Abstract
Insulin resistance, a critical component of type 2 diabetes (T2D), precedes and predicts T2D onset. T2D is also associated with mitochondrial dysfunction. To define the cause-effect relationship between insulin resistance and mitochondrial dysfunction, we compared mitochondrial metabolism in induced pluripotent stem cells (iPSC) from 5 healthy individuals and 4 patients with genetic insulin resistance due to insulin receptor mutations. Insulin-resistant iPSC had increased mitochondrial number and decreased mitochondrial size. Mitochondrial oxidative function was impaired, with decreased citrate synthase activity and spare respiratory capacity. Simultaneously, expression of multiple glycolytic enzymes was decreased, while lactate production increased 80%. These perturbations were accompanied by an increase in ADP/ATP ratio and 3-fold increase in AMPK activity, indicating energetic stress. Insulin-resistant iPSC also showed reduced catalase activity and increased susceptibility to oxidative stress. Thus, insulin resistance can lead to mitochondrial dysfunction with reduced mitochondrial size, oxidative activity and energy production.
Similar content being viewed by others
Introduction
Insulin resistance is a key component of type 2 diabetes (T2D) pathophysiology and an early marker and predictor of diabetes risk1, occurring several decades before hyperglycaemia develops. Due to the difficulty in accessing important tissues for study in humans, the specific molecular mechanisms responsible for common forms of insulin resistance remain unknown but likely include genetic, developmental and environmental contributions.
Insights into human insulin resistance have been provided by rare syndromes due to insulin receptor (INSR) mutations, such as Donohue syndrome and type A insulin resistance2,3,4,5,6,7. Clinically, these syndromes share several features, including acanthosis nigricans, hyperandrogenism and severe insulin resistance. In addition, patients with Donohue syndrome display reduced body weight and postnatal growth. Recent data have also demonstrated that humans with INSR mutations may have abnormalities in mitochondrial function, as indicated by decreased phosphocreatine recovery in muscle after exercise8. While skin fibroblasts derived from these individuals have elucidated key information about the structure and function of the insulin receptor and its downstream signalling pathways9, these cells are already terminally differentiated and are poorly responsive to insulin. To address these limitations, we have utilised induced pluripotent stem cells (iPSC) derived from these patients as a new model system to identify differentiation-independent and cell autonomous molecular drivers of insulin resistance. These cells also allow us to identify mechanisms by which insulin resistance modulates stem cell function and metabolism.
Both human and animal studies highlight key links between insulin resistance and energetic defects10,11. For example, in vivo magnetic resonance spectroscopy studies in patients with T2D demonstrate reduced ATP synthesis12,13,14,15 and tricarboxylic acid (TCA) cycle flux16. Moreover, muscle biopsies from individuals with T2D and obesity-linked insulin resistance show reductions in nuclear-encoded mitochondrial gene expression17,18, mitochondrial DNA (mtDNA) levels19 and oxidative phosphorylation (OXPHOS) complex activity20. Conversely, interventions such as exercise, caloric restriction and weight loss typically improve mitochondrial activity and insulin sensitivity in parallel21,22,23,24.
We have previously shown that insulin resistance affects stem cell function through decreased cell proliferation; however, it remains unknown whether insulin resistance can also affect metabolism in these cells. To test the hypothesis that insulin resistance can modulate stem cell metabolism, we utilised our unique collection of iPSC from patients with severe insulin resistance (IR-Mut) and observed several metabolic defects including decreased mitochondrial size and functional capacity.
Results
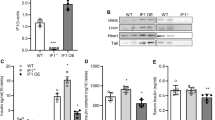
iPS cells from control individuals and patients with insulin resistance were generated and characterised previously25. Clinical information is presented in Supplementary Table S1. To determine if mitochondrial number or structure was altered by insulin resistance, electron microscopy analysis was performed. IR-Mut iPSC had a 22% increase in mitochondrial number (p = 0.02; Fig. 1a,b), but a 16% decrease in mitochondrial area (p = 0.04; Fig. 1a,c and Supplementary Fig. S1). This increase in number of smaller mitochondria was associated with increased expression of mitochondrial fission factor (MFF) and inverted formin 2 (INF2), which both promote mitochondrial fission, whereas expression of genes regulating mitochondrial fusion were unchanged between control and IR-Mut iPS (Supplementary Fig. S2A,B). mtDNA content (Fig. 1d), MitoTracker Green staining (Supplementary Fig. S3) and mRNA expression of mitochondrial transcription factor A (TFAM), which is also a regulator of mtDNA replication (Supplementary Fig. S4A), were all similar between control and IR-Mut iPSC. Likewise, citrate synthase (CS) protein, a marker for mitochondrial mass and CS maximal activity, examined in the presence of exogenous oxaloacetate, did not differ between control and IR-Mut iPSC (Fig. 1e,f), suggesting that total mitochondrial mass was similar in the control and insulin-resistant iPSC, despite the changes in mitochondrial size and shape. Interestingly, in the absence of exogenous substrate, endogenous CS activity was decreased by 40% in insulin-resistant iPSC (p = 0.04; Fig. 1g). Collectively, these data indicate similar mitochondrial mass but reduced CS activity, potentially due to substrate limitation.
Decreased mitochondrial area and CS activity in IR-Mut iPSC.
(a) Representative electron micrographs of control and IR-Mut iPSC. (b) Quantification of the number of mitochondria relative to total cellular area. Images at 4500X; 9 micrographs/cell line. (c) Quantification of the area of mitochondria relative to total cellular area. Images at 34000X; 5 micrographs/cell line. (d) Mitochondrial DNA content, as determined by qRT-PCR using primers for mt-ND1 and mt-ND2, with nuclear beta actin as control (n = 3). (e) Expression of CS protein by representative western blot. (f,g) Assay for maximal (f) and endogenous (g) citrate synthase activity measured as μmole/min and normalised to cellular protein (n = 3). *indicates p < 0.05 for IR-Mut vs. control.
To determine if the change in mitochondrial structure and citrate synthase activity was paralleled by a change in oxidative metabolism, cellular oxygen consumption was measured under basal conditions and after treatment with compounds that block different mitochondrial proteins to interrogate several components of mitochondrial oxidative function (Fig. 2a). There was a trend for increased basal respiration (p = 0.08) in insulin-resistant iPSC, but no difference in ATP-dependent respiration (Fig. 2b,c), calculated by the change in oxygen consumption rate (OCR) upon addition of the ATP synthase inhibitor oligomycin. Treatment with the uncoupler 2,4-dintrophenol (DNP) induced maximum respiration to a similar level in control and IR-Mut iPSC (Fig. 2a). However, spare respiratory capacity, i.e. ability to respond to increased energy demand, was decreased by 56% in the IR-Mut cells (p = 0.01; Fig. 2d), calculated by the difference between maximal and basal respiration. The decreased capacity was not due to uncoupling, as evidenced by similar proton leak (Fig. 2e) and unchanged expression of uncoupling protein 1 (UCP1) (Fig. 2f).
Decreased spare respiratory capacity in IR-Mut iPSC.
(a) Bioenergetic profile for oxygen consumption rate (OCR) of control and IR-Mut iPSC assessed at baseline and following sequential addition of oligomycin (10 μg/ml), DNP (10 μM) and KCN (50 nM). OCR was analysed at 5-minute intervals and normalised by DNA (n = 3). (b) Basal OCR was calculated from the last measurement before oligomycin addition minus non-mitochondrial respiration. (c) ADP-dependent respiration calculated from last measurement before oligomycin minus minimum measurement after oligomycin. (d) Spare respiratory capacity calculated from maximum measurement after DNP minus last measurement before oligomycin. (e) Proton leak calculated from minimum measurement after oligomycin minus non-mitochondrial respiration. (f) Relative mRNA expression of uncoupling protein 1, as assessed by qRT-PCR. Data are normalised to 36B4 and expressed relative to control (n = 3). *p < 0.05 for IR-Mut vs. control.
We next examined expression of mitochondrial proteins, including OXPHOS subunits, as a possible cause of this perturbed oxidative metabolism. Western blot analysis revealed no difference between control and insulin-resistant iPSC in representative components of the TCA cycle (fumarase) or OXPHOS complexes (NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8 (complex 1), complex II subunit 30 kDa, complex III subunit core 2, complex IV subunit I and ATP synthase subunit alpha; Fig. 3a). Gene expression data from prior microarray analysis25 were queried to examine the expression of genes that were significantly altered in IR-Mut cells and also annotated with mitochondrial ontology terms (p < 0.05; Fig. 3b, Supplementary Table S2), with 150 of 1312 mitochondrial ontology genes significantly changed. Expression of 12 genes related to oxidative phosphorylation was significantly altered (with 8 increased and 4 decreased), including succinate dehydrogenase complex, subunit A (SDHA) and several ETC subunits. Furthermore, 6 genes involved in stress response were downregulated, while 17 genes related to mitochondrial ribosomes and protein translation were significantly increased. qPCR confirmed a small but significant decrease in SDHA (Fig. 3c), while representative genes in other electron chain complexes were unchanged. Interestingly, expression of the mitochondrial regulatory coactivator peroxisome proliferator-activated receptor gamma, coactivator 1 alpha PPARGC1A (encoding PGC1α) was reduced by 54% in IR-Mut iPSC (p < 0.001; Fig. 3d); other transcriptional regulators of mitochondria and metabolism, including PPARGC1B, PPRC1, SIRT1, SIRT2 and SIRT3 (Fig. 3d) and GABPA, GABPB1, GABPB2, RB1, E2F1, E2F4, ESRRA and ESRRG (Supplementary Fig. S4A), were unchanged.
IR-Mut iPSC have decreased PGC1α expression.
(a) Representative western blots showing protein expression of fumarase and key enzymes in OXPHOS. VDAC was used as a loading control (n = 3). (b) Heat map of differentially expressed genes (p < 0.05) related to mitochondria using ontology terms “mitoc*” as determined by microarray analysis, with red indicating increased expression and blue indicating decreased expression. (c) Relative mRNA expression of selected nuclear-encoded mitochondrial genes, as assessed by qRT-PCR. Data are normalised to 36B4 and expressed relative to control (n = 3). (d) Relative mRNA expression of selected genes regulating mitochondrial biogenesis and metabolism, as assessed by qRT-PCR. Data are normalised to 36B4 and expressed relative to control (n = 3). *indicates p < 0.05 for IR-Mut vs. control.
To specifically examine genes associated with glucose metabolism that may be modulated along with the impaired oxidative metabolism, microarray data were queried for significant changes in the KEGG glycolysis/gluconeogenesis gene set (p < 0.05; Fig. 4a, Supplementary Table S3). Notably, expression of several genes regulating glucose metabolism, including hexokinase 1 (HK1), aldolase C (ALDOC), phosphoglucomutase 1 (PGM1) and pyruvate kinase, muscle isozyme 2 (PKM2), were significantly decreased in IR-Mut iPSC (Fig. 4b). qRT-PCR revealed reduced mRNA expression of the glucose transporters GLUT1 (SLC2A1) and GLUT4 (SLC2A4) and glycolytic enzymes HK1, ALDOA and PKM2 in IR-Mut iPSC (Fig. 4c). Protein expression of hypoxia-inducible factor 1 alpha (HIF1α), a transcription factor subunit that responds to hypoxia and energetic stress and promotes glycolytic compensation, was undetectable in both the control and IR-Mut iPSC (not shown).
IR-Mut iPSC have increased extracellular lactate.
(a) Heat map of differentially expressed genes (nominal p < 0.05) related to glucose metabolism based on KEGG glycolysis/gluconeogenesis gene set, as determined by microarray analysis, with red indicating increased expression and blue indicating decreased expression. (b) Glycolysis pathway schematic of enzymes with significantly decreased mRNA expression in IR-Mut iPSC (indicated in blue). (c) Relative mRNA expression of glucose metabolism genes, as assessed by qRT-PCR. Data are normalised to 36B4 and expressed relative to control cells (n = 3). (d) [3H]-2-deoxyglucose uptake was assessed at baseline and in response to 100 nM insulin and 10 nM IGF1. Data are expressed relative to basal control values (n = 4). (e) Basal extracellular acidification rate (ECAR) in control and IR-Mut iPSC, expressed relative to control and normalised by DNA concentration (n = 3). (f) Lactate concentrations in conditioned medium at 0 and 4 hours after addition of fresh media and normalised to cellular protein content (n = 4). *indicates p < 0.05 for IR-Mut vs. control.
Given these patterns suggesting that glycolytic metabolism could be altered in insulin resistant iPS cells, we next determined cellular glucose uptake using radiolabelled 2-deoxyglucose (2-DG). Under basal conditions, control and IR-Mut iPSC had similar 2-DG uptake (Fig. 4d, left bars). Upon stimulation with 100 nM insulin or 10 nM IGF1, both control and IR-Mut iPSC showed small increases in 2-DG uptake, which were not significantly different from each other (Fig. 4d, middle and right bars). Anaerobic glucose metabolism was measured by both extracellular acidification rate (ECAR; Seahorse Bioanalyzer) and lactate release into conditioned media. There was no difference in basal ECAR (measured after 60-min incubation in 5 mM glucose in 5-min intervals) between control and IR-Mut iPSC (Fig. 4e). However, with prolonged incubation (4 hrs), insulin-resistant iPSC had an 82% increase in lactate levels in conditioned media (p = 0.002; Fig. 4f), despite unaltered activity of lactate dehydrogenase (not shown). These data suggest that IR-Mut iPSC have increased reliance on glycolytic metabolism.
The above data indicate that multiple metabolic pathways are impaired with insulin resistance, potentially due to coordinated transcriptional regulation. Promoter analysis of all significantly downregulated genes highlights enrichment for gene promoters containing transcription factor Sp1 (SP1), nuclear factor of activated T-cells, cytoplasmic (NFATC) and jun proto-oncogene (JUN) motifs in IR-Mut iPSC (Supplementary Table S4). In addition, expression of NFATC1 (not shown) and JUN25 mRNA were also downregulated in the IR-Mut iPSC. SP1 mRNA was significantly increased by more than 3-fold in IR-Mut iPSC (p = 0.02), while transcriptional modifier YY1 (also known as Ying Yang 1), a PGC1α target, was decreased (p = 0.02; Supplementary Fig. S4B). Furthermore, expression of several tumour protein p53-related genes, which regulate both cell proliferation and metabolism26, was significantly altered in the IR-Mut iPSC (Supplementary Table S5), including decreased expression of p63, p73 and p53-inducible ribonucleotide reductase and increased expression of p53 and p53-binding protein 1.
To ascertain the net impact of decreased aerobic metabolic activity on energy production, ATP and ADP levels were measured. Insulin-resistant iPSC had a trend for decreased ATP content (19%, p = 0.07; Fig. 5a) and a significant 34% increase in ADP/ATP ratio (p = 0.04; Fig. 5b), consistent with inefficient production or maintenance of ATP levels. This energy deficit was not associated with decreased viability (Supplementary Fig. S5A). Reduced energy production can lead to phosphorylation and activation of AMP-activated protein kinase (AMPK), a key monitor of cellular energy status. Indeed, insulin-resistant iPSC showed a 3-fold increase in phosphorylated AMPK (p < 0.001) and a 2-fold increase in the phosphorylation of its downstream target acetyl-CoA carboxylase (ACC; p < 0.001), with no change in total protein levels (Fig. 5c,d).
Decreased energy production in IR-Mut iPSC.
(a) ATP level expressed relative to control (n = 3). (b) Ratio of ADP/ATP for control and IR-Mut iPSC (n = 3). (c) Representative western blot and quantification of phosphorylated to total AMPK expression ratio (n = 3). (d) Representative western blot and quantification of phosphorylated to total ACC expression ratio (n = 3). *p < 0.05 for IR-Mut vs. control.
Dysregulation of mitochondrial function can lead to increased reactive oxygen species (ROS) production, which in turn could be linked to insulin resistance27. Therefore, we examined expression of genes regulating oxidative stress. Insulin-resistant iPSC had modest, but significant, ~20% increases in SOD1 and SOD2 expression (p < 0.001; Fig. 6a). By contrast, there was a robust 50% decrease in catalase (CAT) expression (p < 0.001; Fig. 6a), with a coordinate 66% decrease in CAT activity (p = 0.01; Fig. 6b). There was a trend for increased protein oxidation (assessed by OxyBlot; Supplementary Fig. S5B,C) and lipid peroxidation (TBARS assay) was increased 26% in insulin-resistant IR-Mut iPSC (p = 0.03; Fig. 6c).
IR-Mut iPSC have increased oxidative stress.
(a) Relative mRNA expression of antioxidant enzymes, as assessed by qRT-PCR. Data are normalised to 36B4 and expressed relative to control (n = 3). (b) Catalase activity was measured as μmole/min and normalised to cellular protein (n = 3). (c) Lipid peroxidation was calculated by MDA equivalents using TBARS assay (n = 3). (d) ATP level expressed relative to vehicle (0 μM H2O2) (n = 2 of 4 Con vs 4 IR-Mut and n = 1 of 2 Con vs 3 IR-Mut). *p < 0.05 for IR-Mut vs. control. #p < 0.05 vs. vehicle.
To determine whether insulin resistance increased the susceptibility of iPSC to oxidative stress, cells were treated with hydrogen peroxide for 1 hour. Control cells displayed no significant reduction in ATP levels at the lower doses of hydrogen peroxide tested (100 and 200 μM). By contrast, ATP levels were decreased in IR-Mut iPSC by 7 and 18% in response to 100 and 200 μM H2O2, respectively, as compared with vehicle (p < 0.01; Fig. 6d). These ATP values were also significantly lower than control iPSC levels (p < 0.05). At the highest concentration of H2O2 tested, ATP levels were significantly decreased in both control and IR-Mut cells by 20–27% compared to vehicle, with no difference between groups.
Discussion
Insulin resistance is an important predictor of diabetes risk1 and is associated with impairments in energy metabolism in both mice and humans. In vivo, it is difficult to discern whether the insulin resistance or the oxidative dysfunction is the primary event, given the close interrelationships between these phenotypes. iPS cells are a robust tool for studying human disease pathology. This in vitro human model system can be used for in depth molecular, biochemical and developmental studies of complex diseases. iPS cells also allow identification of cell autonomous phenotypes present in the absence of differentiation-dependent confounders. We demonstrate that stem cells with primary insulin resistance exhibit profound impairments in cellular energy metabolism that result from the collective impact of several metabolic defects.
Firstly, IR-Mut iPS cells have alterations in mitochondrial size and number indicative of increased mitochondrial fission. This is supported by finding of increased expression of MFF and INF2, both of which promote mitochondrial fission. However, total mitochondrial mass, DNA content and citrate synthase protein content are similar between the insulin resistant cells and controls. Such differences in mitochondrial number, despite similar mass, also are consistent with studies showing the ability of insulin signalling to promote mitochondrial fusion28,29,30,31. Enhanced mitochondrial fission has also been observed in terminally differentiated tissues from patients with type 2 diabetes, including skeletal muscle and pancreatic beta cells29,32 and our current data suggest that such changes may be programmed even in the pre-differentiation state. These data are particularly interesting as smaller mitochondria produce ATP less efficiently due to decreased expression of mtDNA-encoded subunits and mitochondrial content mixing33,34,35 and, thus, could contribute to reduced oxidative capacity in IR-Mut iPSC or differentiated tissues such as skeletal muscle36,37.
Secondly, IR-Mut iPS cells demonstrate distinct phenotypes indicating dysregulation of oxidative metabolism. Lactate levels in conditioned media are strikingly increased in insulin resistant iPSC and this pattern was also observed in mesenchymal progenitor cells (MPC) derived from these insulin resistant iPS cells, indicating this is a phenotype that can be identified in at least two distinct populations of progenitor cells38. In parallel, endogenous citrate synthase enzymatic activity was markedly reduced in IR-Mut cells. Again, these findings are similar to the increased plasma lactate levels39,40,41 and decreased citrate synthase activity in skeletal muscle37,42 and adipose tissue43 with insulin resistance and T2D. In the iPS cells, differences in citrate synthase activity resolved after addition of exogenous oxaloacetate, indicating that substrate availability might be limiting for TCA cycle metabolism in insulin resistance. Whether the same is true in vivo still needs to be resolved44,45,46. Similarly, our analysis revealed that spare respiratory capacity was reduced by more than 50% in insulin resistant cells.
Collectively, these results may indicate that primary insulin resistance can result in impaired oxidative capacity, with a shift in balance toward nonoxidative metabolism. Despite this shift, insulin resistant cells are incapable of increasing energy production and thus unable to adapt to conditions promoting energy deficit or increased energetic stress, such as exercise. These data are also consistent with the decreased OXPHOS activity and ATP production in animals with insulin receptor mutations47 and with recent data from humans with insulin receptor mutations demonstrating decreases in phosphocreatine recovery after exercise8.
Though stem cells have functional OXPHOS complexes and mitochondrial networks, they largely depend upon glycolysis for ATP production48,49,50,51 and a shift to glycolysis is necessary for reprogramming and establishment of pluripotency52,53. The observed increase in lactate production in IR-Mut iPSC may indicate an even greater degree of dependence on glycolytic metabolism in these cells, potentially in response to their defective oxidative metabolism. However, the increased ADP/ATP ratio and AMPK activation in IR-Mut cells indicate that glycolytic energy metabolism is insufficient to compensate, resulting in inability to maintain normal energy stores. This inadequate energy availability may also contribute to decreased proliferation (as we have previously reported25), thus impairing an essential component of stem cell function. In turn, such reductions in proliferation of stem cells and other developmentally important progenitors could also potentially play a role in the diminished prenatal growth in patients with Donohue syndrome.
Imbalances between ROS production and antioxidant responses are linked to insulin resistance in both rodents and humans27,54,55. Experimental increases in ROS and induction of mitochondrial dysfunction can provoke insulin resistance47,56 and treatment with antioxidants can alleviate insulin resistance27. Here, we demonstrate that both mitochondrial dysfunction and increased oxidative stress develop in cells that have primary, i.e. genetically-determined, insulin resistance. Insulin resistant iPSC exhibit increased sensitivity to hydrogen peroxide. These defects may be linked, in part, to the profound reduction in expression and activity of the antioxidant enzyme catalase. Interestingly, catalase mRNA was also significantly reduced in MPCs derived from these IR-Mut iPSC38. Since increased expression of catalase in mitochondria prevents aging-induced mitochondrial dysfunction, as well as lipid-induced insulin resistance57, it is possible that insulin resistance itself may both impair mitochondrial function and ability to clear ROS, thus promoting increased oxidative stress and initiating a vicious cycle of metabolic defects.
We do not yet know the specific transcriptional regulatory molecules responsible for the altered metabolism in insulin resistance. Transcriptome and promoter analysis suggests a potential role for genes regulated by PGC1α, SP1, NFATC, JUN and p53. p53 is a possible regulator as it controls not only cell proliferation25, but also reduces transcription of key metabolic regulatory genes including glucose transporters and glycolytic genes hexokinases 1 and 2, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (PFKFB3), phosphofructokinase isozymes M and P and phosphoglucomutase, while increasing glutaminase 2 expression58,59,60. Consistent with this possibility, expression of glucose transporters 1 and 4, HK1 and PGM1 was decreased in IR-Mut iPSC. However, glutaminase 2, which plays an important role in oxidative stress response, was also downregulated in IR-Mut cells. Moreover, mRNA expression of the coactivator PGC1α and its target YY1 were also downregulated in IR-Mut cells – a pattern similar to the decreased PGC1α expression in muscle, adipose and liver of humans with insulin resistance and in INSR-knockdown C2C12 myotubes18,61,62. Since both PGC1α and YY1 are key regulators of mitochondrial metabolism and responsive to cellular growth and nutrient status via interactions with mechanistic target of rapamycin (serine/threonine kinase) (mTOR)63,64,65, perturbations in these pathways could contribute to observed phenotypes. We did not observe changes in MTOR-dependent activity (phosphorylation of S6 ribosomal protein) or p53 protein or acetylation in IR-Mut cells (data not shown), indicating that insulin receptor mutations do not impair all regulatory pathways.
Our study in iPS cells demonstrates an important role for insulin resistance as a driver of mitochondrial oxidative dysfunction, even prior to differentiation of cells into more classical insulin target tissue cell types, such as muscle, fat and liver. At present, our study is limited to severe genetic mutations in the insulin receptor as the cause for insulin resistance and some between-patient differences may be related to the specific insulin receptor mutation, the genetic background of the individual, or residual epigenetic differences maintained during cellular reprogramming66,67. Nevertheless, these data support the importance of future studies of cells from individuals with more common and less severe forms of insulin resistance, such as patients with T2D. Comparative studies in these additional cell lines would be beneficial in determining the primary and fundamental metabolic phenotypes associated with insulin resistance in stem cell populations and their influence on developmental trajectories.
In summary, our results demonstrate that primary insulin resistance can lead to reduced mitochondrial oxidative function, increased reliance on anaerobic metabolism and impaired ability to respond to conditions of maximal respiratory demand. Collectively, these defects contribute to not only impaired energy homeostasis, but also increase oxidative stress. Moreover, these data highlight that phenotypes associated with human insulin resistance can be recapitulated in a cell culture system, allowing future analyses of mechanisms by which defects in insulin signalling and cellular metabolism alter the function of key stem cell populations.
Methods
Generation and use of human iPSC was approved by Joslin Diabetes Center’s Committee on Human Studies. All methods were performed in accordance with the approved guidelines. The pre-existing human fibroblast lines used to generate iPSC are available from commercial sources, with no identifiable private information provided to the study investigators.
Reagents
Primary antibodies included anti-citrate synthase (abcam ab96600), anti-fumarase (Novus Biologicals NBP1-31336), MitoProfile Total OXPHOS Cocktail (abcam ab110413), anti-phospho-AMPKalpha (Thr172; Cell Signaling 2535), anti-AMPKalpha (Cell Signaling 2532), anti-phospho-acetyl-CoA carboxylase (Cell Signaling 3661), anti-acetyl-CoA carboxylase (Cell Signaling 3662) and anti-VDAC (Cell Signaling 4866).
iPSC Culture
iPSC were generated as previously described25 and grown on Geltrex-coated plates in mTesR1 media (StemCell Technologies). Cells were studied at similar passage numbers and at approximately 75% confluency.
Western Blot Analysis
iPSC were lysed in 1% sodium dodecyl sulfate and protein concentrations were determined by BCA (Thermo Scientific). Equal amounts of protein were separated by SDS-PAGE and transferred to nitrocellulose membranes, blocked with 5% nonfat dry milk in Tris-buffered saline with 0.1% Tween, incubated with primary antibodies overnight and washed prior to incubation with HRP-conjugated secondary antibodies. Proteins were detected by enhanced chemiluminescence (PerkinElmer Life Sciences). For protein oxidation analysis, samples were processed using an OxyBlot protein oxidation detection kit (Millipore). For quantitation, band intensities were measured using Adobe Photoshop on inverted scanned images; target bands were selected with rectangular marquee tool and the average pixel intensity was determined using the histogram function.
RNA Expression Analysis
Microarray analyses were performed as previously reported25. Data were analysed using the limma package from Bioconductor, with results selected by nominal p-value (p < 0.05). For qRT-PCR analysis, RNA was isolated using RiboZol (AMRESCO) and 1 μg was used to synthesise cDNA (High Capacity cDNA Reverse Transcription Kit, Life Technologies). Amplification was performed using iTaq Universal SYBR Green Supermix (Bio-Rad) and ABI 7900HT Fast Real-Time PCR System (Life Technologies). The gene 36B4 was used for normalisation and the average of the control cell values was used for calculating relative expression using the ΔΔCT method68. Overrepresentation of transcription factor binding sites was assessed using mSigDB (Broad Institute)69. Supplementary Table S6 lists the gene symbols with the coordinate gene name and primer sequences used for qRT-PCR.
Cellular Metabolism Phenotypic Analysis
Equal numbers of cells were incubated with Kreb’s Ringer buffer (KRB; 110 nM NaCl, 4.7 nM KCl, 2 mM MgSO4, 1.2 mM Na2HPO4, 0.24 mM MgCl2) with 5 mM glucose and 0.5% bovine serum albumin (BSA) for one hour. OCR and ECAR were measured using XF24 Flux Analyzer (Seahorse Biosciences). Data were collected at baseline and at 5-minute intervals following sequential addition of oligomycin (10 μg/ml), 2,4-dinitrophenol (DNP; 10 μM) and potassium cyanide (KCN; 50 μM) and then normalised by DNA content.
Lactate Production
Equal numbers of cells were plated per well in a 12-well plate, washed with PBS and incubated in KRB with 5 mM glucose and 0.5% bovine serum albumin (BSA). Conditioned media were sampled at baseline and at 2 and 4 hours for measurement of lactate concentration using a colourimetric kit (Biomedical Research Service Center); values were normalised to cellular protein content.
Enzymatic Assays
Endogenous and maximal CS activity was measured using a colourimetric assay (Sigma-Aldrich). Levels of adenine nucleotides were measured by a modified coupled enzymatic assay70. Cells were plated in an opaque 96-well plate. To initiate reaction, 100 μL of CellTiter-Glo luminescent assay reagent (Promega) were added to each well. After incubation for 15 minutes, intensity values were determined on a microplate reader. Next, 1.5 mM phosphoenolpyruvate (PEP) and 2.3 units/ml pyruvate kinase (PK) were added to convert ADP to ATP and values were obtained after 2 min. Finally, 36 units/ml adenylate kinase were added to each well and measurements were obtained after 2 min. Relative ADP levels were determined by subtracting the post-PEP/PK from basal values. Relative AMP levels were determined by subtracting the post-AK from post-PEP/PK values. Catalase activity was measured using a colourimetric kit (Biomedical Research Service Center). Lipid peroxidation was measured using a TBARS assay kit (Cayman Chemical).
Mitochondrial DNA Content
Total DNA was prepared by lysis with 50 mM NaOH. After boiling for 5 min at 95 °C, lysates were neutralised by addition of 1 M Tris (pH 6.8). Mitochondrial DNA content was measured by qRT-PCR (Applied Biosystems 7900) using primers for mt-ND1 and mt-ND2; values were normalised to the nuclear gene ACTB.
Glucose Uptake
Following 4-hour serum starvation in KRB buffer with 0.5% BSA, cells were treated with PBS, 100 nM insulin, or 10 nM insulin-like growth factor 1 (IGF1) for 30 minutes and then incubated with3H 2-deoxyglucose (1:20 in cold 5.1 mM 2-deoxyglucose) for 10 minutes. Cells were lysed in 1% Triton X-100 and radioactivity was determined by scintillation counting. Values were normalised to protein content (BCA).
Transmission Electron Microscopy
iPSC in suspension were fixed in 2.5% glutaraldehyde, 0.1 M phosphate buffer (pH 7.4) for 1 hour before several washes in phosphate buffer and 1-hr post-fixation in 2% OsO4 (in phosphate buffer). After ethanol dehydration (50/70/95/100%) and clearing with propylene oxide, propylene oxide and Araldite 502 epoxy resin (1:1) with vacuum desiccation were used for resin infiltration. Pellets were embedded and cured in fresh resin using BEEM capsules. Sections (80 nM) stained with uranyl acetate and lead citrate were imaged on 75-mesh copper grids with a Philips 301 transmission electron microscope. Micrographs were taken for quantification of mitochondrial number (4500X) and area (34000X) using ImageJ the cell counter function to count mitochondria and the freehand selection to measure cellular and mitochondrial areas.
Statistical Analysis
Data are presented as mean ± SEM. Significant differences were analysed by Student’s t-test (p values), with p < 0.05 considered significant.
Additional Information
How to cite this article: Burkart, A. M. et al. Insulin Resistance in Human iPS Cells Reduces Mitochondrial Size and Function. Sci. Rep. 6, 22788; doi: 10.1038/srep22788 (2016).
References
Tabak, A. G. et al. Trajectories of glycaemia, insulin sensitivity and insulin secretion before diagnosis of type 2 diabetes: an analysis from the Whitehall II study. Lancet 373, 2215–2221 (2009).
Savage, D. B. et al. A clinical approach to severe insulin resistance. Endocr. Dev. 11, 122–132 (2007).
Longo, N. et al. Genotype-phenotype correlation in inherited severe insulin resistance. Hum. Mol. Genet. 11, 1465–1475 (2002).
Podskalny, J. M. & Kahn, C. R. Cell culture studies on patients with extreme insulin resistance. I. Receptor defects on cultured fibroblasts. J. Clin. Endocrinol. Metab. 54, 919–930 (1982).
Donohue, W. L. & Uchida, I. Leprechaunism: a euphemism for a rare familial disorder. J. Pediatr. 45, 505–519 (1954).
Kosztolanyi, G. Leprechaunism/Donohue syndrome/insulin receptor gene mutations: a syndrome delineation story from clinicopathological description to molecular understanding. Eur. J. Pediatr. 156, 253–255 (1997).
Semple, R. K., Savage, D. B., Cochran, E. K., Gorden, P. & O’Rahilly, S. Genetic syndromes of severe insulin resistance. Endocr. Rev. 32, 498–514 (2011).
Sleigh, A. et al. Mitochondrial dysfunction in patients with primary congenital insulin resistance. J. Clin. Invest. 121, 2457–2461 (2011).
Taylor, S. I. et al. Mutations in insulin-receptor gene in insulin-resistant patients. Diabetes Care 13, 257–279 (1990).
Patti, M. E. & Corvera, S. The Role of Mitochondria in the Pathogenesis of Type 2 Diabetes. Endocr. Rev. 31, 364–396 (2010).
Ozcan, U. et al. Endoplasmic reticulum stress links obesity, insulin action and type 2 diabetes. Science 306, 457–461 (2004).
Petersen, K. F., Dufour, S., Befroy, D., Garcia, R. & Shulman, G. I. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N. Engl. J. Med. 350, 664–671 (2004).
Petersen, K. F., Dufour, S. & Shulman, G. I. Decreased insulin-stimulated ATP synthesis and phosphate transport in muscle of insulin-resistant offspring of type 2 diabetic parents. PLOS Med. 2, e233 (2005).
Szendroedi, J. et al. Muscle Mitochondrial ATP Synthesis and Glucose Transport/Phosphorylation in Type 2 Diabetes. PLOS Med. 4, e154 (2007).
Kacerovsky-Bielesz, G. et al. Short-term exercise training does not stimulate skeletal muscle ATP synthesis in relatives of humans with type 2 diabetes. Diabetes 58, 1333–1341 (2009).
Cline, G. W., Rothman, D. L., Magnusson, I., Katz, L. D. & Shulman, G. I. 13C-nuclear magnetic resonance spectroscopy studies of hepatic glucose metabolism in normal subjects and subjects with insulin-dependent diabetes mellitus. J. Clin. Invest. 94, 2369–2376 (1994).
Patti, M. E. & Butte, A. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential Role of PGC1 and NRF1. Proc. Natl. Acad. Sci. USA 100, 8466–8471 (2003).
Mootha, V. K. et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Ritov, V. B. et al. Deficiency of subsarcolemmal mitochondria in obesity and type 2 diabetes. Diabetes 54, 8–14 (2005).
Ritov, V. B. et al. Deficiency of electron transport chain in human skeletal muscle mitochondria in type 2 diabetes mellitus and obesity. Am. J. Physiol. Endocrinol. Metab. 298, E49–E58 (2010).
Ostergard, T. et al. Impact of exercise training on insulin sensitivity, physical fitness and muscle oxidative capacity in first-degree relatives of type 2 diabetic patients. Am. J. Physiol. Endocrinol. Metab. 290, E998–1005 (2006).
Menshikova, E. V. et al. Effects of weight loss and physical activity on skeletal muscle mitochondrial function in obesity. Am. J. Physiol. Endocrinol. Metab. 288, E818–E825 (2005).
Toledo, F. G. et al. Effects of physical activity and weight loss on skeletal muscle mitochondria and relationship with glucose control in type 2 diabetes. Diabetes 56, 2142–2147 (2007).
Dube, J. J. et al. Effects of weight loss and exercise on insulin resistance and intramyocellular triacylglycerol, diacylglycerol and ceramide. Diabetologia 54, 1147–1156 (2011).
Iovino, S. et al. Genetic insulin resistance is a potent regulator of gene expression and proliferation in human iPS cells. Diabetes 63, 4130–4142 (2014).
Berkers, C. R., Maddocks, O. D., Cheung, E. C., Mor, I. & Vousden, K. H. Metabolic regulation by p53 family members. Cell Metab. 18, 617–633 (2013).
Houstis, N., Rosen, E. D. & Lander, E. S. Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature 440, 944–948 (2006).
Parra, V. et al. Insulin stimulates mitochondrial fusion and function in cardiomyocytes via the Akt-mTOR-NFkappaB-Opa-1 signaling pathway. Diabetes 63, 75–88 (2014).
Anello, M. et al. Functional and morphological alterations of mitochondria in pancreatic beta cells from type 2 diabetic patients. Diabetologia 48, 282–289 (2005).
Fealy, C. E., Mulya, A., Lai, N. & Kirwan, J. P. Exercise training decreases activation of the mitochondrial fission protein dynamin-related protein-1 in insulin-resistant human skeletal muscle. J. Appl. Physiol. (1985) 117, 239–245 (2014).
Bach, D. et al. Mitofusin-2 determines mitochondrial network architecture and mitochondrial metabolism. A novel regulatory mechanism altered in obesity. J Biol. Chem. 278, 17190–17197 (2003).
Kelley, D. E., He, J., Menshikova, E. V. & Ritov, V. B. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes 51, 2944–2950 (2002).
Mitra, K., Wunder, C., Roysam, B., Lin, G. & Lippincott-Schwartz, J. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc. Natl. Acad. Sci. USA 106, 11960–11965 (2009).
Westermann, B. Bioenergetic role of mitochondrial fusion and fission. Biochim. Biophys. Acta. 1817, 1833–1838 (2012).
Otera, H. & Mihara, K. Molecular mechanisms and physiologic functions of mitochondrial dynamics. J. Biochem. 149, 241–251 (2011).
Befroy, D. E. et al. Impaired mitochondrial substrate oxidation in muscle of insulin-resistant offspring of type 2 diabetic patients. Diabetes 56, 1376–1381 (2007).
Simoneau, J. A. & Kelley, D. E. Altered glycolytic and oxidative capacities of skeletal muscle contribute to insulin resistance in NIDDM. J. Appl. Physiol. 83, 166–171 (1997).
Balhara, B. et al. Severe insulin resistance alters metabolism in mesenchymal progenitor cells. Endocrinology 156, 2039–2048 (2015).
Doar, J. W., Wynn, V. & Cramp, D. G. Blood pyruvate and plasma glucose levels during oral and intravenous glucose tolerance tests in obese and non-obese women. Metabolism 17, 690–701 (1968).
Crawford, S. O. et al. Association of blood lactate with type 2 diabetes: the Atherosclerosis Risk in Communities Carotid MRI Study. Int. J. Epidemiol. 39, 1647–1655 (2010).
Lovejoy, J., Newby, F. D., Gebhart, S. S. & DiGirolamo, M. Insulin resistance in obesity is associated with elevated basal lactate levels and diminished lactate appearance following intravenous glucose and insulin. Metabolism 41, 22–27 (1992).
Ortenblad, N. et al. Reduced insulin-mediated citrate synthase activity in cultured skeletal muscle cells from patients with type 2 diabetes: evidence for an intrinsic oxidative enzyme defect. Biochim. Biophys. Acta. 1741, 206–214 (2005).
Christe, M. et al. Obesity affects mitochondrial citrate synthase in human omental adipose tissue. ISRN Obes. 2013, 826027 (2013).
Szendroedi, J., Phielix, E. & Roden, M. The role of mitochondria in insulin resistance and type 2 diabetes mellitus. Nat. Rev. Endocrinol. 8, 92–103 (2012).
Patti, M. E. & Corvera, S. The role of mitochondria in the pathogenesis of type 2 diabetes. Endocr. Rev. 31, 364–395 (2010).
Adams, S. H. et al. Plasma acylcarnitine profiles suggest incomplete long-chain fatty acid beta-oxidation and altered tricarboxylic acid cycle activity in type 2 diabetic African-American women. J. Nutr. 139, 1073–1081 (2009).
Cheng, Z. et al. Foxo1 integrates insulin signaling with mitochondrial function in the liver. Nat. Med. 15, 1307–1311 (2009).
Zhang, J. et al. UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. EMBO J. 30, 4860–4873 (2011).
Takubo, k. et al. Regulation of Glycolysis by Pdk Functions as a Metabolic Checkpoint for Cell Cycle Quiescence in Hematopoietic Stem Cells. Cell Stem Cell 12, 49–61 (2013).
Zhang, J. et al. Measuring energy metabolism in cultured cells, including human pluripotent stem cells and differentiated cells. Nat. Protoc. 7, 1068–1085 (2012).
Vacanti, N. M. & Metallo, C. M. Exploring metabolic pathways that contribute to the stem cell phenotype. Biochim. Biophys. Acta 1830, 2361–2369 (2013).
Vazquez-Martin, A. et al. Activation of AMP-activated protein kinase (AMPK) provides a metabolic barrier to reprogramming somatic cells into stem cells. Cell Cycle 11, 974–989 (2012).
Folmes, C. D. et al. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 14, 264–271 (2011).
Konrad, T. et al. alpha-Lipoic acid treatment decreases serum lactate and pyruvate concentrations and improves glucose effectiveness in lean and obese patients with type 2 diabetes. Diabetes Care 22, 280–287 (1999).
Fridlyand, L. E. & Philipson, L. H. Reactive species, cellular repair and risk factors in the onset of type 2 diabetes mellitus: review and hypothesis. Curr. Diabetes Rev. 2, 241–259 (2006).
Wang, C. H., Wang, C. C., Huang, H. C. & Wei, Y. H. Mitochondrial dysfunction leads to impairment of insulin sensitivity and adiponectin secretion in adipocytes. FEBS J. 280, 1039–1050 (2013).
Lee, H. Y. et al. Targeted expression of catalase to mitochondria prevents age-associated reductions in mitochondrial function and insulin resistance. Cell Metab. 12, 668–674 (2010).
Schwartzenberg-Bar-Yoseph, F., Armoni, M. & Karnieli, E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 64, 2627–2633 (2004).
Hu, W. et al. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc. Natl. Acad. Sci. USA 107, 7455–7460 (2010).
Zawacka-Pankau, J. et al. Inhibition of glycolytic enzymes mediated by pharmacologically activated p53: targeting Warburg effect to fight cancer. J. Biol. Chem. 286, 41600–41615 (2011).
Pagel-Langenickel, I. et al. PGC-1alpha integrates insulin signaling, mitochondrial regulation and bioenergetic function in skeletal muscle. J. Biol. Chem. 283, 22464–22472 (2008).
Patti, M. E. et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc. Natl. Acad. Sci. USA 100, 8466–8471 (2003).
Ramanathan, A., Wang, C. & Schreiber, S. L. Perturbational profiling of a cell-line model of tumorigenesis by using metabolic measurements. Proc. Natl. Acad. Sci. USA 102, 5992–5997 (2005).
Chen, E. I. et al. Adaptation of energy metabolism in breast cancer brain metastases. Cancer Res. 67, 1472–1486 (2007).
Cunningham, J. T. et al. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature 450, 736–740 (2007).
Kim, K. et al. Epigenetic memory in induced pluripotent stem cells. Nature 467, 285–290 (2010).
Tompkins, J. D. et al. Epigenetic stability, adaptability and reversibility in human embryonic stem cells. Proc. Natl. Acad. Sci. USA 109, 12544–12549 (2012).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Detimary, P., Jonas, J. C. & Henquin, J. C. Possible links between glucose-induced changes in the energy state of pancreatic B cells and insulin release. Unmasking by decreasing a stable pool of adenine nucleotides in mouse islets. J. Clin. Invest. 96, 1738–1745 (1995).
Acknowledgements
The authors thank Deborah Lee, Hitomi Murao and Mai Shirozu for technical assistance, Christopher Cahill of the Joslin DRC Advanced Microscopy Core for assistance with electron microscopy and Christine Miller and Andrew Minotti of the Joslin DRC iPS Core. Funding was gratefully received from the NIH T32DK007260 (to AMB), the Harold Whitworth Pierce Charitable Trust Postdoctoral Fellowship (to AMB), the American Diabetes Association (to MEP), the Novo Foundation (to MEP and CRK), R37DK031036 (to CRK) and the Joslin DRC grant P30 DK036836.
Author information
Authors and Affiliations
Contributions
A.M.B. and M.E.P. designed the research; A.B. and K.T. performed and analysed the experiments, with S.I., L.W. and K.J.H. providing experimental assistance. A.B. wrote the paper with M.E.P., S.I. and C.R.K. contributed useful discussion and suggestions.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Burkart, A., Tan, K., Warren, L. et al. Insulin Resistance in Human iPS Cells Reduces Mitochondrial Size and Function. Sci Rep 6, 22788 (2016). https://doi.org/10.1038/srep22788
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep22788
This article is cited by
-
iPSCs derived from insulin resistant offspring of type 2 diabetic patients show increased oxidative stress and lactate secretion
Stem Cell Research & Therapy (2022)
-
β-cell mitochondria in diabetes mellitus: a missing puzzle piece in the generation of hPSC-derived pancreatic β-cells?
Journal of Translational Medicine (2022)
-
Beyond genomics—technological advances improving the molecular characterization and precision treatment of heart failure
Heart Failure Reviews (2021)
-
Current status and future prospects of patient-derived induced pluripotent stem cells
Human Cell (2021)
-
Modeling different types of diabetes using human pluripotent stem cells
Cellular and Molecular Life Sciences (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.