets and outlets defined. To have the device endure high fluidic stress, patterned SU-8 was exposed to O2 plasma (30 W, 160 mTorr, 30 s) followed by dipping in 5% (3-Aminopropyl)triethoxysilane for 10 minutes at 80 °C, then it was attached with the PDMS that was treated with O2 plasma (30 W, 160 mTorr, 30 s) resulting in firm Si-O-Si covalent bonds35. The width of the horizontal and vertical channels was 500 μm, and 35 × 25 μm2 round-edged pillars with specific intervals were aligned at 45° to the channel walls (Fig. 4a). The fabricated micro-pillars had intervals of 12 μm for the polymer microsphere sieving microfluidic chip (Fig. 4b), and 7 μm for cancer cell sieving from whole blood (Fig. 4c).
Results and Discussion
Characteristics of particle oscillation
We evaluated the movement of different sized particles within the oscillating fluid through a high speed camera. Figure 5a shows the oscillation amplitude of a 20 μm diameter PS microsphere observed for 0.5 ml/h and 1 ml/h flow speeds with oscillation frequencies between 70 Hz to 230 Hz, in 20 Hz steps. The oscillation amplitude was measured from the moment when the PS particles were blocked by the filter, to its maximum receding displacement position. The inset images in Fig. 5a shows the optical images of the filtered PS particles during the receding phase of its oscillation, from a to e in sequence, with the time interval of 1 ms. It can be seen that for both of the fluid flow speeds the maximum particle oscillation length was at 130 Hz. We believe that this frequency is near the resonant frequency of the total fluid mass contained within the microfluidic channel and the connecting tubes. This should vary with the microfluidic channel dimensions, the lengths of the inlet and outlet tubes connected with it, and the distance from the channel inlet to the piezoelectric actuation along the inlet tube. The dependence of the oscillation amplitudes on the fluid speeds was a result of the difference in the average forward motion of the particles during the receding phase of their oscillation. We then examined the oscillation according to the size of the particles, and discovered that the particle displacement was exactly the same regardless of their sizes for the same flow speed (Fig. 5b). Judging from their low particle Reynolds number (<0.1), this is due to the small sizes of the microspheres not creating enough of a difference in their relative movements14.
Demonstration of μ-sieving and retrieval of PS micro-particles
To quantitatively analyze the μ-sieving technique and test the feasibility of μ-sieving CTCs from whole blood, PS microspheres of 20 μm and 5 μm were utilized representing the size of CTCs and red blood cells (RBCs), respectively. Initially, the μ-sieving chip was operated without fluid oscillation resulting in the smaller PS particles aggregated among the larger PS particles filtered by the micro-pillars (Fig. 6a). Figure 6b shows the fluorescent image of smaller PS particles in Fig. 6a. Immediately after operating the piezoelectric actuator, the longitudinal oscillation started to release the small PS particles, allowing them to pass through the micro-pillar openings (Fig. 6c, see Supplementary Video S1). After only a second of oscillation, it can be seen in Fig. 6d and 6e that all of the small PS particles were released. Upon detailed evaluation (Fig. 6f), we find that most of the particles were released within the first 20 cycles of oscillations (0.128 s), and the rest of the particles were released gradually afterwards by advancing forward in the filter with each oscillation cycle. After 60 cycles (0.395 s), we observed that more than 97% of the trapped particles were released. Finally, the last two particles were released after 0.8 s of oscillation. The μ-sieving with fluid oscillation did not allow any larger PS particles to pass through the filter, showing 100% separation efficiency. The filtered larger PS particles were then retrieved by changing the flow direction. The retrieval experiment was repeated for 4 μ-sieving chips, and the average retrieval efficiency was 99.2% (Fig. 6g). Three of the four experiments showed 100% retrieval of the filtered PS particles. There were four larger PS particles trapped between the micro-pillars for device #1 which may be due to the slightly wider micro-pillar openings. It is generally reported that in micro-filter or membrane based microfluidic separation devices, retrieval rate of filtered particles have not been satisfactory due to some particles being squeezed into the filter openings. This is from the hydraulic pressure gradually increasing as the number of open filters becomes reduced by clogging. In our μ-sieving device, clogging was absent due to the fluidic oscillation; target particles were released from the filter repeatedly by backward flow, and so they did not experience such pressure between the pillars. This resulted in higher release efficiency.
(a) Bright field and (b) fluorescent images before the activation of sieving. Red dots indicate the smaller non-target particles (5 μm) which were surrounded by the larger particles (20 μm). (c) Image of the smaller particles being released with the activation of fluid oscillation. (d) Bright field and (e) fluorescent images after fluid oscillation. (f ) The recorded number of smaller particles released during μ-sieving depending on the oscillation cycle. Inset table shows the time duration and the particle release efficiency (measured as the percentage of the number of smaller particles released) up to the given oscillation cycle. (g) Larger particle retrieval efficiency of the μ-sieving systems. The average value was 99.2%.
Cancer cell μ-sieving from whole blood
As proof-of-concept of cancer cell separation, we applied the μ-sieving technique to the separation of fixed cancer cells from whole blood. As with the previous experiments, fixed MDA-MB-231 cells were initially trapped prior to whole blood injection (see Supplementary Figure S2a). To reduce the hydraulic pressure on the filtered tumor cells, the sample flow rate was reduced to 0.2 ml/h, and the buffer flow rate was reduced to 0.1 ml/h. In normal flow condition, the shear rate at the filters increases with the trapping of cancer cells due to the reduction of fluid cross-section inducing platelet activation and raising thrombogenicity of whole blood36,37,38. The coagulation of the hemocytes with the cancer cells led to complete blocking of the filters preventing further operation. In most filter based whole blood CTC separation systems, to avoid this type of coagulation, lysis of RBCs, centrifuge separation of RBCs and the anti coagulant, or both was performed prior to the experiments7,13,33. However, this process may lead to the loss of CTCs which may critically hinder its application due to the relevant number of CTCs being very small (few out of 109 hemocytes)39. Here, we avoided most of the pretreatments to remove the RBCs to demonstrate that it was possible to filter cancer cells directly from whole blood with minimum chemical treatment. We observed that the coagulated cancer cells were freed from the filter and started oscillating with the fluid when the fluid oscillation (130 Hz) was applied. This enabled the RBCs to pass through the filters allowing continued operation of the μ-sieve (see Supplementary Figure S2b and Supplementary Video S3).
Figure 7 shows the result of continuous μ-sieving applied to cancer cell spiked blood. We began the experiment without fluid oscillation which led to several cancer cells being captured by the filters. It can be seen that the fluid flow through the filter pore with cancer cells was reduced significantly increasing the flow through the rest of the unblocked pores (Fig. 7a). If the high shear rate continues, it may lead to coagulation, making the filtered cancer cells irretrievable. However, as fluid oscillation was applied at an early stage of the experiment, we see that the filtered cancer cells began to oscillate with the fluid freeing the filter pores (Fig. 7b, see Supplementary Video S4). This allowed the whole blood specimen (0.2 ml) to be filtered continuously for one hour, and resulted in the filtration of 49 cancer cells (Fig. 7c). No cells conforming to the size and morphology of the fixed cancer cells was observed in the outlet resulting in complete separation. By reversing the fluid flow as shown in Fig. 2c, it was possible to collect the filtered cancer cells with the average retrieval efficiency of 99.2% (Fig. 8a). Optical microscope image of the retrieved particles shows many different types, ranging from circular cells to sharp debris (Fig. 8b). From the microscope image taken under fluorescence, most of the circular cells with proper morphology were identified as green fluorescent protein (GFP) overexpressed cancer cells that were spiked into the whole blood (Fig. 8c). Other smaller particles visible in Fig. 8b may be hemocytes that were not completely drained flowing back through the filter during flow reversal for particle retrieval, which does lower purity. Sufficient rinsing after sieving should reduce reverse flow contamination and improve purity.
(a) The filters partially blocked by captured cancer cells prior to fluid oscillation. White dotted circles indicate the cancer cells. (b) Continuous μ-sieving with filter pores unblocked by fluid oscillation. (c) The filtered cancer cells after completing μ-sieving with continuous fluid oscillation.
The simplicity of the method of μ-sieving and its effectiveness is noteworthy. By simply contacting a piezoelectric actuator with the inlet tube, existing filtration systems can also have greatly improved selectivity without structural modification, freedom from clogging by small particle impurities, increased throughput and retrieval efficiency. Such improvements achieved by μ-sieving will lead to effective post-processing and downstream analysis. However, further research is required to utilize μ-sieving in clinical application. The key issue for live cell manipulation is in their deformability making size-based separation more difficult. Viability of the cells exposed to oscillation is also an important issue that should be addressed.
Conclusion
We have developed a μ-sieving system which is capable of size-based separation of PS particles and cancer cells. Different from conventional filtration systems, we added fluid oscillations that enabled continuous operation without filter clogging. The fluid oscillation was induced by a piezoelectric actuator making contact with the inlet tube. The maximum amplitude of particle oscillation was at the actuation frequency of 130 Hz. PS particles of 20 μm were successfully filtered from 5 μm PS particles showing 100% separation efficiency and purity. Sieved particles were retrievable with 99.2% efficiency by changing the flow direction. We also demonstrated μ-sieving of MDA-MB-231 spiked whole blood, and showed continuous sieving of cancer cells without blood coagulation and cell loss. With the ability to allow continuous operation of size-based filtered separation, our device may bring closer application of microfluidic separation devices to biochemical analyte separation, clinical liquid biopsy and industrial micro-particle separation.
Materials
PS microspheres were utilized to characterize the μ-sieving. Two different diameters of PS, 20 μm (Sigma-Aldrich) and 5 μm (Micromod), were chosen. The smaller PS particles have red fluorescence when exposed to UV radiation for effective visualization of the particle separation. The PS microspheres were suspended in a 20% sucrose solution to expand the time of sinking during the experiments, and surfactant, 0.1% F-127 (Sigma-Aldrich), was also included to disperse the microspheres preventing aggregation.
For μ-sieving cancer cells, MDA-MB-231, a human breast cancer cell line, spiked blood was utilized. Written informed consent was obtained from healthy donors at Hanyang University Seoul Hospital with an approval granted by the Hanyang University Institutional Review Board. All methods were carried out in accordance with the approved guidelines. The blood was diluted twice with 1× phosphate buffer saline. MDA-MB-231 was established from the American Type Culture Collection (Manassas, VA). The cells were tagged by GFP and grown in Dulbeco’s modified eagle’s medium supplemented with 10% fetal bovine serum, penicillin (100 units/ml), streptomycin (100 g/ml) and cultured in a humidified 5% CO2 atmosphere at 37 °C. Cultured tumor cells were harvested and fixed in 70% ethanol prior to the experiments.
Additional Information
How to cite this article: Yoon, Y. et al. Clogging-free microfluidics for continuous size-based separation of microparticles. Sci. Rep. 6, 26531; doi: 10.1038/srep26531 (2016).
References
Desitter, I. et al. A new device for rapid isolation by size and characterization of rare circulating tumor cells. Anticancer Res 31, 427–441 (2011).
Hayes, D. F. et al. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin Cancer Res 12, 4218–4224, doi: 10.1158/1078-0432.CCR-05-2821 (2006).
Hua, M. et al. Heavy metal removal from water/wastewater by nanosized metal oxides: a review. J Hazard Mater 211–212, 317–331, doi: 10.1016/j.jhazmat.2011.10.016 (2012).
Choi, S. & Park, J. K. Microfluidic system for dielectrophoretic separation based on a trapezoidal electrode array. Lab Chip 5, 1161–1167, doi: 10.1039/b505088j (2005).
Fong, E. J. et al. Acoustic focusing with engineered node locations for high-performance microfluidic particle separation. Analyst 139, 1192–1200, doi: 10.1039/c4an00034j (2014).
Jung, Y., Choi, Y., Han, K. H. & Frazier, A. B. Six-stage cascade paramagnetic mode magnetophoretic separation system for human blood samples. Biomed Microdevices 12, 637–645, doi: 10.1007/s10544-010-9416-3 (2010).
Li, P. et al. Acoustic separation of circulating tumor cells. Proc Natl Acad Sci USA 112, 4970–4975, doi: 10.1073/pnas.1504484112 (2015).
Lin, Y.-H. & Lee, G.-B. Optically induced flow cytometry for continuous microparticle counting and sorting. Biosensors and Bioelectronics 24, 572–578, doi: 10.1016/j.bios.2008.06.008 (2008).
Pamme, N., Eijkel, J. C. T. & Manz, A. On-chip free-flow magnetophoresis: Separation and detection of mixtures of magnetic particles in continuous flow. Journal of Magnetism and Magnetic Materials 307, 237–244, doi: 10.1016/j.jmmm.2006.04.008 (2006).
Doh, I. et al. Viable capture and release of cancer cells in human whole blood. Applied Physics Letters 101, 043701, doi: 10.1063/1.4737936 (2012).
Huang, L. R., Cox, E. C., Austin, R. H. & Sturm, J. C. Continuous particle separation through deterministic lateral displacement. Science 304, 987–990, doi: 10.1126/science.1094567 (2004).
Lim, L. S. et al. Microsieve lab-chip device for rapid enumeration and fluorescence in situ hybridization of circulating tumor cells. Lab Chip 12, 4388–4396, doi: 10.1039/c2lc20750h (2012).
Lv, P., Tang, Z., Liang, X., Guo, M. & Han, R. P. Spatially gradated segregation and recovery of circulating tumor cells from peripheral blood of cancer patients. Biomicrofluidics 7, 34109, doi: 10.1063/1.4808456 (2013).
Park, J. S. & Jung, H. I. Multiorifice flow fractionation: continuous size-based separation of microspheres using a series of contraction/expansion microchannels. Anal Chem 81, 8280–8288, doi: 10.1021/ac9005765 (2009).
Seo, J., Lean, M. H. & Kole, A. Membrane-free microfiltration by asymmetric inertial migration. Applied Physics Letters 91, 033901, doi: 10.1063/1.2756272 (2007).
Sochol, R. D. et al. Dual-mode hydrodynamic railing and arraying of microparticles for multi-stage signal detection in continuous flow biochemical microprocessors. Lab Chip 14, 1405–1409, doi: 10.1039/c4lc00012a (2014).
Yamada, M., Nakashima, M. & Seki, M. Pinched flow fractionation: continuous size separation of particles utilizing a laminar flow profile in a pinched microchannel. Anal Chem 76, 5465–5471, doi: 10.1021/ac049863r (2004).
Zheng, S. et al. 3D microfilter device for viable circulating tumor cell (CTC) enrichment from blood. Biomed Microdevices 13, 203–213, doi: 10.1007/s10544-010-9485-3 (2011).
Li, X., Chen, W., Liu, G., Lu, W. & Fu, J. Continuous-flow microfluidic blood cell sorting for unprocessed whole blood using surface-micromachined microfiltration membranes. Lab Chip 14, 2565–2575, doi: 10.1039/c4lc00350k (2014).
Fan, X. et al. A microfluidic chip integrated with a high-density PDMS-based microfiltration membrane for rapid isolation and detection of circulating tumor cells. Biosens Bioelectron 71, 380–386, doi: 10.1016/j.bios.2015.04.080 (2015).
Kuo, J. S. et al. Deformability considerations in filtration of biological cells. Lab Chip 10, 837–842, doi: 10.1039/b922301k (2010).
Suresh, S. et al. Connections between single-cell biomechanics and human disease states: gastrointestinal cancer and malaria. Acta Biomater 1, 15–30, doi: 10.1016/j.actbio.2004.09.001 (2005).
Vaziri, A. & Gopinath, A. Cell and biomolecular mechanics in silico. Nat Mater 7, 15–23, doi: 10.1038/nmat2040 (2008).
Vona, G. et al. Isolation by Size of Epithelial Tumor Cells. The American Journal of Pathology 156, 57–63, doi: 10.1016/s0002-9440(10)64706-2 (2000).
Zabaglo, L. et al. Cell filtration-laser scanning cytometry for the characterisation of circulating breast cancer cells. Cytometry A 55, 102–108, doi: 10.1002/cyto.a.10071 (2003).
Wakeman, R. J. & Williams, C. J. Additional techniques to improve microfiltration. Sep Purif Technol 26, 3–18, doi: Doi 10.1016/S1383-5866(01)00112-5 (2002).
Dong, X. et al. Patient-derived first generation xenografts of non-small cell lung cancers: promising tools for predicting drug responses for personalized chemotherapy. Clin Cancer Res 16, 1442–1451, doi: 10.1158/1078-0432.CCR-09-2878 (2010).
Beattie, W., Qin, X., Wang, L. & Ma, H. Clog-free cell filtration using resettable cell traps. Lab Chip 14, 2657–2665, doi: 10.1039/c4lc00306c (2014).
Gironès i Nogué, M., Akbarsyah, I. J., Bolhuis-Versteeg, L. A. M., Lammertink, R. G. H. & Wessling, M. Vibrating polymeric microsieves: Antifouling strategies for microfiltration. Journal of Membrane Science 285, 323–333, doi: 10.1016/j.memsci.2006.09.001 (2006).
Huang, S.-B., Wu, M.-H. & Lee, G.-B. A tunable micro filter modulated by pneumatic pressure for cell separation. Sensors and Actuators B: Chemical 142, 389–399, doi: 10.1016/j.snb.2009.07.046 (2009).
Li, H. Y., Bertram, C. D. & Wiley, D. E. Mechanisms by which pulsatile flow affects cross-flow microfiltration. Aiche J 44, 1950–1961, doi: DOI 10.1002/aic.690440903 (1998).
Liu, W. et al. Dynamic trapping and high-throughput patterning of cells using pneumatic microstructures in an integrated microfluidic device. Lab Chip 12, 1702–1709, doi: 10.1039/c2lc00034b (2012).
McFaul, S. M., Lin, B. K. & Ma, H. Cell separation based on size and deformability using microfluidic funnel ratchets. Lab Chip 12, 2369–2376, doi: 10.1039/c2lc21045b (2012).
Cheng, Y., Ye, X., Ma, Z., Xie, S. & Wang, W. High-throughput and clogging-free microfluidic filtration platform for on-chip cell separation from undiluted whole blood. Biomicrofluidics 10, 014118, doi: 10.1063/1.4941985 (2016).
Li, P., Lei, N., Sheadel, D. A., Xu, J. & Xue, W. Integration of nanosensors into a sealed microchannel in a hybrid lab-on-a-chip device. Sensors and Actuators B: Chemical 166–167, 870–877, doi: 10.1016/j.snb.2012.02.047 (2012).
Haynes, L. M., Dubief, Y. C., Orfeo, T. & Mann, K. G. Dilutional control of prothrombin activation at physiologically relevant shear rates. Biophys J 100, 765–773, doi: 10.1016/j.bpj.2010.12.3720 (2011).
Miyazaki, Y. et al. High shear stress can initiate both platelet aggregation and shedding of procoagulant containing microparticles. Blood 88, 3456–3464 (1996).
Sheriff, J. et al. Evaluation of shear-induced platelet activation models under constant and dynamic shear stress loading conditions relevant to devices. Ann Biomed Eng 41, 1279–1296, doi: 10.1007/s10439-013-0758-x (2013).
Hyun, K. A. & Jung, H. I. Advances and critical concerns with the microfluidic enrichments of circulating tumor cells. Lab Chip 14, 45–56, doi: 10.1039/c3lc50582k (2014).
Acknowledgements
This research was supported by the Converging Research Center Program funded by the Ministry of Science, ICT & Future Planning (Project No. 2015054348), and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2013R1A1A2011532 & 2012R1A6A1029029).
Author information
Authors and Affiliations
Contributions
Y.Y. and S.K. designed the device and conducted experiments together. S.K., J.L. and J.C. fabricated μ-sieving devices. R.-K.K. provided the tumor cells under supervision of S.-J.L. and S.K. performed the experiments, while data interpretation and analysis were carried out by S.K. and Y.Y. under supervision of S.-B.L. Computational simulation was performed by Y.Y. and S.K. Initial manuscript was written by Y.Y. and reviewed by O.S. Final manuscript was written by Y.Y. under guidance of S.-B.L. All authors reviewed the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Yoon, Y., Kim, S., Lee, J. et al. Clogging-free microfluidics for continuous size-based separation of microparticles. Sci Rep 6, 26531 (2016). https://doi.org/10.1038/srep26531
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep26531
This article is cited by
-
Electrically-driven modulation of flow patterns in liquid crystal microfludics
Scientific Reports (2024)
-
Visualization-enhanced under-oil open microfluidic system for in situ characterization of multi-phase chemical reactions
Nature Communications (2024)
-
Isolation of exosome from the culture medium of Nasopharyngeal cancer (NPC) C666-1 cells using inertial based Microfluidic channel
Biomedical Microdevices (2022)
-
Pillar based microfluidic approach to sorting of microparticle mixtures at various particle ratios
Microfluidics and Nanofluidics (2021)
-
Optimizing Flux Capacity of Dead-end Filtration Membranes by Controlling Flow with Pulse Width Modulated Periodic Backflush
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.