Abstract
Coral reef success is largely dependent on the symbiosis between coral hosts and dinoflagellate symbionts belonging to the genus Symbiodinium. Elevated temperatures can result in the expulsion of Symbiodinium or loss of their photosynthetic pigments and is known as coral bleaching. It has been postulated that the expression of light-harvesting protein complexes (LHCs), which bind chlorophylls (chl) and carotenoids, are important in photobleaching. This study explored the effect a sixteen-day thermal stress (increasing daily from 25–34 °C) on integral LHC (chlorophyll a-chlorophyll c2-peridinin protein complex (acpPC)) gene expression in Symbiodinium within the coral Acropora aspera. Thermal stress leads to a decrease in Symbiodinium photosynthetic efficiency by day eight, while symbiont density was significantly lower on day sixteen. Over this time period, the gene expression of five Symbiodinium acpPC genes was quantified. Three acpPC genes exhibited up-regulated expression when corals were exposed to temperatures above 31.5 °C (acpPCSym_1:1, day sixteen; acpPCSym_15, day twelve; and acpPCSym_18, day ten and day sixteen). In contrast, the expression of acpPCSym_5:1 and acpPCSym_10:1 was unchanged throughout the experiment. Interestingly, the three acpPC genes with increased expression cluster together in a phylogenetic analysis of light-harvesting complexes.
Similar content being viewed by others
Introduction
Photosynthetic eukaryotic dinoflagellates belonging to the genus Symbiodinium form symbiotic relationships with a variety of marine taxa. Endosymbiotic associations observed between these photosynthetic dinoflagellates and corals are generally classed as mutualistic, as both the host and symbiont benefit from the relationship1,2. The photosynthetic symbiont acquires nutrients such as inorganic carbon, nitrogen and phosphate from host cells3, in turn symbionts then provide up to 90% of the energy required by corals to grow and reproduce. These relationships between scleractinian corals and Symbiodinium are critical for the proliferation of reefs supporting diverse marine ecosystems. Coral bleaching, the loss of photosynthetic pigments or endosymbionts from host cells occurs under stress conditions such as elevated sea temperatures of only a few degrees above long-term maxima4,5. While ocean temperature fluctuations occur on a daily basis, the mean sea surface temperature is predicted to rise by approximately 1–2 °C over the next century and is expected to lead to more mass bleaching events6,7.
Experimentation on Symbiodinium and the coral holobiont has focussed on many environmental factors implicated in the onset of coral bleaching including elevated seawater temperatures, eutrophication and disease. The effect of high sea-surface temperatures have been a key focus due to mass coral bleaching events (~42% GBR reefs bleached in 1998 and ~54% reefs bleached in 20028), attributed to global climate change7 with the 1998 bleaching event coinciding with an El Niño Southern Oscillation event4,9.
Differential thermal stress sensitivity is observed across the diverse Symbiodinium species complex, with both heat tolerant and heat sensitive species observed within the same clade10. Differences in photoinhibition sensitivity have either been acquired independently by thermally tolerant types or have been acquired in the common ancestor of all Symbiodinium types and since lost in thermally sensitive species10. Elucidation of sites of thermal sensitivity within Symbiodinium has focussed on potential points where damage results in a decrease in photosynthetic efficiency11,12. These potential points include damage to the D1 protein of photosystem II (PSII)12, inhibition of the de novo synthesis of the D1 protein13, the enzyme Rubisco14, thylakoid membrane integrity10, the carbon concentrating mechanism15 and LHCs16. However, none of these sites have conclusively been demonstrated as the initial site of thermal damage.
LHCs, also called antenna proteins, are found in photoautotrophic organisms and are an array of protein, chl and accessory pigment molecules with roles in light-harvesting and photoprotection17,18. In Symbiodinium the light-harvesting system can be divided into two associated complexes, the highly conserved core LHCs and variable periphery LHCs19. Studies of green plant LHCs have elucidated structural information, which has further improved the understanding of light capture and transfer and the arrangement of peripheral LHCs in photosynthetic eukaryotes18,20. Peripheral LHCs may be categorized within the large gene super-family based on associated pigments into three related groups of pigment binding proteins21,22. The first group binds chl a and b, the second binds chl a and c and the third group binds chl a and phycobilins. The chl a/c lineage LHCs are additionally divided into the fucoxanthin-chl a/c and peridinin–chl a/c (PCP) complexes, which are found in dinoflagellates21,22,23.
In Symbiodinium two types of peripheral LHCs are found, PCP and acpPC19. Dinoflagellate PCPs share no sequence similarity with other known LHCs and are water soluble complexes found on the luminal periphery of thylakoid membranes24. In contrast, dinoflagellate acpPCs are integral thylakoid membrane complexes that share sequence similarity with the chl a/c subfamily of LHCs24. Further, characterisation of Symbiodinium C3 acpPCs and Symbiodinium A1.1 LHCs cluster sequences with three clades within the chl a/c binding LHC family, indicating high diversity of these proteins within species21. Dinoflagellate and Symbiodinium PCPs are well studied due to their unique features25,26,27,28. In Symbiodinium the LHCs have been shown to decrease energy transfer and dissociate from the photosystem reaction centres following photoinhibition in order to protect cells during stress events12,29,30,31,32. Decreasing the number of peripheral LHCs available to absorb and transfer energy is a proposed photoprotection mechanism, as this reduces the amount of light reaching the reaction centres and limits the risk of possible photodamage to the D1 reaction centre proteins30. Further, kinetic studies have revealed that LHC proteins disassociate and reattach to thylakoid membranes under increased light levels to ameliorate stress32. However, few studies have examined the effect of thermal stress on Symbiodinium acpPC expression16,32.
Targeted studies of Symbiodinium transcript levels have shown that changes in gene expression occur on a relatively small scale. Quantitative-PCR has been used to determine changes in a variety of genes of interest related to stress response33,34,35,36. The validation of housekeeping genes for use in Symbiodinium has allowed for a reference to be established in order to determine differential gene expression under various conditions36,37. Although significant changes have been observed in Symbiodinium physiology and large fold changes in host gene expression have been recorded in cells exposed to stress, changes in Symbiodinium gene expression occur at a far smaller scale (±<5-fold)33,34,35.
This study focussed on the response of acpPC genes in Symbiodinium cells during a prolonged thermal stress event (sixteen days) in Acropora aspera. We find that the response of five acpPC genes in Symbiodinium under stress varies across a LHC phylogeny. This is the first study to investigate the effect of thermal stress on gene expression patterns in the LHC superfamily within a coral host.
Results
Symbiodinium density
Over a period of sixteen days, branches of A. aspera were exposed to temperature increasing from ambient levels (~25 °C) to a bleaching temperature of ~34 °C (Fig. 1). As has previously been found, this temperature increase led to a significant decrease in Symbiodinium cell densities (p < 0.001) over the course of the experiment, with average densities of 6.0 × 105 cells cm−2 in the treatment corals on day sixteen, compared to 1.6 × 106 cells cm−2 in the control corals on the same day (Fig. 2a).
(a) Symbiodinium cell density per cm2 in A. aspera (b) Symbiodinium chl a pigment concentrations in A. aspera (c) Symbiodinium chl c pigment concentrations in A. aspera. (d) Ratio of chl c to chl a per Symbiodinium cell in A. aspera nubbins. A. aspera nubbins subjected to control conditions (solid line) and heated treatment (dashed line). Error bars represent ± s.e.m., n = 9–12, some error bars obscured by data point markers. The statistical difference (post hoc sequential Bonferroni analysis) between treatment and control is indicated as *p < 0.05 or **p < 0.01.
Chl pigment content
Chl a content per Symbiodinium cell increased over the experimental period in the heated treatments from day eight of the experimental period (Fig. 2b). Analysis of the chl a content found that there were significant differences between day (p < 0.001, df = 5) and treatment (p < 0.05, df = 1) but not in the interaction treatment × day (p > 0.05, df = 5) (Fig. 2b). Similarly chl c content per Symbiodinium cell increased from day ten onwards in the experiment period (Fig. 2c). Analysis of the chl c content found that there were significant differences between day (p < 0.01, df = 5) and treatment (p < 0.01, df = 1) but not in the interaction treatment × day (p > 0.05, df = 5) (Fig. 2c). The ratio of chl c to chl a was unchanged between control and treatment conditions throughout the experiment (Fig. 2d).
Chl fluorescence and photosynthetic efficiency
Maximum quantum yield of photosynthesis (Fv/Fm) was measured following sunset during the experiment. For corals maintained at control temperatures, Fv/Fm was between 0.602 and 0.692 (average 0.657) (Fig. 3a). Analysis of Fv/Fm found that there was a significant effect between day and treatment (p < 0.001, df = 5) and differences between day (p < 0.001, df = 5) and treatment (p < 0.001, df = 1) (Fig. 3a). A sequential Bonferroni post hoc analysis found that Fv/Fm decreased in the heated treatment on days eight, ten, twelve and sixteen of the experiment compared to controls from the same days (p < 0.01) (Fig. 3a).
(a) Symbiodinium Fv/Fm within A. aspera during the experiment. (b) Symbiodinium NPQ within A. aspera at the last point of the induction phase during the Imaging-PAM analysis. A. aspera nubbins exposed to control conditions (solid line) and heated treatment (dashed line). Values represent average obtained from twelve biological replicates across four replicate tanks. Error bars represent ± s.e.m., n = 12, some error bars obscured by data point markers. The statistical difference (post hoc sequential Bonferroni analysis) between treatment and control is indicated as *p < 0.05 or **p < 0.01.
Non-photochemical quenching (NPQ) was also measured over the course of the experiment. As with Fv/Fm, there were significant interaction effects between treatment × day (p < 0.001, df = 5) and between day (p < 0.001, df = 5) and treatment (p < 0.05, df = 1) (Fig. 3b). Post hoc analysis demonstrated NPQ was significantly increased by heating on days ten, twelve and fourteen before declining to zero on the final day of the experiment (Fig. 3b).
Gene expression under thermal stress
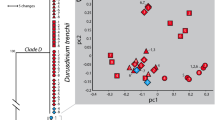
The expression of five acpPC genes (acpPCSym_1:1, acpPCSym_5:1, acpPCSym_10:1, acpPCSym_15 and acpPCSym_18) from three distinct LHC clades (Fig. 4) was determined. Over the course of the experiment three acpPC genes were found to have significant increases in gene expression, acpPCSym_1:1 on day sixteen (1.74-fold, p = 0.001), acpPCSym_15 on day twelve (1.33-fold, p = 0.014) and acpPCSym_18 on days ten (2.44-fold, p = 0.012) and sixteen (2.08-fold, p = 0.020) (Fig. 5a,d,e). These three genes belong to two distinct LHC clades (Fig. 4), both acpPCSym_15 and acpPCSym_18 belong to Clade 1, while acpPCSym_1:1 belongs to Clade 2. The largest fold change seen in these genes was a 2.44-fold increase in acpPCSym_18 compared to control. For the remaining two acpPCs (acpPCSym_5:1 and acpPCSym_10:1) which both belong to Clade 3b (Fig. 4) no significant changes in gene expression were detected (Fig. 5b,c). In addition to the five acpPCs the expression of the psbA gene was also determined. During the course of the experiment no significant differences in psbA expression between control and treatment conditions were found (Fig. 5f).
Phylogenetic analysis of with LHCs from chl a/b and chl a/c containing organisms.
Chl a/b binding protein complexes cluster together while the chl a/c binding protein complexes form a second cluster. Symbiodinium sp. C3 acpPC sequences and Symbiodinium type A1.1 LHCs are found throughout the four clades (Clade 1-3b) of the chl a/c binding protein complexes. Reproduced from Boldt et al.21.
Relative expression of Symbiodinium genes of interest when exposed to prolonged thermal stress.
Values expressed as relative expression of treatment (dashed line) to control (solid line) for each time point: (a) acpPCSym_1:1, (b) acpPCSym_5:1, (c) acpPCSym_10:1, (d) acpPCSym_15, (e) acpPCSym_18 and (f) psbA. Error bars represent ± s. e. m., n = 4–10, some error bars obscured by data point markers. The statistical differences (post hoc sequential Bonferroni analysis) between treatment transcript abundance and control is indicated as *p < 0.05 or **p < 0.01.
Discussion
This study investigated the effect of increased temperatures on the expression of five acpPC genes in Symbiodinium under prolonged thermal stress of A. aspera and is the first to examine the expression of acpPC genes from different LHC Clades. The sixteen-day thermal regime was selected to enable sampling at temperatures leading up to and inclusive of, a bleaching event and is the first experiment to investigate differential expression of integral antenna proteins in Symbiodinium within a coral host under thermal stress. Quantitative PCR was used to quantify the expression of five acpPC genes that are dispersed though out three clades of the chl a/c lineage of the LHC phylogeny (Fig. 4).
Over the course of the experiment temperature significantly effected Symbiodinium density and physiology. Symbiont cells decreased to approximately half the density in thermally stressed corals compared to control corals (Fig. 2a) as has been found in variety of other studies4,35,38. In addition chl a and chl c levels were elevated over the course of the experiment (Fig. 2b,c) in a manner seen before in this species (Gierz and Leggat, unpublished data)35. However, a statistical difference was only observed on day sixteen in chl c (Fig. 2c), this is consistent with other studies where an increase in chl pigments were observed35. In corals, heat-related increases in chl a have previously been recorded at low symbiont densities39,40, though in other experiments Symbiodinium pigmentation may be unchanged or decreased41,42. Increases in chl pigments have been attributed to repackaging of chls in the chloroplast membrane, with evidence that specific pigment-protein complexes may absorb more light at specific wavelengths43. In phytoplankton, chl a-specific absorption of different pigment-protein complexes from the same organism can be highly variable43. Therefore, it is possible that the increases in Symbiodinium pigments observed in heated corals may be attributed to alterations in the type of pigment-protein complexes expressed under thermal stress.
Imaging-pulse amplitude modulated (PAM) fluorometry analysis demonstrated that Symbiodinium cells exposed to elevated temperatures exhibited decreased photosynthetic efficiency (Fig. 3a), this is consistent with previous studies demonstrating the response of cells to elevated temperatures. Decreases in dark-adapted yield occurred throughout the experiment despite small changes in symbiont density, Fv/Fm levels of ~0.00 were recorded on day sixteen despite cell density being approximately five hundred thousand per cm2, indicating cells were incapable of photosynthesis at the end of the stress period. Increased NPQ response in cells at days eight, ten and twelve (Fig. 3b) illustrates that the cells were dissipating excess light energy. However, this NPQ response was not present on day sixteen of thermal stress indicating that the symbionts had passed a threshold where photosynthetic processes were no longer functioning. Together, the photosynthetic efficiency results, Symbiodinium densities and changes to pigment levels, demonstrate that in this experiment Symbiodinium were subjected to the full range of temperatures that are seen in a bleaching event, with responses from initial thermal stress through to Symbiodinium expulsion. As such it is reasonable to conclude that acpPC expression patterns are representative of what would be seen in a natural bleaching event.
Expression of acpPC genes was found to vary in Symbiodinium cells throughout the experiment. Functionally little is known about the diversity of LHCs, for example whether complexes only associate with specific photosystems, are some more efficient at light capture or energy transfer, are others favoured for photoprotection or do some display increased stability under high temperatures. Characterisation of Symbiodinium acpPCs has shown that there is large diversity within the gene super-family21. Analysis of the Symbiodinium genome has provided more of an insight into the diversification of the LHC family23, reinforcing theories on gene duplication and deletion events leading to the current structure of the Symbiodinium genome. The complexity observed in the integral LHC family has been attributed to multiple rounds of intra- and inter-genic gene duplication events20,23. A large gene super-family encodes integral LHCs and a significant level of sequence similarity has been detected between the protein complexes21,23. Phylogenetic analysis of LHCs and LHC-like protein super-families indicate that the ancestor is most likely a central group of two-helix stress-enhanced proteins that had previously evolved from a gene-duplication event of the high-light induced proteins of cyanobacteria44,45. However, based upon sequence divergence, it is reasonable to assume that different clades of acpPC may have different functions. As such, the differences in expression between those acpPC (Fig. 5a–e) from Clade 1 and 2 versus Clade 3b (Fig. 4), may be indicative of functional roles, with Clade 1 and 2 possibly being involved in stress response while those of Clade 3b are constitutively expressed under the conditions used here.
Some ways in which acpPC may functionally vary is in the binding of varied pigment ratios, specificity for association to photosystems and response to stress events. For example it has been found that a variety of acpPC transcripts are missing key chl and pigment binging residues21. In addition it is not clear to which photosystems different acpPCs bind. In green plants, ten highly conserved genes encoding chl a/b binding proteins have been identified, associated with photosystem I (PSI) are four pigment - protein complexes (encoded by genes Lhca1, Lhca2, Lhca3 and Lhca4) and associated with PSII are six pigment – protein complexes (encoded by genes Lhcb1, Lhcb2, Lhcb3, Lhcb4, Lhcb5, Lhcb6)20. This can be contrasted to Symbiodinium where there is high sequence diversity coupled with high copy number and as yet it is not clear which proteins bind to PSI or PSII21,23. It has been suggested that this sequence diversity allows for functional diversity such as, stress response21, attachment/dissociation32 and enhanced photoprotection46. As such, it will only be with the linkage of more transcriptome and genome studies and the analysis of chl and accessory pigments binding residues, linked to functional studies, that we will be able to elucidate the reason for the expansion of this gene family in dinoflagellates.
Core photosystem genes, psaA and psbA have previously been investigated in Symbiodinium under thermal stress. Decreases of psaA and psbA are hypothesised to significantly impair the mechanisms associated with coping with thermal stress47. In this study, the expression of the psbA gene, which encodes the core PSII D1 protein, was also quantified (Fig. 5f). Over the course of the experiment psbA expression increased on days eight and ten (Fig. 5f) although expression in treatment samples was not statistically significant. However, on day sixteen, psbA expression decreased (Fig. 5f), potentially to reduce light absorption to limit the amount of energy captured under stress conditions as a photoprotective mechanism.
As in previous studies, investigating transcript abundance in Symbiodinium very small changes in gene expression were observed in this study. In the five acpPC genes quantified, the largest observed change was a 2.44 fold increase (acpPCSym_18) on day ten of the thermal stress experiment. In Symbiodinium in hospite, these small changes in transcripts have been observed previously33,34,35,36 and it is postulated that regulation is most likely post-translational and not at the transcriptional level33,47,48.
This study exploited a bleaching experiment to investigate the effect of thermal stress on photosynthetic genes. Quantitative PCR was used to determine the expression of five integral LHC genes. Three LHC genes (acpPCSym_1:1, acpPCSym_15 and acpPCSym_18) were found to have increased expression over the duration of the experiment and interestingly, grouped in Clade 1 and Clade 2 of the LHC phylogeny. Additionally two LHC genes (acpPCSym_5:1 and acpPCSym_10:1) grouped with Clade 3b did not exhibit differences in expression. Though transcriptional changes were detected, expression changes observed were less than 2.5 fold throughout the experiment. This is consistent with previous studies where small-scale changes in gene expression were also observed. Given that we currently do not know how the diverse range of LHCs are associated with the photosystems, both PSII and PSI, their specific functional roles, (e.g., light harvesting efficiency versus photoprotection), or the importance of Symbiodinium photosynthesis to the survival of corals, it is imperative that future research focuses on the specific roles of LHCs.
Methods
Thermal stress experimental design
Coral fragments (n = 312) were collected (under Great Barrier Reef Marine Park Authority permit G13/36402.1) from four different colonies of Acropora aspera (tan morph, approximately 78 from each colony) on the reef flat of Heron Island at low tide in May 2013. Heron Island colonies of A. aspera have been demonstrated to associate only with Symbiodinium clade C335,49,50. Coral fragments were taken to the Heron Island Research Station and placed in a holding tub supplied with filtered water from the reef flat. Fragments were randomly selected and placed upright in racks. Nubbin racks were then transferred to eight 65 L replicate tanks and allowed to acclimate for five-days, at the end of this period tissue regrowth was observed on the cut section of the nubbins. Each of the replicate tanks were supplied with a flow of sand-filtered water pumped from the reef flat into two sump tanks, each sump tank then supplied water to four experimental tanks forming a semi-closed system.
The eight tanks were assigned to one of two treatments, control conditions (ambient temperature) and thermal-stress treatment conditions. The control conditions remained at the ambient seawater temperature (~24 °C) for the duration of the experiment (Fig. 1). Fluctuations observed in control conditions are natural daily temperature fluctuations (Fig. 1). The thermal-stress treatment temperature was increased by 0.7 °C for eleven days (25–32.3 °C) and then held at 33 °C for three days and then at 34 °C for a further three days (Fig. 1) to simulate a bleaching event. To achieve the required temperatures for the thermal treatment a three hundred Watt Eheim Jager (Eheim, Deisizou, Germany) heater was used in the heated sump as well as four 25 W Aqua One glass heaters. To ensure diurnal temperature variation in thermal treatment tanks the Eheim heater was turned off overnight to reflect natural fluctuations. Temperatures in each tank were recorded every 10 minutes with HOBO® temperature/alarm pendant data loggers (Onset, Massachusetts, USA). Light levels were monitored over the course of the experiment, every 10 minutes with Odyssey Photosynthetic Active Radiation (PAR) recorders (Dataflow Systems Limited, New Zealand).
Imaging-PAM fluorometry
Imaging-PAM fluorometry (MAXI Imaging-PAM, Waltz, Effletrich, Germany) was used to measure photosynthetic efficiency of Symbiodinium within A. aspera. Imaging-PAM analysis was performed on the first day and every day from the third day following sunset. Nubbins were dark-adapted for twenty minutes prior to imaging-PAM analysis. Three replicate nubbins from each of the eight replicate tanks were designated for Imaging-PAM analysis and used throughout the experiment. Corals were measured in the same order at each imaging-PAM analysis. Dark-adapted yield and maximal fluorescence were determined using a weak pulse of light, followed by a saturating pulse of 2,700 μmol quanta m−2 s−1 of photosynthetically active radiation (PAR) for 800 ms. Induction recovery curves were used to examine the photosynthetic efficiency and ability of symbionts to recover from light stress throughout the experiment. The induction recovery analysis utilised fifteen pulses of saturating light (2,700 μmol quanta m−2 s−1) with a constant actinic light (111 μmol quanta m−2 s−1) over a 5-minute period, followed by a dark recovery period of 14 min. Data from the induction recovery curve was used to determine photo-kinetic parameters, such as Fv/Fm and NPQ. Following analysis, nubbins were returned to experimental tanks.
Pigment quantification and Symbiodinium cell count
At 2 pm on days zero, eight, ten, twelve and sixteen, three replicate nubbins were taken from each replicate tank (n = 9–12) and were stripped of tissue using a Waterpik™ dental irrigator using seawater. On day zero the blastate was centrifuged at 3,076 g for 5 min immediately following tissue removal. On days eight, ten, twelve and sixteen the blastate was homogenised with an immersion blender for 5 s and centrifuged at 3,076 g for 3 min to pellet algal cells. Total pelleted cells were resuspended in 50 mL of seawater. Aliquots (1 mL) were taken for cell density approximation. The remaining cells were centrifuged at 3,076 g for 3 min to pellet cells and stored at −80 °C for chl a and c quantification. Chl was extracted in 90% acetone for 20 h in the dark at 4 °C and quantified using the equations of Jeffrey and Humphrey (1975)51. Cell number was determined using a Neubauer haemocytometer, with replicate cell counts performed (n = 5). Surface area of waterpiked nubbins was determined using the wax dipping method52.
Gene expression analysis
At 2 pm on days five, eight, ten, twelve and sixteen, three replicate nubbins were taken from each replicate tank (total twelve nubbins per treatment) and snap-frozen in liquid nitrogen and stored at −80 °C for later mRNA isolation. Coral branches that had been snap-frozen in liquid nitrogen were crushed with a hydraulic press before transfer to a mortar chilled with liquid nitrogen and ground finely with a chilled pestle. The powder was then divided into cryotubes and stored at −80 °C. Messenger RNA was isolated from cells using the Dynabeads® mRNA DIRECT™ kit as per the protocol outlined in Leggat et al.33. Extracted mRNA was quantified spectrophotometrically using the Nucleic acid: RNA-40 setting on a NanoDrop-1000 (NanoDrop Technologies, Wilmington USA). DNase treatment of 0.1 μg of mRNA per reaction was carried out using RQ1 RNase-Free DNase (Promega) in a total volume of 9 μl. cDNA was reverse transcribed from DNase treated mRNA isolated from Symbiodinium cells for quantitative real-time-PCR (qRT-PCR). Reverse transcription was performed using the SuperScript™ III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen). The 2x RT Reaction Mix contains the following components oligo (dT)20 (2.5 μM), random hexamers (2.5 ng.μl−1), 10 mM MgCl2 and dNTPs. Template cDNA dilution series were prepared to optimize quantification accuracy. For analysis, cDNA was diluted 1:40 prior to use as a template in the qRT-PCR analysis. Quantitative RT-PCR was performed using a Rotor-Gene™ 6000 (Corbett Life Science, Australia). The qRT-PCR was performed in a final volume of 15 μl, containing 7.5 μl of GoTaq® qPCR Master Mix (Promega, USA), 4 μl of diluted template and gene specific primers (266 nM). qRT-PCR conditions were as follows: 95 °C for 2 min; 40 cycles of 95 °C for 15 s and 61 °C for 60 s. All qRT-PCR were followed by a melt curve analysis from 55 to 95 °C to ensure single product amplification (Supplementary Fig. S1). Each Rotor-Disc™-100 included three technical replicates for ten biological samples with three genes. Non-template controls for each primer set were performed in triplicate in each run.
Housekeeping genes used in our analysis were selected from previously established reference genes, which included Proliferating Cell Nuclear Antigen (PCNA)36, cyclophin (Cyc)37, glyceraldehyde 3-phosphate dehydrogenase (GAPDH)37, S4 ribosomal protein (Rp-S4)37 and S-adenosyl-L-methionine synthetase (SAM)37. Five acpPC (acpPCSym_1:1, acpPCSym_5:1, acpPCSym_10:1, acpPCSym_15 and acpPCSym_18) Symbiodinium C3 primers were designed against acpPC sequences from an EST library53 obtained from the NCBI Genbank database (www.ncbi.nlm.nih.gov). Primers were designed for acpPC genes in Symbiodinium using the software DNASTAR Primer Select (Lasergene 11) (Table 1). The psbA primers were previously established for use in Symbiodinium47. Relative expression analysis was performed using qBASE plus 2.5 software (Biogazelle; http://www.biogazelle.com/products/qbasePLUS)54,55. Validation of housekeeping genes (expression stability and the optimum number of genes) in this experiment was performed using geNorm (qBASE plus)54,55.
Data analyses
Statistics software package (SPSS Statistics v 22.0, IBM, USA) was used for all statistical analyses. A generalized linear model with ‘day’ and ‘treatment’ as main effects and ‘day × treatment’ as an interaction was used for pairwise comparisons of cell density, chl a and c, imaging-PAM and gene expression data. The sequential Bonferroni post hoc test was used to adjust for the false discovery rate (or type I error).
Additional Information
How to cite this article: Gierz, S. L. et al. Integral Light-Harvesting Complex Expression In Symbiodinium Within The Coral Acropora aspera Under Thermal Stress. Sci. Rep. 6, 25081; doi: 10.1038/srep25081 (2016).
References
Coffroth, M. A. & Santos, S. R. Genetic diversity of symbiotic dinoflagellates in the genus Symbiodinium. Protist 156, 19–34, 10.1016/j.protis.2005.02.004 (2005).
Stat, M., Morris, E. & Gates, R. D. Functional diversity in coral–dinoflagellate symbiosis. P Natl Acad Sci USA 105, 9256–9261, 10.1073/pnas.0801328105 (2008).
Yellowlees, D., Rees, T. A. V. & Leggat, W. Metabolic interactions between algal symbionts and invertebrate hosts. Plant Cell Environ. 31, 679–694, 10.1111/j.1365-3040.2008.01802.x (2008).
Fujise, L. et al. Moderate thermal stress causes active and immediate expulsion of photosynthetically damaged zooxanthellae (Symbiodinium) from corals. PLoS ONE 9, e114321, 10.1371/journal.pone.0114321 (2014).
Goreau, T. J. & Hayes, R. L. Coral bleaching and ocean “hot spots”. Ambio 23, 176–180 (1994).
IPCC. Climate Change 2007 - The physical science basis. Contribution of working group I to the fourth assessment report of the IPCC. (Cambridge University Press, 2007).
Hoegh-Guldberg, O. Climate change, coral bleaching and the future of the world’s coral reefs. Mar Freshwater Res 50, 839–866, 10.1071/MF99078 (1999).
Berkelmans, R., De’ath, G., Kininmonth, S. & Skirving, W. J. A comparison of the 1998 and 2002 coral bleaching events on the Great Barrier Reef: spatial correlation, patterns and predictions. Coral Reefs 23, 74–83, 10.1007/s00338-003-0353-y (2004).
Bruno, J., Siddon, C., Witman, J., Colin, P. & Toscano, M. El Niño related coral bleaching in Palau, Western Caroline Islands. Coral Reefs 20, 127–136, 10.1007/s003380100151 (2001).
Tchernov, D. et al. Membrane lipids of symbiotic algae are diagnostic of sensitivity to thermal bleaching in corals. P Natl Acad Sci USA 101, 13531–13535, 10.1073/pnas.0402907101 (2004).
Iglesias-Prieto, R., Matta, J. L., Robins, W. A. & Trench, R. K. Photosynthetic response to elevated temperature in the symbiotic dinoflagellate Symbiodinium microadriaticum in culture. P Natl Acad Sci USA 89, 10302–10305, 10.1073/pnas.89.21.10302 (1992).
Warner, M. E., Fitt, W. K. & Schmidt, G. W. The effects of elevated temperature on the photosynthetic efficiency of zooxanthellae in hospite from four different species of reef coral: a novel approach. Plant Cell Environ 19, 291–299, 10.1111/j.1365-3040.1996.tb00251.x (1996).
Warner, M. E., Fitt, W. K. & Schmidt, G. W. Damage to photosystem II in symbiotic dinoflagellates: A determinant of coral bleaching. P Natl Acad Sci USA 96, 8007–8012, 10.1073/pnas.96.14.8007 (1999).
Jones, R. J., Hoegh-Guldberg, O., Larkum, A. W. D. & Schreiber, U. Temperature-induced bleaching of corals begins with impairment of the CO2 fixation mechanism in zooxanthellae. Plant Cell Environ 21, 1219–1230, 10.1046/j.1365-3040.1998.00345.x (1998).
Leggat, W., Whitney, S. & Yellowlees, D. Is coral bleaching due to the instability of the zooxanthellae dark reactions? Symbiosis 37, 137–153 (2004).
Takahashi, S., Whitney, S., Itoh, S., Maruyama, T. & Badger, M. Heat stress causes inhibition of the de novo synthesis of antenna proteins and photobleaching in cultured Symbiodinium. P Natl Acad Sci USA 105, 4203–4208, 10.1073/pnas.0708554105 (2008).
Horton, P., Ruban, A. V. & Walters, R. G. Regulation of light harvesting in green plants Annu Rev Plant Phys 47, 655–684, 10.1146/annurev.arplant.47.1.655 (1996).
Kuhlbrandt, W., Wang, D. N. & Fujiyoshi, Y. Atomic model of plant light-harvesting complex by electron crystallography. Nature 367, 614–621, 10.1038/367614a0 (1994).
Iglesias-Prieto, R., Govind, N. S. & Trench, R. K. Isolation and characterization of three membrane-bound chlorophyll-protein complexes from four dinoflagellate species. Philos T Roy Soc B 340, 381–392, 10.1098/rstb.1993.0080 (1993).
Green, B. R. & Pichersky, E. Hypothesis for the evolution of three-helix Chl a/b and Chl a/c light-harvesting antenna proteins from two-helix and four-helix ancestors. Photosynth Res 39, 149–162, 10.1007/BF00029382 (1994).
Boldt, L., Yellowlees, D. & Leggat, W. Hyperdiversity of genes encoding integral light-harvesting proteins in the dinoflagellate Symbiodinium. PLoS ONE 7, e47456, 10.1371/journal.pone.0047456 (2012).
Green, B. R. & Durnford, D. G. The chlorophyll-carotenoid proteins of oxygenic photosynthesis. Annu Rev Plant Phys 47, 685–714 (1996).
Maruyama, S., Shoguchi, E., Satoh, N. & Minagawa, J. Diversification of the light-harvesting complex gene family via intra- and intergenic duplications in the coral symbiotic alga Symbiodinium. PLos One 10, e0119406, 10.1371/journal.pone.0119406 (2015).
Hiller, R. G., Wrench, P. M., Gooley, A. P., Shoebridge, G. & Breton, J. The major intrinsic light-harvesting protein of Amphidinium: characterization and relation to other light-harvesting proteins. Photochem Photobiol 57, 125–131, 10.1111/j.1751-1097.1993.tb02267.x (1993).
Hiller, R. G., Crossley, L. G., Wrench, P. M., Santucci, N. & Hofmann, E. The 15-kDa forms of the apo-peridinin-chlorophyll a protein (PCP) in dinoflagellates show high identity with the apo-32kDa PCP forms and have similar N-terminal leaders and gene arrangements. Mol Genet Genomics 266, 254–259, 10.1007/s004380100551 (2001).
Hofmann, E. et al. Structural basis of light harvesting by carotenoids: peridinin-chlorophyll-protein from Amphidinium carterae. Science 272, 1788–1791, 10.1126/science.272.5269.1788 (1996).
Jiang, J. et al. Characterization of the peridinin–chlorophyll a-protein complex in the dinoflagellate Symbiodinium. BBA - Bioenergetics 1817, 983–989, 10.1016/j.bbabio.2012.03.027 (2012).
Norris, B. J. & Miller, D. J. Nucleotide sequence of a cDNA clone encoding the precursor of the peridinin-chlorophyll a-binding protein from the dinoflagellate Symbiodinium sp. Plant Mol Biol 24, 673–677, 10.1007/BF00023563 (1994).
Allakhverdiev, S. et al. Heat stress: an overview of molecular responses in photosynthesis. Photosynth Res 98, 541–550, 10.1007/s11120-008-9331-0 (2008).
Hill, R. & Ralph, P. J. Photosystem II heterogeneity of in hospite zooxanthellae in scleractinian corals exposed to bleaching conditions. Photochem Photobiol 82, 1577–1585, 10.1111/j.1751-1097.2006.tb09814.x (2006).
Iglesias-Prieto, R. & Trench, R. K. Acclimation and adaptation to irradiance in symbiotic dinoflagellates. II. Response of chlorophyll–protein complexes to different photon-flux densities. Mar Biol 130, 23–33, 10.1007/s002270050221 (1997).
Hill, R. et al. Light-induced dissociation of antenna complexes in the symbionts of scleractinian corals correlates with sensitivity to coral bleaching. Coral Reefs 31, 963–975, 10.1007/s00338-012-0914-z (2012).
Leggat, W. et al. Differential responses of the coral host and their algal symbiont to thermal stress. PLos One 6, e26687, 10.1371/journal.pone.0026687 (2011).
Rosic, N. N., Pernice, M., Dove, S., Dunn, S. & Hoegh-Guldberg, O. Gene expression profiles of cytosolic heat shock proteins Hsp70 and Hsp90 from symbiotic dinoflagellates in response to thermal stress: possible implications for coral bleaching. Cell Stress Chaperon 16, 69–80, 10.1007/s12192-010-0222-x (2011).
Ogawa, D., Bobeszko, T., Ainsworth, T. & Leggat, W. The combined effects of temperature and CO2 lead to altered gene expression in Acropora aspera. Coral Reefs 32, 895–907, 10.1007/s00338-013-1046-9 (2013).
Boldt, L., Yellowlees, D. & Leggat, W. Measuring Symbiodinium sp. gene expression patterns with quantitative real-time PCR. Proceedings of the 11th ICRS. 118–122 (2009).
Rosic, N. N., Pernice, M., Rodriguez-Lanetty, M. & Hoegh-Guldberg, O. Validation of housekeeping genes for gene expression studies in Symbiodinium exposed to thermal and light stress. Mar Biotechnol 13, 355–365, 10.1007/s10126-010-9308-9 (2011).
Middlebrook, R., Hoegh-Guldberg, O. & Leggat, W. The effect of thermal history on the susceptibility of reef-building corals to thermal stress. J Exp Biol 211, 1050–1056, 10.1242/jeb.013284 (2008).
Fitt, W. K., Spero, H. J., Halas, J., White, M. W. & Porter, J. W. Recovery of the coral Montastrea annularis in the Florida Keys after the 1987 Caribbean “bleaching event”. Coral Reefs 12, 57–64, 10.1007/bf00302102 (1993).
Jones, R. J. Changes in zooxanthellar densities and chlorophyll concentrations in corals during and after a bleaching event. Mar Ecol-Prog Ser 158, 51–59, 10.3354/meps158051 (1997).
Strychar, K. B. & Sammarco, P. W. Effects of heat stress on phytopigments of zooxanthellae (Symbiodinium spp.) symbiotic with the corals Acropora hyacinthus, Porites solida and Favites complanata. Int J Biol 4, 3, 10.5539/ijb.v4n1p3 (2012).
Dove, S. et al. Response of holosymbiont pigments from the scleractinian coral Montipora monasteriata to short-term heat stress. Limnol Oceanogr 51, 1149–1158, 10.4319/lo.2006.51.2.1149 (2006).
Bissett, W. P., Patch, J. S., Carder, K. L. & Lee, Z. P. Pigment packaging and Chl a-specific absorption in high-light oceanic waters. Limnol Oceanogr 42, 961–968, 10.4319/lo.1997.42.5.0961 (1997).
Engelken, J., Brinkmann, H. & Adamska, I. Taxonomic distribution and origins of the extended LHC (light-harvesting complex) antenna protein superfamily. BMC Evol Biol 10, 233, 10.1186/1471-2148-10-233 (2010).
Green, B. R. In Light-harvesting antennas in photosynthesis Advances in photosynthesis and respiration (eds Green, B. R. & Parson, W. W. ) Ch. 4, 129–168 (Springer Netherlands, 2003).
Reynolds, J. M., Bruns, B. U., Fitt, W. K. & Schmidt, G. W. Enhanced photoprotection pathways in symbiotic dinoflagellates of shallow-water corals and other cnidarians. P Natl Acad Sci USA 105, 13674–13678, 10.1073/pnas.0805187105 (2008).
McGinley, M. P. et al. Transcriptional response of two core photosystem genes in Symbiodinium spp. exposed to thermal stress. PLos ONE 7, e50439, 10.1371/journal.pone.0050439 (2012).
Bachvaroff, T. R. & Place, A. R. From stop to start: Tandem gene arrangement, copy number and trans-splicing sites in the dinoflagellate Amphidinium carterae. PLos One 3, 10.1371/journal.pone.0002929 (2008).
LaJeunesse, T. C. et al. Low symbiont diversity in southern Great Barrier Reef corals, relative to those of the Caribbean. Limnol Oceanogr 48, 2046–2054, 10.4319/lo.2003.48.5.2046 (2003).
Dove, S. Scleractinian corals with photoprotective host pigments are hypersensitive to thermal bleaching. Mar Ecol-Prog Ser 272, 99–116, 10.3354/meps272099 (2004).
Jeffery, S. W. & Humphrey, G. F. New spectrophotometric equations for determining chlorophylls a, b, c1 and c2 in higher plants, algae and natural phytoplankton. Biochem Physiol Pfl 167, 191–194 (1975).
Stimson, J. & Kinzie, R. A. The temporal pattern and rate of release of zooxanthellae from the reef coral Pocillopora damicornis (Linnaeus) under nitrogen-enrichment and control conditions. J Exp Mar Biol Ecol 153, 63–74, 10.1016/S0022-0981(05)80006-1 (1991).
Leggat, W., Hoegh-Guldberg, O., Dove, S. & Yellowlees, D. Analysis of an EST library from the dinoflagellate (Symbiodinium sp.) symbiont of reef-building corals. J Phycol 43, 1010–1021, 10.1111/j.1529-8817.2007.00387.x (2007).
Hellemans, J., Mortier, G., De Paepe, A., Speleman, F. & Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8, R19–R19, 10.1186/gb-2007-8-2-r19 (2007).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3, Research0034, 10.1186/gb-2002-3-7-research0034 (2002).
Acknowledgements
This study was supported by the Australian Research Council Centre of Excellence for Coral Reef Studies (CE0561435) and the Australian Research Council Discovery Grant (DP130101421).
Author information
Authors and Affiliations
Contributions
S.L.G., B.R.G. and W.L. designed thermal stress experiment. S.L.G. and B.R.G. performed experiment. S.L.G. performed cell density and chlorophyll pigment analysis. S.L.G. analysed Imaging-PAM data. S.L.G. prepared samples, designed primers and performed quantitative real-time PCR. S.L.G. and W.L. analysed the data. S.L.G. and W.L. wrote the paper. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Gierz, S., Gordon, B. & Leggat, W. Integral Light-Harvesting Complex Expression In Symbiodinium Within The Coral Acropora aspera Under Thermal Stress. Sci Rep 6, 25081 (2016). https://doi.org/10.1038/srep25081
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep25081
This article is cited by
-
Symbiodiniaceae photophysiology and stress resilience is enhanced by microbial associations
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.