Abstract
Biotin protein ligase is universal in three domains of life. The paradigm version of BPL is the Escherichia coli BirA that is also a repressor for the biotin biosynthesis pathway. Streptococcus suis, a leading bacterial agent for swine diseases, seems to be an increasingly-important opportunistic human pathogen. Unlike the scenario in E. coli, S. suis lacks the de novo biotin biosynthesis pathway. In contrast, it retains a bioY, a biotin transporter-encoding gene, indicating an alternative survival strategy for S. suis to scavenge biotin from its inhabiting niche. Here we report functional definition of S. suis birA homologue. The in vivo functions of the birA paralogue with only 23.6% identity to the counterpart of E. coli, was judged by its ability to complement the conditional lethal mutants of E. coli birA. The recombinant BirA protein of S. suis was overexpressed in E. coli, purified to homogeneity and verified with MS. Both cellulose TLC and MALDI-TOFF-MS assays demonstrated that the S. suis BirA protein catalyzed the biotinylation reaction of its acceptor biotin carboxyl carrier protein. EMSA assays confirmed binding of the bioY gene to the S. suis BirA. The data defined the first example of the bifunctional BirA ligase/repressor in Streptococcus.
Similar content being viewed by others
Introduction
Biotin (vitamin H) is one of two known sulfur-containing fatty acid derivatives (biotin1,2 and lipoic acid3) and acts as an enzyme cofactor universal in three domains of the life. Although the biotin-requiring enzymes are rare proteins (in that mammals have only four such proteins whereas Escherichia coli has only a single biotinylated protein)2,4, they play critical roles in certain important reactions (like carboxylation, decarboxylation and trans-carboxylation) implicated into fatty acid synthesis, gluconeogenesis and amino acid degradation in both prokaryotes and eukaryotes5,6. Right now, it is aware that most microorganisms (bacteria and fungi) and plants possess the ability to synthesize biotin, whereas mammals and birds cannot4. The earlier steps of biotin synthesis are involved in a modified type II fatty acid synthesis pathway in E. coli7,8, whereas the latter of biotin synthesis route refers to a highly-conserved four-step reactions catalyzed by BioF, BioA, BioD and BioB, respectively6,9. In light that biotin is an energetically-expansive molecule in that generally 15–20 ATP equivalents are estimated to be consumed via its paths of de novo synthesis for each biotin4, it seems reasonable that bacteria have developed diversified mechanisms to tightly monitor the level of biotin production in vivo1,4,6,10,11. In addition to the paradigm E. coli BirA regulatory system that also retains the activity of biotin-protein ligase12,13,14,15, at least two more regulatory machineries have been reported1,4,10,11,16. Among them, one is the two-protein system of BirA coupled with BioR, the GntR-family transcription factor1,4,11 and the other denotes the two-protein system of the BirA linked to BioQ, a TetR family of transcription factor10,16.
In fact, bacteria have evolved two different mechanisms to obtain the biotin cofactor for the metabolic requirement, one of which is de novo synthesis route2,11, the other is a system of BioY transporter-mediated uptake5. Unlike the human pathogen Brucella, a member of α-proteobacteria that encodes the above two systems for the availability of biotin11, it seems likely that the species of Streptococcus/Lactococcus family only have the BioY-based scavenging route and compensates the lack of the de novo biotin synthesis pathway17. Among microorganisms, the paradigm enzyme with the biotin requirement refers to biotin carboxyl carrier protein (abbreviated as BCCP, i.e., the AccB subunit of acetyl-CoA carboxylase (ACC)) that catalyzes the first committed reaction for type II fatty acid synthesis pathway18. Biotin protein ligase (BPL) is widespread in three domains of the life in that it transfers/attaches the biotin cofactor to the specific domain of the relevant subunits of key enzymes from the certain central metabolisms19,20. Most of bacteria including E. coli14,18 and Bacillus21 only encode a single BPL to account for such kind of physiological requirement, while the pathogen Fracisella novicida developed an additional BPL to gain the competitive advantage in the infected host environment2. In general, the BPL members are categorized into the following two groups (Group I and Group II) that can be easily distinguished by the presence of N-terminal DNA-binding domain that allow the BirA protein to bind the cognate genes (e.g., bio operon) and thereafter inhibit expression of biotin metabolism19,21. Unlike the paradigm Group II BPL proteins, the E. coli birA gene product retaining the DNA-binding activity, the Group I BPL that lacks the N-terminal helix-turn-helix domain solely function as an enzyme responsible for protein biotinylation6,9. In particular, the regulatory role of the Group II BPL depends on the participation of the physiological ligand/effector (biotinoyl-5′-AMP), the product of the first ligase half reaction for biotin utilization/protein biotinylation12.
Streptococcus suis (S. suis) is a leading agent of bacterial diseases including meningitis, arthritis and septicemia) in swine industry worldwide22,23 and also appears to be an opportunistic zoonotic pathogen responsible for human S. suis infections such as meningitis and even streptococcal toxic shock-like syndrome (STSS)24. Given the difference of bacterial capsule, 35 kinds of serotypes (1–34, 1/2) have been attributed to S. suis25. Among them, the serotype 2 of S. suis is frequently isolated from diseased piglets and highly-related to the strong virulence22,23,25. S. suis 2 (SS2) is a previously-neglected, but newly-emerging human pathogen, claiming a series of occupational/ opportunistic infections. Since the first discovery of human SS2 meningitis was recoded in Denmark, in 196825, SS2 has spread to nearly 30 countries (and/or regions) and caused no less than 1,600 human cases26. In particular, two big-scale outbreaks of fatal human SS2 infections had ever emerged in China (one in Jiangsu Province, 1998 and the other in Sichuan Province, 2005), posing a great concern to public health24,27,28. In 2007, we also reported three sporadic cases of human SS2 meningitis in China (two cases in Shenzhen City and one case in Chongqing City)29, implying the co-existence of outbreaks and sporadic cases in China24,26,28,29. It is unusual that the epidemic strain of Chinese virulent SS2 harbors a pathogenicity island (PAI) referred to 89 K30,31. Subsequent functional exploration suggested that this 89 K PAI behaves as a transposon-like genetic element32 and encodes type IV secretion system33 and SezAT toxin-antitoxin system34. Our epidemiological investigation argued that the 89 K PAI might be undergoing unknown selective pressure in that some variants losing 89 K PAI are emerging35. The remodeling of bacterial surface structure significantly was found to attenuate full virulence of the epidemic SS2 strain36. Very recently, we observed that regulation of the D-galactosamine (GalN)/N-acetyl-D-galactosamine (GalNAc) catabolism pathway is linked to its infectivity37. It seems likely that the regulatory network of bacterial metabolism is complicated into SS2 virulence26,38. Given the fact that biotin metabolism and utilization is associated with Francisella pathogenesis2,39,40, it is much interest to define the biotin utilization pathway in the human pathogen S. suis 2.
In this paper, the epidemic SS2 strain in China, S. suis 05ZYH33 with the known genome sequence was subjected to the context analyses of the bacterial biotin metabolism and its possible regulation. Unlike the scenarios seen with the paradigm organism E. coli, the possible biotin machinery in the S. suis we detected comprises a single BioY (SSU05_0509) transporter regulated by the BirA bifunctional protein (05SSU_1625) and the biotin-requiring substrate protein AccB (SSU05_1801). By employing integrative approaches that ranged from comparative genomics, bioinformatics, biochemistry/biophysics, metabolomics, to bacterial genetics, we attempted to present a full picture of biotin utilization pathway in the zoonotic pathogen S. suis.
Results and Discussion
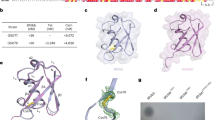
S. suis BirA Protein is a Group II BPL Member
It seems likely that S. suis is biotin auxotrophic in that it is deficient in biotin synthesis and depends on the mechanism of BioY-BirA to scavenge biotin from the inhabiting niche and/or infected host environment (Fig. 1). System biology by Rodionov et al.41 revealed that bi-functional BPL enzymes (Group II) exemplified with the E. coli BirA, are universal in both Eubacteria and Archaea, implying the group II form might be the ancestor of the BPL. Unlike the group I without DNA-binding domain BPL (e.g., BirA orthologues of Agrobacterium4 and Brucella11), the multiple sequence alignment analyses suggested that S. suis BirA is generally similar to the paradigm E. coli birA product (Fig. 2). However the situation seemed unusual in the close-relative of S. suis, Lactococcus lactis in that this probiotic bacterium contained two versions of BPL, one of which refers to BirA1_LL (Group II BPL) and the other is BirA2_LL lacking the DNA-interacting motif (Group I BPL) (Fig. 2). To address the BPL biochemistry of the S. suis BirA, we applied protein engineering to produce the recombinant protein. As anticipated, we harvested the BirA_ss protein with the mass of around 37 kDa (Fig. 3A). Also, the purity was judged with SDS-PAGE (Fig. 3A). To further assure the identity, the polypeptide fragments digested from the recombinant BirA protein were subjected to the analyses of MALDI-TOFF. The MS result suggested that the recombinant protein matched well the native form of the S. suis BirA in that it exhibited the coverage of 54% (Fig. 3B). Structural modelling assigned the S. suis BirA as a typical version of the group II BPL with the perfect architecture (Fig. 3C). It comprised the following three functional motifs: N-terminal DNA-binding domain, Central domain and C-terminal domain (Fig. 3C). Obviously, the above data defined the S. suis BirA as a member of Group II BPL.
Current model for biotin transport and utilization in Streptococcus suis.
The biotin transporter BioY was illustrated with modeled ribbon structure (in purple) and integrated into the scheme of bacterial membrane. The biotin transported from environment was activated into biotinoyl-5′-AMP and then transferred to BCCP acceptor protein giving the biotinoyl-5′-BCCP. Sulphur was labeled in orange and AMP (and/or ATP/PPi) was highlighted in red. The biotin acceptor protein BCCP was indicated with a blue rectangle.
Sequence comparison of Streptococcus suis BirA orthologue with the prototypical E. coli version.
The multiple sequence alignment of BirA protein was performed using the program of Clustal Omega, an updated version of ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalo) and the final output was given with the program ESPript 3.0 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). Identical residues are white letters with red background, similar residues are red letters in white background, the varied residues are in grey letters and gaps are denoted with dots. The protein secondary structure was shown in cartoon (on top), α: α-helix; β: β-sheet; T: Turn; η: coil.
Characterization of S. suis BirA protein.
(A) SDS-PAGE profile of the purified recombinant BirA protein from Streptococcus suis Gradient SDS-PAGE (4–20%) was applied to separate the protein. (B) MS-based determination of the recombinant protein of S. suis BirA The peptide fragments that matched the native form of S. suis BirA were given with bold and underlined letters (in red). Totally, 54% coverage was detected. (C) Modeled ribbon structure of S. suis BirA protein S. suis BirA protein included three domains: the N-terminal DNA-binding domain (in purple), the central domain (in blue) and the C-terminal domain with enzymatic activity (in yellow).
Activity of Biotin Protein Ligase of S. suis BirA
We employed the in vitro and in vivo approaches to address the BPL activity of S. suis BirA. The two E. coli birA mutants used for functional complementation included the temperature-sensitive mutant of birAts (BM4062) and the birA1 Km mutant (BM4092). As expected, the BM4062 Strain with/without the empty vector pBAD24 cannot grow on the M9 agar plates under the non-permissive temperature of 42 °C (Fig. 4A). In contrast, the arabinose-induced expression (and even basal expression) of the plasmid-borne birA_ss supported the growth of the birAts mutant BM4062 at 42 °C (Fig. 4A). The measurement of bacterial growth curves for BM4062 strains in liquid media also reproduced the similar results to those obtained from the agar plates (Fig. 4B). The presence of pBAD24 plasmid-borne birA_ss allowed the birA1 Km mutant of E. coli, BM4092 to grow on the minimal media supplemented with low level of biotin (25 nM) (Fig. 4C), which agreed well with the scenario seen in the growth curves (Fig. 4D). Obviously, it confirmed that S. suis BirA has a role of being the BPL ligase in the alternative model, E. coli.
Functional complementation of E. coli birA mutants with the putative biotin protein ligase-encoding gene birA.
(A) Growth of E. coli BM4062 birAts mutant carrying plasmid-borne S. suis birA gene on the M9 agar plates (B) Growth curves of E. coli BM4062 birAts mutant and its derivatives E. coli strains were maintained at 42 °C (the non-permitted temperature of BM4062) on the defined media M9 with/without 0.2% arabinose for around 36 hours. M9 agar plates were supplemented with 0.5 mM X-gal plus 100 nM biotin. (C) Growth of E. coli BM4092 birA1 Km mutant carrying plasmid-borne S. suis birA on the M9 agar plates. (D) Growth curves of E. coli BM4092 birA1 Km mutant and its derivatives. E. coli strains were maintained at 30 °C on the defined media M9 with/without 0.2% arabinose for around 36 hours. M9 agar plates were supplemented with 0.5 mM X-gal plus 25 nM biotin. The bacterial growth was measured by optical density at 600 nm, which is automatically recorded using a BioScreen C instrument. Each growth curve assay was carried out in triplicate and the average was used in this plot50. Designations: Vec, vector (c); ts: temperature-sensitive; Ara, Arabinose.
To further prove the BPL function of S. suis BirA, we established the assays of enzymatic reaction in vitro. In this system of BirA-catalyzed reaction, the substrate used is biotinylated domain (designated as AccB87 or BCCP87) of the AccB protein (Fig. 5A,B), which carries a conserved biotinylation site of lysine at the position 122 (K122) (Fig. 5A,C). The method of thin layer chromatography (TLC) was applied to assay conversion of α-32P-labeled ATP and biotin to biotinoyl-AMP (Fig. 5D). In principle, it represents direct evidence for the first ligase partial reaction (Fig. 5D) and upon an addition of the acceptor protein AccB87 provides an indirect proof of the second ligase partial reaction (i.e., transferring of biotin from biotinoyl-5′-AMP to the AccB87 acceptor protein) (Fig. 5D). As expected, the S. suis BirA protein was shown to convert biotin and [α-32P]-ATP to the canonical biotinoyl-5′-AMP intermediate (Fig. 5D) and transferred the biotin moiety to the AccB-87 acceptor protein (Fig. 5D).
Evidence that S. suis BirA biotinylates the AccB substrate protein.
(A) Sequence analyses for the biotinylation domain of the BCCP acceptor protein (also referred to AccB). (B) SDS-PAGE profile of the biotin substrate domain (BCCP87 or AccB87). (C) Structural modeling of the biotinylated domain of the S. suis AccB protein The biotinylation site of AccB is lysine at the position 122 (K122). (D) TLC assays for the BPL enzymatic activity of S. suis BirA. The abbreviations: M, protein marker (Biorad); kDa, kilo-dalton. Plus (+) and Minus (−) denotes presence and absence of BirA protein, Biotin or AccB87 substrate protein.
Subsequently, we utilized the matrix-assisted laser desorption/ionization (MALDI) to measure the level of BirA ligase-catalyzed biotinylation of AccB-87 as we recently described4. The MS results illustrated that the calculated mass for AccB87 is 10324.2~10327.5 (Fig. 6A) and the expected mass for biotinoyl-AccB87 is 10550.6 (Fig. 6B). Collectively, the integrative data demonstrated that S. suis BirA acts as a functional member of the BPL family.
MS-based verification for S. suis BirA-catalyzed biotinylation of the AccB87 substrate protein.
(A) MALDI-TOF determination of the molecular weight for the un-biotinylated AccB87 polypeptide. (B) MALDI-TOF identification for the biotinylation of the AccB87 substrate by S. suis BirA. The calculated mass for AccB87 is 10324.2~10327.5 and the expected mass for biotinoyl-AccB87 by S. suis BirA (Panel B) is 10550.6. Designations: minus denotes no addition of BirA enzyme, plus denotes addition of the BirA protein.
Binding of S. suis BirA to the cognate bioY gene
It is unusual that S. suis does not have the ability of de novo biotin synthesis in that this zoonotic pathogen lacks the bio operon found in E. coli6 and other organisms like Agrobacterium4 and Paracoccus1. However, it seemed likely that the inability of S. suis to make biotin is compensated with the BioY-mediated biotin uptake/scavenging pathway (Figs 1 and 7A). Also, the bioY lous is present in other two closely-relatives of the human pathogen (Enterococcus faecalis and Lactococcus lactis) (Fig. 7A). In particular, the L. lactis encoded two versions of bioY genes as well as two orthologues of BirA (Fig. 7A), implying the complexity of biotin metabolism in the certain species of low-GC contents, gram-positive bacteria.
Genetic organization of birA (and bioY) and S. suis bioY promoter.
(A) Genetic organization of birA (and bioY). The locus of birA (or referred to birA1) and bioY (referred to bioY1) is highlighted in blue and yellow, respectively. The blue spot denotes the predicted BirA-binding site. The locus of birA2 and bioY2 is indicated with an arrow in grey and purple. (B) S. suis bioY promoter. “S” denotes the putative transcriptional start site and “ATG” in red is translation initiation site. The predicted BirA-binding site is underlined.
The transcription start site of the S. suis bioY gene is estimated to be “T” that is 29 bp ahead of the translation initiation site “ATG”. Bioinformatics analyses suggested that a putative BirA-binding site (TTT TGT TAA CCA TAA AAT TTT AAG AGG ATA ACA A) covering the transcription start site is present in the S. suis bioY promoter region (Fig. 7B). Given the above observations, we proposed that the bioY might be negatively regulated by the S. suis BirA. While this hypothesis required experimental evidence. We tested the ability of BirA to bind bioY promoter using a 54 bp probe containing the predicted site using the electrophoretic mobility shift (gel shift) assays (Fig. 8A) as we recently performed2 with minor modifications. Gel shift assays showed that S. suis BirA efficiently bound the bioY probe in a dose-dependent manner (Fig. 8B) in that nearly 100% of bioY probe was transferred into the DNA-protein complex in the presence of 0.5 pmol BirA (Fig. 8B). In light of the appreciable conservation of the BirA sites in the bioY promoter from the related organisms (Fig. 8A), we also examined possible crosstalk of the BirA to bioY of various origins. In fact, the S. suis BirA was found to exhibit comparable binding to the bioY probes of L. lactis (Fig. 8C) and E. faecalitis (Fig. 8D). It demonstrated that physical interaction is present between the BirA bifunctional protein and the biotin transporter-encoding gene bioY.
Interplay between BioY and BirA.
(A) Multiple sequence alignment of the BirA-binding sites. (B) BirA protein of S. suis binds to its own bioY promoter. (C) Binding of S. suis BirA to the L. lactis bioY promoter. (D) Interaction of S. suis BirA with the E. faecalitis bioY promoter. Using 7% native PAGE, gel-shift assays were conducted and a representative photograph is given. In each assay, levels of BirA are denoted with a triangle on right hand (0.1, 0.5, 2 and 5 pmol), whereas all the DIG-labeled probes (bioY_SS, bioY_LL and bioY_EF) are added to 0.2 pmol. Minus sign denotes no addition of BirA protein. Designations: SS, Streptococcus suis; LL, Lactococcus lactis; EF, Enterococcus faecalitis.
Physiological Implications for Biotin Utilization Pathway
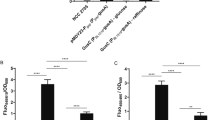
It is reasonable that S. suis deficient in biotin synthesis evolved the mechanism of BioY-BirA to utilize the biotin scavenged from the inhabiting niche and/or infected host environment (Fig. 1). In the epidemic strain of S. suis serotype 2, 05YH33, three genes with the involvement of biotin metabolism denote bioY (SSU05_0509), birA (05SSU_1625) and accB (05SSU1801), respectively. Following the biotinylation by BirA, the AccB was converted from its apo-form into holo-form and participated into the initiation of fatty acid biosynthesis. Given the fact that BirA binds the bioY promoter with the help of biotinoyl-5′-AMP (the intermediate of biotin biotinylation), the regulatory function of the BirA protein is supposed to guarantee that the wasteful production of the BioY transporter is avoided/minimized upon the biotin uptake from the outside environment. Probably, it is a physiological advantage for certain species of Streptococcus/Lactococcus in the context of lipid metabolism. To test above anticipation, we constructed the birA(ΔN) mutant of S. suis of which the DNA-binding domain was in-frame deleted (Fig. 9A,B). The removal of N-terminal DNA-binding motif affect bacterial growth on THB media, but this growth defection can be rescued by supplementation of the 5% defibrinated blood (or blood sera) (Fig. 9C,D). However, the expression of the plasmid-borne bioY promoter-driven lacZ is not altered significantly in the birA(ΔN) mutant in relative to the wild type (not shown). It might suggest a possibility that the interplay between BirA and bioY represent a developing and/or degenerating system for S. suis.
Conclusions
Our data shown here defined a working model for the route of biotin uptake/utilization in the zoonotic pathogen S. suis 2 (Fig. 1). Unlike the scenarios observed in both Brucella11 and Paracoccus1 in that the bioY gene interacts with the BioR regulator, our finding represents a first example for the interplay between the bioY and BirA in the Streptococcus/Lactococcus. Of note, no reaction is present between the bioY gene and BioR in the plant pathogen Agrobacterium4, a close relative of the human pathogen Brucella11. It suggested the complexity and diversity of bacterial biotin metabolism and regulation. Given the fact that the biotin synthetic genes bioJ39 and bpl2 are involved in bacterial virulence of the intracellular pathogen Francisella novicida, it is of much interest to probe the possible role of biotin metabolism in Streptococcus pathogenesis. While the fact that both bioY and birA are essential for bacterial viability of Streptococcus suis argued the technical feasibility in the genetic removal of the two biotin-related genes. As we knew, biotin and lipoic acid both are sulfur-containing vitamins required for the three domains of the life. Similarly, the scavenging of lipoic acids by LplA was also required for the intracellular growth/survival and virulence42,43. Thereby we screened the genome sequence of S. suis 05ZYH33 for the presence of the lplA gene that encodes lipoate-protein ligase, giving the perfect hit (SSU05_1836). We are planning to examine its relevance to bacterial infectivity. Right now, it seemed true that both biotin and lipoic acid are nutritional virulence factors for certain species of bacterial pathogens. Given the fact that S. suis 2 is an emerging/reemerging infectious agent threatening public health26, our finding might be helpful to better understanding biology and even infection of this zoonotic pathogen.
Methods
Bacterial strains and growth conditions
Bacterial strains used here included E. coli and Streptococcus suis (Table 1) and all the E. coli strains are derived from the wild-type K-12 (Table 1). The two media (Luria Bertani (LB) and rich broth (RB)) were utilized for E. coli, whereas the Todd Hewitt Broth (THB) medium was used for S. suis37. Antibiotics were supplemented as follows (in mg/liter): sodium ampicillin, 100; kanamycin sulfate, 50; and Spectinomycin, 100.
Plasmids and genetic manipulations
The birA gene (SSU05_1625) was amplified by PCR with genomic DNA of S. suis 05ZYH33 as the template and cloned into the expression vector pET28(a), giving the recombinant plasmid pET28-birA_ss (Table 1). To prepare the BirA protein, the expression plasmid pET28-birA_ss was transformed into the strain BL21(DE3), giving the strain FYJ280 (Table 1)44.
Also, the birA_ss gene was cloned into the arabinose-inducible expression vector pBAD244, giving the plasmid pBAD24-birA_ss. To evaluate the in vivo activity of BirA, two birA mutants of E. coli were applied, which referred to the birA km mutant strain BM4092 and the temperature-sensitive mutant BM4062, respectively (Table 1). Given the fact the birA is a bifunctional gene and is required for bacterial viability, it is reasonable to delete the partial function of birA at 5′-end. Therefore we employed an approach of homologous recombination to remove the N-terminal DNA-binding domain from the birA gene of S. suis 05ZYH33, giving the mutant birA(ΔN) (Table 1). In this case, a thermos-sensitive suicide vector pSET4s45 was applied. The promoter of S. suis bioY was fused to the promoter-less lacZ gene, creating the plasmid-borne PbioY-lacZ fusion (Table 1). To examine role of birA in vivo, the PbioY-lacZ fusion was separately introduced into the wild-type strain and the birA(ΔN) mutant of S. suis. All the acquired plasmids were verified by the PCR assay and direct DNA sequencing.
Expression and purification of BirA protein
The E. coli carrying the 28-birAss was used for preparation of the recombinant protein of S. suis BirA. The bacterial cultures were induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 30 °C for 3 h. The clarified bacterial supernatant was loaded onto a nickel affinity column (Qiagen). The 6x His-tagged protein of interest was eluted in elution buffer containing 150 mM imidazole and the purity was judged with SDS-PAGE.
Liquid chromatography quadrupole time-of-flight mass spectrometry
A Waters Q-Tof API-US Quad-ToF mass spectrometer was applied to determine the identity of S. suis BirA (BirA_ss) protein1,46. The purified protein band was cut from the gel and digested with Trypsin (G-Biosciences St. Louis, MO), giving a pool of overlapping peptides loaded on a Waters Atlantis C-18 column (0.03 mm particle, 0.075 mm × 150 mm). The acquired data were subjected to the ms/ms analyses.
In vitro Bio-5′-AMP synthesis and thin-layer chromatography
The in vitro assay was established to determine the protein biotinylation activity of BirA ligase as we described previously47 with some modifications. The system of enzymatic reactions included 50 mM Tris-HCl (pH 8), 5 mM tris-(2-carboxyethyl) phosphine, 5 mM MgCl2, 20 μM biotin, 5 μM ATP plus 16.5 nM [α-32P]ATP, 100 mM KCl and 2 μM ligase protein. The reaction mixtures were maintained at 37 °C for 30 min. To figure out the role of the BirA ligase, two tubes of reaction were kept in parallel, only one of which was supplemented with AccB-87 (50 μM). Subsequently, 1 μl of each reaction mixture was spotted on a cellulose thin-layer chromatography plate of microcrystalline cellulose and the plates were developed in isobutyric acid-NH4OH-water (66:1:33 by volume)48. The thin-layer chromatograms were dried overnight, exposed to a phosphor-imaging plate and visualized using a Fujifilm FLA-3000 Phosphor Imager.
MALDI-based determination for the biotinylation activity of BirA
The reaction of BirA-catalyzed biotinylation comprised the following components (100 μM AccB-87, 3 μM ligase, 100 μM biotin, 1 mM ATP, 10 mM MgCl2, 100 mM KCl, 5 mM tris-(2-carboxyethyl) phosphine in 50 mM Tris-HCl (pH 8.0)). The reactions were kept at 37 °C for 16 h then dialyzed against 25 mM ammonium acetate, lyophilized to dryness. Subsequently, the biotinylated form of the AccB87 from the above samples was assayed using the approache of matrix-assisted laser desorption/ionization (MALDI)4.
Electrophoretic mobility shift assays
Gel shift experiments were conducted to test interaction of BirA protein with the bioY promoters of different origins44,46,49. Three sets of DNA probes ((bioY_SS, bioY_LL and bioY_EF) were prepared by annealing two complementary oligonucleotides (Table 2). In the EMSA trials, the digoxigenin-labeled DNA probes (~0.2 pmol) were incubated with the purified BirA_ss protein in the binding buffer (Roche). When necessary, the biotinyl-5′-AMP ligand was supplemented. The DNA/protein mixtures were separated with the native 7% PAGE and transferred onto nylon membrane by the direct contact gel transfer, giving the chemical-luminescence signals captured via the exposure of the membrane to ECL films (Amersham).
β-Galactosidase assays
Overnight cultures of S. suis carrying the lacZ fusion grown in THB medium were subjected to measure direct measurement of β-galactosidase activity44. When necessary, the blood sera were added to augment bacterial growth of the mutant S. suis. The bacterial lysates were prepared using French pressure. The data were recorded in triplicate more than three independent assays.
Bioinformatics analyses
The orthologues of BirA protein were from E. coli, Lactococcus lactis and S. suis 05ZYH33, respectively. The BirA-binding sites were collected from RegPrecise database (http://regprecise.lbl.gov/RegPrecise/regulon.jsp?regulon_id=53141). Using the program of ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html), the multiple alignment of protein (and/or DNA) were carried out and the final outputs were given with the program ESPript 2.2 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). The transcription start site was predicted using the server of Neutral Network Promoter Prediction (http://www.fruitfly.org/seq_tools/promoter.html). Structural modelling was proceeded with CPHmodels 3.2 Server (http://www.cbs.dtu.dk/services/CPHmodels).
Additional Information
How to cite this article: Ye, H. et al. Functional definition of BirA suggests a biotin utilization pathway in the zoonotic pathogen Streptococcus suis. Sci. Rep. 6, 26479; doi: 10.1038/srep26479 (2016).
References
Feng, Y., Kumar, R., Ravcheev, D. A. & Zhang, H. Paracoccus denitrificans possesses two BioR homologs having a role in regulation of biotin metabolism. Microbiology Open 4, 644–59 (2015).
Feng, Y. et al. The atypical occurrence of two Biotin protein ligases in Francisella novicida is due to distinct roles in virulence and biotin metabolism. MBio 6, e00591 (2015).
Zhang, H., Luo, Q., Gao, H. & Feng, Y. A new regulatory mechanism for bacterial lipoic acid synthesis. Microbiology Open 4, 282–300 (2015).
Feng, Y., Zhang, H. & Cronan, J. E. Profligate biotin synthesis in α-Proteobacteria-A develoing or degenerating regulatory system? Mol Microbiol 88, 77–92 (2013).
Hebbeln, P., Rodionov, D. A., Alfandega, A. & Eitinger, T. Biotin uptake in prokaryotes by solute transporters with an optional ATP-binding cassette-containing module. Proc Natl Acad Sci USA 104, 2909–14 (2007).
Beckett, D. Biotin sensing: universal influence of biotin status on transcription. Annu Rev Genet 41, 443–64 (2007).
Lin, S. & Cronan, J. E. Closing in on complete pathways of biotin biosynthesis. Mol Biosyst 7, 1811–21 (2011).
Lin, S., Hanson, R. E. & Cronan, J. E. Biotin synthesis begins by hijacking the fatty acid synthetic pathway. Nat Chem Biol 6, 682–8 (2010).
Beckett, D. Biotin sensing at the molecular level. J Nutr 139, 167–70 (2009).
Tang, Q. et al. Mycobacterium smegmatis BioQ defines a new regulatory network for biotin metabolism. Mol Microbiol 94, 1006–1023 (2014).
Feng, Y., Xu, J., Zhang, H., Chen, Z. & Srinivas, S. Brucella BioR regulator defines a complex regulatory mechanism for bacterial biotin metabolism. J Bacteriol 195, 3451–67 (2013).
Weaver, L. H., Kwon, K., Beckett, D. & Matthews, B. W. Corepressor-induced organization and assembly of the biotin repressor: a model for allosteric activation of a transcriptional regulator. Proc Natl Acad Sci USA 98, 6045–50 (2001).
Wilson, K. P., Shewchuk, L. M., Brennan, R. G., Otsuka, A. J. & Matthews, B. W. Escherichia coli biotin holoenzyme synthetase/bio repressor crystal structure delineates the biotin- and DNA-binding domains. Proc Natl Acad Sci USA 89, 9257–61 (1992).
Barker, D. F. & Campbell, A. M. Genetic and biochemical characterization of the birA gene and its product: evidence for a direct role of biotin holoenzyme synthetase in repression of the biotin operon in Escherichia coli. J Mol Biol 146, 469–92 (1981).
Barker, D. F. & Campbell, A. M. The birA gene of Escherichia coli encodes a biotin holoenzyme synthetase. J Mol Biol 146, 451–67 (1981).
Brune, I., Gotker, S., Schneider, J., Rodionov, D. A. & Tauch, A. Negative transcriptional control of biotin metabolism genes by the TetR-type regulator BioQ in biotin-auxotrophic Corynebacterium glutamicum ATCC 13032. J Biotechnol 159, 225–34 (2012).
Zhang, H. et al. Deciphering a unique biotin scavenging pathway with redundant genes in the probiotic bacterium Lactococcus lactis. Scientific Reports, 6:25680. doi: 10.1038/srep25680 (2016).
Chakravartty, V. & Cronan, J. E. Altered regulation of Escherichia coli biotin biosynthesis in birA superrepressor mutant strains. J Bacteriol 194, 1113–26 (2012).
Chapman-Smith, A. & Cronan, J. E. Jr. The enzymatic biotinylation of proteins: a post-translational modification of exceptional specificity. Trends Biochem Sci 24, 359–63 (1999).
Cronan, J. E. Biotin and Lipoic Acid: Synthesis, Attachment and Regulation. Ecosal Plus 6, doi: 10.1128/ecosalplus.ESP-0001-2012 (2014).
Henke, S. K. & Cronan, J. E. Successful conversion of the Bacillus subtilis BirA Group II biotin protein ligase into a Group I ligase. PLoS One 9, e96757 (2014).
Feng, Y., Zhang, H., Ma, Y. & Gao, G. F. Uncovering newly emerging variants of Streptococcus suis, an important zoonotic agent. Trends Microbiol 18, 124–31 (2010).
Gottschalk, M., Segura, M. & Xu, J. Streptococcus suis infections in humans: the Chinese experience and the situation in North America. Anim Health Res Rev 8, 29–45 (2007).
Tang, J. et al. Streptococcal toxic shock syndrome caused by Streptococcus suis serotype 2. PLoS Med 3, e151 (2006).
Staats, J. J., Feder, I., Okwumabua, O. & Chengappa, M. M. Streptococcus suis: past and present. Vet Res Commun 21, 381–407 (1997).
Feng, Y. et al. Streptococcus suis infection: an emerging/reemerging challenge of bacterial infectious diseases? Virulence 5, 477–97 (2014).
Ye, C. et al. Streptococcus suis sequence type 7 outbreak, Sichuan, China. Emerg Infect Dis 12, 1203–8 (2006).
Yu, H. et al. Human Streptococcus suis outbreak, Sichuan, China. Emerg Infect Dis 12, 914–20 (2006).
Feng, Y. et al. Recurrence of human Streptococcus suis infections in 2007: three cases of meningitis and implications that heterogeneous S. suis 2 circulates in China. Zoonoses Public Health 56, 506–14 (2009).
Chen, C. et al. A glimpse of streptococcal toxic shock syndrome from comparative genomics of S. suis 2 Chinese isolates. PLoS One 2, e315 (2007).
Li, M. et al. SalK/SalR, a two-component signal transduction system, is essential for full virulence of highly invasive Streptococcus suis serotype 2. PLoS One 3, e2080 (2008).
Li, M. et al. GI-type T4SS-mediated horizontal transfer of the 89 K pathogenicity island in epidemic Streptococcus suis serotype 2. Mol Microbiol 79, 1670–1683 (2011).
Zhao, Y. et al. Role of a Type IV-Like Secretion System of Streptococcus suis 2 in the Development of Streptococcal Toxic Shock Syndrome. J Infect Dis 204, 274–81 (2011).
Yao, X. et al. The chromosomal SezAT toxin-antitoxin system promotes the maintenance of the SsPI-1 pathogenicity island in epidemic Streptococcus suis. Mol Microbiol 98, 243–57 (2015).
Shi, X. et al. Loss of 89 K pathogenicity island in epidemic Streptococcus suis, China. Emerging Infectious Diseases, Accepted (2016).
Feng, Y. et al. Attenuation of Streptococcus suis virulence by the alteration of bacterial surface architecture. Sci Rep 2, 710 (2012).
Zhang, H. et al. Two novel regulators of N-acetyl-galactosamine utilization pathway and distinct roles in bacterial infections. Microbiology Open 4, 983–1000 (2015).
Feng, Y., Zhang, H. M. C. & Wang, C. Regulation of Virulence in Streptococcus suis. J Bacteriol Parasitol 3, e108 (2012).
Feng, Y. et al. A Francisella virulence factor catalyses an essential reaction of biotin synthesis. Mol Microbiol 91, 300–14 (2014).
Napier, B. A. et al. Link between intraphagosomal biotin and rapid phagosomal escape in Francisella. Proc Natl Acad Sci USA 109, 18084–9 (2012).
Rodionov, D. A., Mironov, A. A. & Gelfand, M. S. Conservation of the biotin regulon and the BirA regulatory signal in Eubacteria and Archaea. Genome Res 12, 1507–16 (2002).
O’Riordan, M., Moors, M. A. & Portnoy, D. A. Listeria intracellular growth and virulence require host-derived lipoic acid. Science 302, 462–4 (2003).
Spalding, M. D. & Prigge, S. T. Lipoic acid metabolism in microbial pathogens. Microbiol Mol Biol Rev 74, 200–28 (2010).
Feng, Y. & Cronan, J. E. A new member of the Escherichia coli fad regulon: transcriptional regulation of fadM (ybaW). J Bacteriol 191, 6320–8 (2009).
Takamatsu, D., Osaki, M. & Sekizaki, T. Thermosensitive suicide vectors for gene replacement in Streptococcus suis. Plasmid 46, 140–8 (2001).
Feng, Y. & Cronan, J. E. Complex binding of the FabR repressor of bacterial unsaturated fatty acid biosynthesis to its cognate promoters. Mol Microbiol 80, 195–218 (2011).
Chakravartty, V. & Cronan, J. E. The wing of a winged helix-turn-helix transcription factor organizes the active site of BirA, a bifunctional repressor/ligase. J Biol Chem 288, 36029–39 (2013).
Prakash, O. & Eisenberg, M. A. Biotinyl 5′-adenylate: corepressor role in the regulation of the biotin genes of Escherichia coli K-12. Proc Natl Acad Sci USA 76, 5592–5 (1979).
Feng, Y. & Cronan, J. E. Overlapping repressor binding sites result in additive regulation of Escherichia coli FadH by FadR and ArcA. J Bacteriol 192, 4289–99 (2010).
Feng, Y. & Cronan, J. E. The Vibrio cholerae fatty acid regulatory protein, FadR, represses transcription of plsB, the gene encoding the first enzyme of membrane phospholipid biosynthesis. Mol Microbiol 81, 1020–33 (2011).
Guzman, L. M., Belin, D., Carson, M. J. & Beckwith, J. Tight regulation, modulation and high-level expression by vectors containing the arabinose pBAD promoter. J Bacteriol 177, 4121–30 (1995).
Acknowledgements
This work was supported by Zhejiang Provincial Natural Science Foundation for Distinguished Young Scholars (Grant No. LR15H190001), the National Natural Science Foundation of China (Grant No. 31570027) and the start-up package from Zhejiang University (Y.F.). Dr. Feng is a recipient of the “Young 1000 Talents” Award.
Author information
Authors and Affiliations
Contributions
Y.F. and R.W. designed this project; Y.F., H.Y., M.C., H.Z., Z.L. and R.W. performed experiments and analyzed the data; Y.F. and R.W. contributed reagents and tools; Y.F. wrote this manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Ye, H., Cai, M., Zhang, H. et al. Functional definition of BirA suggests a biotin utilization pathway in the zoonotic pathogen Streptococcus suis. Sci Rep 6, 26479 (2016). https://doi.org/10.1038/srep26479
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep26479
This article is cited by
-
The type II histidine triad protein HtpsC facilitates invasion of epithelial cells by highly virulent Streptococcus suis serotype 2
Journal of Microbiology (2021)
-
Biotin-mediated growth and gene expression in Staphylococcus aureus is highly responsive to environmental biotin
Applied Microbiology and Biotechnology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.